Abstract
Five new microsatellite loci were described and characterized for use as molecular markers for the identification and genetic differentiation of Candida albicans strains. Following the typing of 72 unrelated clinical isolates, the analysis revealed that they were all polymorphic, presenting from 5 to 30 alleles and 8 to 46 different genotypes. The discriminatory power obtained by combining the information generated by three microsatellites used in a multiplex PCR amplification strategy was 0.99, the highest ever reported. The multiplex PCR was later used to test a total of 114 C. albicans strains, including multiple isolates from the same patient collected from different body locations and along episodes of vulvovaginal infections. Three different scenarios for strain relatedness were identified: (i) different isolates that were revealed to be the same strain, (ii) isolates that were the same strain but that apparently underwent a process of microevolution, and (iii) isolates that corresponded to different strains. Analysis of the microevolutionary changes between isolates from recurrent infections indicated that the genotype alterations observed could be the result of events that lead to the loss of heterozygosity (LOH). In one case of recurrent infection, LOH was observed at the CAI locus, and this could have been related to exposure to fluconazole, since such strains were exposed to this antifungal during treatment. The analysis of microsatellites by a multiplex PCR strategy was found to be a highly efficient tool for the rapid and accurate differentiation of C. albicans strains and adequate for the identification of fine microevolutionary events that could be related to strain microevolution in response to environmental stress conditions.
Candida albicans, the most common fungal pathogen, is a commensal yeast that belongs to the normal microbial population of the mouth, vagina, and gastrointestinal tract in humans. However, in people with a variety of transient or permanent immunocompromised conditions, including transplant recipients, chemotherapy patients, underweight neonates, and human immunodeficiency virus-infected individuals, it may become an invasive pathogen (30, 56). Infections by opportunistic fungal agents are a major medical problem due to the growing number of immunocompromised patients with risk factors for such infections. Moreover, this problem is exacerbated by the fact that only a limited array of antifungal drugs is available and by the growing resistance among clinical isolates (39, 46, 55). The development of techniques and strategies that can accurately differentiate clinical isolates is of great relevance. These techniques should provide the ability to differentiate among strains responsible for clinical infections, as well as to trace their epidemiological pathways. Several molecular methods have been used to differentiate C. albicans strains, including electrophoretic karyotyping (2), the use of species-specific probes such as Ca3 or 27A in restriction enzyme analysis (28, 35, 36, 43), PCR-based methods (1, 7, 15, 51), and more recently, multilocus sequence typing, which uses nucleotide sequences from several genes and offers good discrimination (4, 49). Microsatellites or simple tandem repeats (STRs) consist of stretches of tandemly repeated mono- to hexanucleotide motifs dispersed throughout the genome and have a high level of polymorphism compared with those of other molecular markers. In yeast, microsatellite loci have considerable length variations, and this polymorphism quickly made them attractive markers for a variety of types of analyses, including strain typing (9, 16, 45), population structure analysis (13, 18, 25), and epidemiological studies (33, 42). Microsatellite polymorphism is manifested as allelic length differences due to the different numbers of repeated units present in the alleles and is easily assayed by PCR amplification (48).
Several polymorphic microsatellite loci have been identified in the C. albicans genome, most of them located near EF3 (5); CDC3 and HIS3 (3); or inside the coding regions ERK1, 2NF1, CCN2, CPH2, and EFG1 (31), although the discriminatory powers (DPs) calculated for such loci were relatively low (between 0.77 and 0.91). A higher value, 0.97, was estimated by Sampaio et al. (42) for microsatellite CAI, located in a noncoding region; and this discriminatory power was identical to that which had previously been reported for a combination of three microsatellites amplified in a multiplex reaction (3). A higher polymorphism is expected in microsatellite loci from noncoding regions; therefore, in order to obtain a greater resolution, a set of new microsatellite markers was selected from these regions and characterized for use in a multiplex PCR. This multiplex typing system was applied to C. albicans clinical isolates to test its efficiency for strain differentiation. The potential capacity of this system to identify fine microevolutionary events was also evaluated.
MATERIALS AND METHODS
Yeast strains and DNA extraction.
A total of 112 clinical isolates of C. albicans, obtained from two hospitals and a health care center located in Braga and Porto (northern Portugal), and reference strain WO-1, as well as type strain PYCC 3436 (ATCC 18804), were used in this study. The isolates were obtained from primary cultures, and one colony of each different phenotype was selected. The type strains C. parapsilosis PYCC 2545 (ATCC 22019), C. krusei PYCC 3341 (ATCC 6258), C. tropicalis PYCC 3097 (ATCC 750), C. glabrata PYCC 2418 (ATCC 2001), C. guilliermondii PYCC 2730 (ATCC 6260), C. lusitaniae PYCC 2705 (ATCC 34449), and C. dubliniensis CBS 7987 (ATCC MYA-646) were also included. All reference strains were obtained from the Portuguese Yeast Culture Collection (PYCC), New University of Lisbon, Lisbon, Portugal, except C. dubliniensis, which was obtained from Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. The isolates were previously identified by PCR fingerprinting with primer T3B (7). Prior to DNA isolation, yeast cells were grown overnight on Sabouraud broth medium at 30°C. A Zymolyase-based method was used to extract DNA by following procedures described previously (22). After DNA extraction, the cultures were frozen in 30% (wt/wt) glycerol.
Microsatellite selection and PCR primers design.
A search of the C. albicans genome sequences, available in databases from Stanford's DNA Sequencing and Technology Center (http://www-sequence.stanford.edu/group/candida), was performed in order to identify sequences containing microsatellite repeats. The search was performed with the aim of identifying microsatellite units of tri-, tetra-, and pentanucleotides according to criteria described previously (42). The sequences that were obtained and selected for locus specific amplification are presented in Table 1. Primers for each locus were designed by using the software Primer3, available from http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi.
TABLE 1.
Microsatellite DNA sequences selected from the database searcha
| Microsatellite designation | Sequence codeb | Primer sequencec | No. of repeated units | PCR product size (bp) |
|---|---|---|---|---|
| CAI | 396062C04.sl.seq | F-5′-ATGCCATTGAGTGGAATTGG-3′ | 41 | 252 |
| R-5′-AGTGGCTTGTGTTGGGTTTT-3′ | ||||
| CAIII | 265080D05.sl.seq | F-5′-TTGGAATCACTTCACCAGGA-3′ | 11 | 113 |
| R-5′-TTTCCGTGGCATCAGTATCA-3′ | ||||
| CAIV | 265080D05.sl.seq | F-5′-TGCCAAATCTTGAGATACAAGTG-3′ | 24 | 263 |
| R-5′-CTTGCTTCTCTTGCTTTAAATTG-3′ | ||||
| CAV | 265147C06.yl.seq | F-5′-TGCCAAATCTTGAGATACAAGTG-3′ | 50 | 237 |
| R-5′-CTTGCTTCTCTTGCTTTAAATTG-3′ | ||||
| CAVI | 385031H11.xl.seq | F-5′-ACAATTAAAGAAATGGATTTTAGTCAG-3′ | 21 | 294 |
| R-5′-TGCTGGTGCTGCTGGTATTA-3′ | ||||
| CAVII | 396033C06.sl.seq | F-5′-GGGGATAGAAATGGCATCAA-3′ | 10 | 222 |
| R-5′-TGTGAAACAATTCTCTCCTTGC-3′ |
The sequence codes and primers used for PCR amplification are presented. The number of repeated units and the predicted size of PCR amplification products were calculated according to the sequence of the strain SC5314 used in the sequencing project.
Code attributed to the sequence in the Stanford database.
For each primer pair, the letter F indicates the forward primer and the letter R indicates the reverse primer.
Chromosomal localization of microsatellite markers.
The sequences selected were searched by BLAST against the latest release (assembly 19) of C. albicans genome sequence, to give a location to a sequence contig (http://www-sequence.stanford.edu/group/candida).
PCR amplification conditions. (i) Singleplex amplification.
For all microsatelite loci, singleplex PCRs were performed with several different strains in order to evaluate the locus-specific amplification and to obtain alleles for sequencing. The locus CAI was amplified by following the conditions described by Sampaio et al. (42). The CAIII, CAV, CAVI, and CAVII loci were amplified by using 25 ng of genomic DNA in a 25-μl reaction volume containing 1× PCR buffer, 0.2 mM of each of the four deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.25 μM of each primer, and 1 U of Taq polymerase (GIBCO). After a 95°C preincubation step of 2 min, PCR amplifications were performed for a total of 30 cycles by using the following conditions: denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min, with a final extension step of 7 min at 72°C.
(ii) Multiplex amplification.
Multiplex PCR was performed by combining 1× PCR buffer (20 mM Tris HCl, pH 8.4, 50 mM KCl), 0.2 mM of each four of the deoxynucleoside triphosphates, 2 mM MgCl2, 40 ng of genomic DNA, and 2 U of Taq Gold polymerase (Applied Biosystems) carried in a 25-μl final volume. The PCR program consisted of an initial denaturation step at 95°C for 10 min, followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, with a final extension step of 60 min at 68°C. The multiplex reaction was designed by combining microsatellites CAI, CAIII, and CAVI; and the primer concentrations used are depicted in Table 2.
TABLE 2.
Characteristics of the microsatellite loci selected
| STR | No. of alleles | No. of genotypes | Size (bp) | Repetitive motif | DP | Chromosome no. | Primer concn (μM) | Dye labela |
|---|---|---|---|---|---|---|---|---|
| CAI | 27 | 46 | 189-303 | (CAA)2CTG(CAA)n | 0.967 | 4 | 0.050 | FAM |
| CAIII | 6 | 12 | 95-110 | (GAA)n | 0.853 | 5 | 0.050 | FAM |
| CAIV | —b | — | — | (ATT)n | — | — | — | — |
| CAV | 21 | 21 | 102-198 | (ATT)n | 0.574 | 3 | ||
| CAVI | 30 | 39 | 224-388 | (TAAA)n | 0.933 | 2 | 0.225 | HEX |
| CAVII | 5 | 8 | 200-235 | (CAAAT)n | 0.670 | 1 |
FAM, 6-carboxyfluorescein; HEX, hexachlorofluorescein.
—, not determined.
DNA sequencing and fragment size determination.
All alleles observed for each locus were sequenced. Before sequencing, the alleles were separated and reamplified as described previously (42). The reamplified DNA fragments were purified with Microspin S-300 HR columns (Amersham Pharmacia Biotech, Quebec, Canada) and subjected to a dideoxy sequencing reaction with the BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems). Following the sequencing reactions, the products were purified with AutoSeqG-50 columns (Amersham Pharmacia Biotech). Finally, the samples were dried, re-suspended in 15 μl of formamide, and run on an ABI PRISM 310 DNA automatic sequencer (Applied Biosystems). The results were analyzed by using Sequencing 3.7 analysis software. Determination of the fragment sizes of the PCR products and the allele sizes were done automatically with GeneScan 3.7 analysis software. The alleles have been designated by the number of repeated units determined after sequencing.
Reproducibility and stability.
The reproducibility of the method and the stability of the microsatellite markers were assessed according to the procedures described previously (42). The reproducibility of the method was also tested by comparing the results obtained by the singleplex analysis with the ones observed by the multiplex analysis.
RESULTS
Screening and selection of repeat regions in Candida albicans sequence database.
The search of the genomic DNA database of C. albicans performed in this study provided a total of 1,086 sequences, 368 obtained with the CAA query, 245 obtained with the GAA query, 220 obtained with the ATT query, 201 obtained with the TAAA query, and 52 obtained with the CAAAT query. Selection of the sequences was primarily based on the number of simple units repeated and their location in the genome. There should be at least 10 repeats, given that these sequences have a higher probability of showing greater genetic variability (11, 37), and they should be located outside known coding regions, since a higher polymorphism is expected in regions less prone to selective forces (32). Subsequently, a third selection was made for the sequences to allow the design of primers with an annealing temperature of about 60°C to ensure specificity and good reproducibility. A total of five sequences were then selected, and specific primers were designed for their amplification. Sequence 265080D05.s1.seq presented two different regions containing microsatellite motifs that were used separately to develop CAIII and CAIV (Table 1). The nomenclature chosen for the new markers was CA, after C. albicans, followed by a roman numeral notation that corresponded to the order of the analysis.
Microsatellite locus analysis.
The microsatellites selected were used to type 72 unrelated C. albicans strains, isolated from different patients, in order to test their specific amplification and polymorphism. This analysis revealed that they were all polymorphic and presented from 5 to 30 alleles and 8 to 46 different genotypes (Table 2). Analysis of the CAIV locus revealed the presence of null alleles; therefore, this marker was discarded.
The different alleles observed at all the loci were sequenced in order to determine the nature of the polymorphisms observed. The sequencing results confirmed that the consensus sequences of all new microsatellite loci analyzed were in accordance with the ones deposited in Stanford's DNA Sequencing and Technology Center database, confirming the amplification of the correct loci. CAIII, CAIV, CAV, CAVI, and CAVII were simple STRs with one variable repetitive motif, and only CAI was a compound microsatellite with two different variable units (Table 2). For all microsatellites the differences in the molecular weights of the distinct alleles reflected the differences in the number of the repeated motifs, which allowed the nomenclature for the alleles to be based upon the number of repeated units. This nomenclature prevents problems if further variation is found in the constant stretches of the alleles when other C. albicans strain typing studies are performed (12, 42).
The distribution of genotypes according to the Hardy-Weinberg equilibrium was investigated for all the microsatellite loci analyzed in the 72 unrelated isolates. A significant departure from Hardy-Weinberg equilibrium expectations was found (P < 0.001), which supported previous findings that the inheritance in infecting C. albicans populations appears to be clonal (15, 25, 50).
The discriminatory power was calculated for each marker according to the Simpson index of diversity:
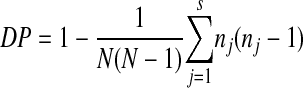 |
where N is the number of strains, s is the total number of different genotypes, and nj is the number of strains of j genotype (20). The results indicated that CAI is the microsatellite with the highest DP value, 0.97, while CAV presented the lowest DP value, 0.57 (Table 2).
Stability and specificity.
The in vitro stability of the microsatellite markers was assessed by growing four independent strains over approximately 300 generations. For all strains tested, the genotypes were always the same after the 300 generations, suggesting an expected mutation rate of less than 3.33 × 10−3 for all microsatellite loci. These markers were also species specific, since no amplification products were obtained when the primers and PCR conditions described above were used for the amplification of other pathogenic Candida species, namely, C. glabrata, C. krusei, C. parapsilosis, C. tropicalis, C. guilliermondii, C. lusitaniae, and C. dubliniensis. The specificity regarding C. dubliniensis, which is very closely related to C. albicans and which was just recently recognized as a different species, is noteworthy (47). Similar results were found in previous studies with microsatellites by testing other Candida species (5, 10, 31, 42).
Optimization of multiplex amplification conditions.
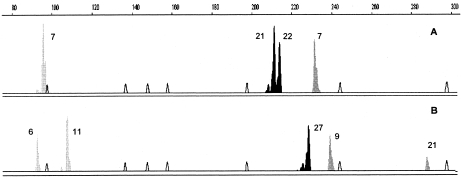
To increase the discriminatory capacity, simultaneous analysis of the different loci was carried out in a multiplex PCR. The CAI, CAIII, and CAVI loci were the ones selected for the multiplex PCR, since they presented the highest DP values in the singleplex assay and a DP value of 0.998 when analyzed together, while they are located on different chromosomes (Table 2). The differences in the sizes of the alleles amplified at each of these loci and the possibility of combining different fluorescent dyes made possible their simultaneous amplification (Fig. 1). For this optimization step, 10 C. albicans strains were used, and the results observed in the multiplex PCRs were always compared with the ones observed in the singleplex assay. The primer concentrations used for the multiplex reaction had to be adjusted, and the best results of multiplex coamplification with the three primer pairs were obtained by using the concentrations described in Table 2. Different DNA template concentrations were also tested, from 1 ng to 50 ng, and the optimal DNA concentration needed to coamplify these loci was 40 ng of template DNA in a final PCR volume of 25 μl. If a smaller amount of DNA was used, the preferential amplification of the CAI locus was observed. Comparison of the alleles obtained in the optimized multiplex reaction with the ones observed in the corresponding singleplex showed identical molecular weights, confirming the accuracy and reproducibility of the technique.
FIG. 1.
GenScan profiles depicting the results of automatic fragment sizing for the microsatellite multiplex analysis for (A) one strain from patient E (Table 4) and (B) strain 19C (Table 3). Each marker is represented by a different shade: black, CAI; dark gray, CAVI; light gray, CAIII.
Use of the multiplex assay for strain differentiation.
The CAI microsatellite was the one presenting the highest discriminatory power. However, several independent or unrelated isolates presented the same allele combination at this locus. These isolates were analyzed with the multiplex system in order to determine if they could indeed be considered the same strain. The genotypes observed with the multiplex assay indicated that, in several cases, the isolates with the same allelic combination at CAI corresponded to different strains or to strains undergoing a microevolutionary process (Table 3). The isolates were defined as identical when the same genotypes were observed at all loci, including the ones not presented in Table 3 (CAV and CAVII), and this was observed in 10 cases. On the contrary, 14 isolates were considered distinct strains since they presented different genotypes at several loci. The remaining three cases could be considered possible microevolutionary events, since minor changes in only one locus were observed. These results clearly showed the need for multilocus analysis, even when a highly discriminatory molecular marker is used.
TABLE 3.
Genotypes observed with microsatellites CAI, CAIII, and CAVI in a selection of strains that showed the same allele combination with CAI
| Isolate | Genotype observeda |
Institutionb | Body location | ||
|---|---|---|---|---|---|
| CAI | CAIII | CAVI | |||
| IGC3436 | 17-23 | 6-6 | 14-14 | ||
| 37M | 17-23 | 8-8 | 7-14 | HSM | Urine |
| 3 | 17-23 | 6-8 | 7-13 | HSJ | Vaginal exudate |
| 61M | 17-21 | 6-8 | 15-15 | HSM | Upper respiratory tract |
| 24C | 17-21 | 6-8 | 9-21 | CSC | Vaginal exudate |
| 1C | 17-21 | 6-8 | 7-17 | CSC | Vaginal exudate |
| 39M | 18-18 | 6-6 | 7-7 | HSM | Upper respiratory tract |
| 77M | 18-18 | 6-6 | 7-7 | HSM | Catheter |
| 5C | 18-18 | 6-6 | 7-7 | CSC | Vaginal exudate |
| 36C | 11-18 | 6-8 | 6-42 | CSC | Vaginal exsudate |
| 45C | 11-18 | 6-8 | 6-42 | CSC | Vaginal exudate |
| WO1 | 16-27 | 6-6 | 7-7 | ||
| 27C | 16-27 | 6-11 | 9-21 | CSC | Vaginal exudate |
| 26M | 20-28 | 6-9 | 35-45 | HSM | Upper respiratory tract |
| 39C | 20-28 | 6-9 | 39-44 | CSC | Vaginal exudate |
| 88M | 20-28 | 6-9 | 42-44 | HSM | Urine |
| 74M | 20-28 | 6-9 | 21-47 | HSM | Urine |
| 31 | 21-26 | 8-9 | 7-10 | HSJ | Vaginal exudate |
| 6C | 21-26 | 8-9 | 7-10 | CSC | Vaginal exudate |
| 49C | 21-26 | 6-7 | 9-15 | CSC | Vaginal exudate |
| 16C | 21-26 | 6-7 | 9-16 | CSC | Vaginal exudate |
| 69M | 21-25 | 6-7 | 9-15 | HSM | Upper respiratory tract |
| 8M | 21-25 | 6-7 | 9-16 | HSM | Urine |
| 1M | 21-25 | 6-7 | 9-16 | HSM | Urine |
| 31C | 21-25 | 6-7 | 9-17 | CSC | Vaginal exudate |
| 11C | 21-25 | 7-7 | 11-16 | CSC | Vaginal exudate |
| 17 | 21-21 | 6-7 | 8-15 | HSJ | Vaginal exudate |
| 52M | 21-21 | 6-7 | 7-7 | HSM | Urine |
| 39 | 21-22 | 6-7 | 7-7 | HSJ | Vaginal exudate |
| SER5 | 21-22 | 6-7 | 7-7 | HSJ | |
| 41M | 21-22 | 7-7 | 7-7 | HSM | Urine |
| H58 | 22-22 | 6-7 | 7-7 | HSJ | |
| 64M | 22-22 | 6-7 | 7-7 | HSM | Upper respiratory tract |
| 20 | 26-26 | 6-6 | 7-7 | HSJ | Vaginal exudate |
| 4M | 26-26 | 6-6 | 7-7 | HSM | Peritoneal exudate |
| H37 | 25-27 | 6-8 | 16-22 | HSJ | Upper respiratory tract |
| 63M | 25-27 | 6-6 | 9-17 | HSM | Urine |
| 22 | 23-27 | 6-8 | 21-21 | HSJ | Vaginal exudate |
| 41 | 23-27 | 6-8 | 18-20 | HSJ | Vaginal exudate |
| 35 | 25-25 | 7-7 | 9-15 | HSJ | Vaginal exudate |
| 45 | 25-25 | 7-7 | 9-15 | HSJ | Vaginal exudate |
| 19C | 27-27 | 6-11 | 9-21 | CSC | Vaginal exudate |
| 46C | 27-27 | 6-8 | 22-24 | CSC | Vaginal exudate |
Cases of microevolution are marked in boldface.
HSM, Hospital S. Marcos; HSJ, Hospital S. João; CSC, Centro de Saúde do Carandá.
When this multiplex assay was applied to the differentiation of multiple isolates from the same patient, it could be observed that all isolates showed exactly the same genotypes, confirming our beliefs that only one strain was present in the infecting population (Table 4). To verify that the infecting population was the same at different body locations, isolates were collected from the same patient with infections at multiple body sites. Isolates were collected from patients F, H, I, and J. The results showed that the isolates from the upper respiratory tract of patient I were identical strains but were different from the urine isolate. The same occurred for patient J, for whom distinct genotypes were observed for the two isolates, one from the vagina and the other from urine. However, isolates from patients F and H showed the same genotypes regardless of their body location. These results show that patients could have different clones at different body sites but that the infecting population at each body site is monoclonal, with some isolates presenting the capacity of invading different microhabitats.
TABLE 4.
Characteristics of multiple strains isolated from the same patient and cultured simultaneously
| Patienta | No. of isolates | Body location | Genotype observed |
||
|---|---|---|---|---|---|
| CAI | CAIII | CAVI | |||
| A | 3 | Urine | 21-25 | 6-7 | 9-16 |
| B | 4 | Peritoneal exudate | 26-26 | 6-6 | 7-7 |
| C | 2 | Upper respiratory tract | 17-17 | 6-8 | 7-14 |
| D | 2 | Urine | 21-21 | 6-7 | 7-7 |
| E | 5 | Urine | 21-22 | 7-7 | 7-7 |
| F | 1 | Urine | 18-47 | 6-11 | 9-19 |
| F | 1 | Upper respiratory tract | 18-47 | 6-11 | 9-19 |
| G | 2 | Urine | 36-36 | 6-11 | 6-41 |
| H | 1 | Upper respiratory tract | 21-25 | 6-7 | 9-15 |
| H | 2 | Urine | 21-25 | 6-7 | 9-15 |
| I | 2 | Upper respiratory tract | 22-22 | 6-7 | 7-7 |
| I | 1 | Urine | 20-28 | 6-9 | 42-44 |
| J | 1 | Vaginal exudate | 23-24 | 6-6 | 16-27 |
| J | 1 | Urine | 17-23 | 8-8 | 7-14 |
| R | 2 | Vaginal exudate | 21-25 | 6-7 | 9-17 |
Each patient is referred to as a separate letter (A to J and R).
Cases of recurrent vaginitis in eight patients were also analyzed with this multiplex system. The results showed that the infecting C. albicans strains isolated sequentially in different relapses of the illness displayed the same genotype for all loci, except in three cases, patients L, N, and P (Table 5). In patient N, a case of strain replacement occurred, since one of the three strains isolated presented different genotypes at all loci analyzed. In patient P, a microevolutionary event seems to have occurred, since the strains presented only a minor change at the CAVI locus, from genotype 18-21 to genotype 21-21. Analysis of the isolates from patient L showed a change at the CAI locus, from allele 30 to allele 32. In a previous study we suggested that these changes from allele 30 to allele 32 could have been due to expansion of the microsatellite (42). However, the analysis of this case with the remaining microsatellite loci suggests the occurrence of genetic changes that could lead to the loss of heterozygosity (LOH). Perepnikhatka et al. (34) studied the incubation of C. albicans isolates on fluconazole-containing medium and observed the loss of one homologue of chromosome 4 after 7 days of incubation. Interestingly, the CAI microsatellite is located on the same chromosome, and these patients were treated with fluconazole; therefore, this hypothesis could not be ruled out. The possible loss of chromosome 4, or part of it, which includes the CAI locus, would turn genotype 30-32 into genotype 30-30 in only one mutational step.
TABLE 5.
Genotypes of sequential isolates from vulvovaginal recurrent infections
| Patienta | No. of recurrent strains | Genotype observedb |
||
|---|---|---|---|---|
| CAI | CAIII | CAVI | ||
| K | 4 | 17-23 | 6-8 | 7-13 |
| L | 3 | 30-32 | 6-6 | 19-23 |
| 30-32 | 6-6 | 19-23 | ||
| 30-30 | 6-6 | 19-23 | ||
| M | 5 | 18-25 | 6-6 | 7-7 |
| N | 3 | 21-21 | 6-7 | 8-15 |
| 20-29 | 6-9 | 39-46 | ||
| 20-29 | 6-9 | 39-46 | ||
| O | 2 | 26-26 | 6-6 | 7-7 |
| P | 2 | 23-27 | 6-8 | 18-21 |
| 23-27 | 6-8 | 21-21 | ||
| Q | 2 | 22-23 | 6-8 | 24-24 |
| S | 2 | 20-20 | 9-9 | 31-37 |
Each patient is referred to as a separate letter (K to S).
Cases of microevolution are marked in boldface.
DISCUSSION
Candida albicans is an opportunistic yeast that can cause severe invasive infections, especially in immunocompromised patients, thus making the development of methodologies for the accurate discrimination of strains essential. Knowledge of the relatedness of strains involved in infections would be of extreme relevance to the development and application of the correct therapeutic strategy as well as to obtaining a better understanding of the epidemiology of this yeast. Moreover, some strains are capable of invading different body locations, and microevolution may occur as an adaptive response to new environments.
Microsatellites are among the most frequent markers used for differentiation purposes due to their hypervariability, the ease of PCR amplification and interpretation, their codominant profiles, and their potential for use in automated assays. These loci are relatively abundant in C. albicans, and several markers have been developed (5, 12, 26). However, the majority of microsatellites appear to present low levels of heterozygosity, which may be explained by the fact that they are situated in coding regions (12). In these situations, the discriminatory power is rather low, but it can be compensated for by surveying a larger number of loci, facilitated by multiplexing the PCR. The best approach was obtained by combining three loci, CDC3, EF3, and HIS3, thus yielding a discriminatory power of 0.97 (3). In the present work, a multiplex strategy was developed with three new microsatellite markers, located in noncoding regions, which presented the highest DP observed for C. albicans differentiation, 0.99.
The analysis of 72 independent isolates with CAI identified 16 different genotypes shared by two or more isolates. Application of this multiplex assay to assessment of the genetic relatedness of those isolates allowed the discrimination of 14 of 27 (51.8%) strains that otherwise would have been considered identical. It also allowed the confirmation of the identities of 10 isolates, which were, in fact, the same strain, and three cases of microevolution. Microevolution resulted from minor changes of the strain genotypes, i.e., a change at only one locus that could be explained by a single mutational step. Overall, the improved discriminatory potential of the methodology described here revealed the presence of three basic scenarios: (i) isolates that were the same strain, (ii) isolates that were the same strain(s) but that were apparently undergoing a process of microevolution, and (iii) isolates that corresponded to distinct strains. These results clearly indicate the need for multilocus analysis even when isolates are studied with a highly discriminatory molecular marker. Application of this methodology to the analysis of isolates from recurrent infections also revealed the same set of basic scenarios described previously (23, 24, 44). Of the 15 recurrent cases analyzed, 12 were due to the same strain, 2 were due to the same strain which was undergoing microevolution, and only 1 was due to strain replacement. This multiplex system demonstrated that C. albicans strains can undergo microevolutionary events at body sites of carriage during colonization between recurrent episodes of infection and that these events seem to be relatively more frequent than strain replacement (6, 23, 24).
Analysis of the microevolutionary events observed in this study suggested that two different mutational events could be occurring: strand slippage of the DNA polymerase in the microsatellite region and LOH as the result of chromosomal rearrangements; both of these mutational events take place at similar rates. The reported rates for chromosomal rearrangements were from 1.2 × 10−3 to 3.0 × 10−3 (8, 41), while for microsatellite loci the values varied between 10−2 and 10−6 (14, 19, 53, 54). Chromosomal alterations have been widely documented in C. albicans, with some being associated with the loss of entire or parts of chromosomes (27, 40, 41, 50, 52). Several conditions are known to induce LOH by chromosomal alterations, i.e., heat shock (17); exposure to different carbon sources, such as l-sorbose and d-arabinose (21, 41); and exposure to fluconazole (34). Metzgar et al. (31) also reported changes in microsatellites in vivo between the pretreatment isolate and the isolate obtained after treatment with different doses of fluconazole. The association of highly polymorphic STRs to different chromosomes and subchromosomal locations could be of great use for the evaluation of the loss of the entire chromosome or parts of chromosomes in C. albicans. Although LOH was observed at CAI, additional experiments are needed to confirm if it can be associated with rearrangements at chromosome 4 in response to fluconazole pressure.
Several studies have supported the concept that C. albicans contains a source of potentially beneficial genes that are activated by changes in chromosome number and that this elaborate mechanism regulates the utilization of food supplies and possibly other important functions in response to environmental stress. As C. albicans clinical populations are usually clonal, we suggest that in the absence of genetic exchange through sexual reproduction, microevolution could conceivably confer a selective advantage under adverse environmental conditions.
It is clear that the microsatellite multiplex PCR-based system described here enables high-speed typing, which makes it useful in large epidemiological studies. Furthermore, its capacity to detect microevolutionary events can make it useful for the detection of strain microevolution in response to environmental stress conditions, providing support for therapeutic adjustments, especially in patients with recurrent infections.
The standardization of microsatellite typing systems, including the primers and the separation techniques used and the allele nomenclature, is an issue that should be accomplished to allow interlaboratory comparisons. The creation of public databases that would make microsatellite allele data available worldwide, similar to those already in use for human microsatellites (29, 38), is another essential topic that deserves attention.
REFERENCES
- 1.Barchiesi, F., L. F. Di Francesco, P. Compagnucci, D. Arzeni, O. Cirioni, and G. Scalise. 1997. Genotypic identification of sequential Candida albicans isolates from AIDS patients by polymerase chain reaction techniques. Eur. J. Clin. Microbiol. Infect. Dis. 16:601-605. [DOI] [PubMed] [Google Scholar]
- 2.Barchiesi, F., R. J. Hollis, M. Del Poeta, D. A. McGough, G. Scalise, M. G. Rinaldi, and M. A. Pfaller. 1995. Transmission of fluconazole-resistant Candida albicans between patients with AIDS and oropharyngeal candidiasis documented by pulsed-field gel electrophoresis. Clin. Infect. Dis. 21:561-564. [DOI] [PubMed] [Google Scholar]
- 3.Botterel, F., C. Cesterke, C. Costa, and S. Bretagne. 2001. Analysis of microsatellite markers of Candida albicans used for rapid typing. J. Clin. Microbiol. 39:4076-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bougnoux, M. E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretagne, S., J. M. Costa, C. Besmond, R. Carsique, and R. Calderone. 1997. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as the basis for a typing system. J. Clin. Microbiol. 35:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong, P. P., Y. L. Lee, B. C. Tan, and K. P. Ng. 2003. Genetic relatedness of Candida strains isolated from women with vaginal candidiasis in Malaysia. J. Med. Microbiol. 52:657-666. [DOI] [PubMed] [Google Scholar]
- 7.Correia, A., P. Sampaio, J. Almeida, and C. Pais. 2004. Study of molecular epidemiology of candidiasis in Portugal by PCR fingerprinting of Candida clinical isolates. J. Clin. Microbiol. 42:5899-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalle, F., N. Franco, J. Lopez, O. Vagner, D. Caillot, P. Chavanet, B. Cuisenier, S. Aho, S. Lizard, and A. Bonnin. 2000. Comparative genotyping of Candida albicans bloodstream and nonbloodstream isolates at a polymorphic microsatellite locus. J. Clin. Microbiol. 38:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field, D., and C. Wills. 1996. Long polymorphic microsatellite in simple organisms. Proc. R. Soc. London B Biol. Sci. 263:209-215. [DOI] [PubMed] [Google Scholar]
- 11.Field, D., and C. Wills. 1998. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl. Acad. Sci. USA 95:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field, D., L. Eggert, D. Metzgar, R. Rose, and C. Wills. 1996. Use of polymorphic short and clustered coding-region microsatellites to distinguish strains of Candida albicans. FEMS Immunol. Med. Microbiol. 15:73-79. [DOI] [PubMed] [Google Scholar]
- 13.Fundyga, R. E., T. J. Lott, and J. Arnold. 2002. Population structure of Candida albicans, a member of the human flora, as determined by microsatellite loci. Infect. Genet. Evol. 2:57-68. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, M. G., C. M. Bull, S. J. B. Cooper, and G. A. Duffield. 2000. Microsatellite mutations in litters of the Australian lizard, Egernia stokesii. J. Evol. Biol. 13:551-560. [Google Scholar]
- 15.Graser, Y., M. Volovsek, J. Arrington, G. Schonian, W. Presber, T. G. Mitchell, and R. Vilgalys. 1996. Molecular markers reveal the population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93:12473-12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennequin, C., A. Thierry, G. F. Richard, G. Lecointre, H. V. Nguyen, C. Gaillardin, and B. Dujon. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilton, C., D. Markie, B. Corner, E. Rikkerink, and R. Poulter. 1985. Heat shock induces chromosome loss in the yeast Candida albicans. Mol. Gen. Genet. 200:162-168. [DOI] [PubMed] [Google Scholar]
- 18.Howell, K. S., E. J. Bartowsky, G. H. Fleet, and P. A. Henschke. 2004. Microsatellite PCR profiling of Saccharomyces cerevisiae strains during wine fermentation. Lett. Appl. Microbiol. 38:315-320. [DOI] [PubMed] [Google Scholar]
- 19.Huang, Q. Y., F. H. Xu, H. Shen, H. Y. Deng, Y. J. Liu, Y. Z. Liu, J. L. Li, R. R. Recker, and H. W. Deng. 2002. Mutation patterns at dinucleotide microsatellite loci in humans. Am. J. Hum. Genet. 70:625-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson′s index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janbon, G., F. Sherman, and E. Rustchenko. 1999. Appearance and properties of l-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 153:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Lockhart, S. R., B. D. Reed, C. L. Pierson, and D. R. Soll. 1996. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “subshuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J. Clin. Microbiol. 34:767-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart, S. R., J. J. Fritch, A. S. Meier, K. Schroppel, T. Srikantha, R. Galask, and D. R. Soll. 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lott, T. J., R. E. Fundyga, M. E. Brandt, L. H. Harrison, A. N. Sofair, R. A. Hajjeh, and D. W. Warnock. 2003. Stability of allelic frequencies and distributions of Candida albicans microsatellite loci from U.S. population-based surveillance isolates. J. Clin. Microbiol. 41:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lunel, F. V., L. Licciardello, S. Stefani, H. A. Verbrugh, W. J. Melchers, J. F. Meis, S. Scherer, and A. van Belkum. 1998. Lack of consistent short sequence repeat polymorphisms in genetically homologous colonising and invasive Candida albicans strains. J. Bacteriol. 180:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 28.Magee, B. B., T. M. Souza, and P. T. Magee. 1987. Strain and species identification by restriction length polymorphism in the ribosomal DNA repeats of Candida species. J. Bacteriol. 169:1639-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, P. D., H. Schmitter, and P. M. Schneider. 2001. A brief history of the formation of DNA databases in forensic science within Europe. Forensic Sci. Int. 119:225-231. [DOI] [PubMed] [Google Scholar]
- 30.Matthews, R. C. 1994. Pathogenicity determinants of Candida albicans: potential targets for immunotherapy? Microbiology 140:1505-1511. [DOI] [PubMed] [Google Scholar]
- 31.Metzgar, D., A. van Belkum, D. Field, R. Haubrich, and C. Wills. 1998. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J. Clin. Microbiol. 36:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metzgar, D., J. Bytof, and C. Wills. 2000. Selection against frameshift mutations limits expansion in coding DNA. Genome Res. 10:72-80. [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer, W., K. Marszewska, M. Amirmostofian, R. P. Igreja, C. Hardtke, K. Methling, M. A. Viviani, A. Chindamporn, S. Sukroongreung, M. A. John, D. H. Ellis, and T. C. Sorrell. 1999. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20:1790-1799. [DOI] [PubMed] [Google Scholar]
- 34.Perepnikhatka, V., F. J. Fischer, M. Niimi, R. A. Baker, R. D. Cannon, Y. K. Wang, F. Sherman, and E. Rustchenko. 1999. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J. Bacteriol. 181:4041-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaller, M. A., S. R. Lockhart, C. Pujol, J. A. Swails-Wenger, S. A. Messer, M. B. Edmond, R. N. Jones, R. P. Wenzel, and D. R. Soll. 1998. Hospital specificity, region specificity and fluconazole resistance of Candida albicans bloodstream isolates. J. Clin. Microbiol. 36:1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pupko, T., and D. Graur. 1999. Evolution of microsatellites in the yeast Saccharomyces cerevisiae: role of length and number of repeated units. J. Mol. Evol. 48:313-316. [DOI] [PubMed] [Google Scholar]
- 38.Roewer, L., M. Krawczak, S. Willuweit, M. Nagy, C. Alves, A. Amorim, K. Anslinger, C. Augustin, A. Betz, E. Bosch, A. Cagliá, A. Carracedo, D. Corach, T. Dobosz, B. M. Dupuy, S. Füredi, C. Gehrig, L. Gusmão, J. Henke, L. Henke, M. Hidding, C. Hohoff, B. Hoste, M. A. Jobling, H. J. Kärgel, P. de Knijff, R. Lessig, E. Liebeherr, M. Lorente, B. Martínez-Jarreta, P. Nievas, M. Nowak, W. Parson, V. L. Pascali, G. Penacino, R. Ploski, B. Rolf, A. Sala, U. Schmidt, C. Schmitt, P. M. Schneider, R. Szibor, J. Teifel-Greding, and M. Kayser. 2001. Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci. Int. 118:106-113. [DOI] [PubMed] [Google Scholar]
- 39.Ruhnke, M., A. Eigler, I. Tennagen, B. Geiseler, E. Engelmann, and M. Trautmann. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J. Clin. Microbiol. 32:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rustchenko, E. P. 1991. Variations of Candida albicans electrophoretic karyotypes. J. Bacteriol. 173:6586-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rustchenko, E. P., D. H. Howard, and F. Sherman. 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J. Bacteriol. 176:3231-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sampaio, P., L. Gusmão, C. Alves, C. Pina-Vaz, A. Amorim, and C. Pais. 2003. Highly polymorphic microsatellite for identification of Candida albicans strains. J. Clin. Microbiol. 41:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid, J., S. Herd, P. R. Hunter, R. D. Cannon, M. S. Yasin, S. Samad, M. Carr, D. Parr, W. McKinney, M. Schousboe, B. Harris, R. Ikram, M. Harris, A. Restrepo, G. Hoyos, and K. P. Singh. 1999. Evidence for a general purpose genotype in Candida albicans, highly prevalent in multiple geographic regions, patient types and types of infection. Microbiology 145:2405-2413. [DOI] [PubMed] [Google Scholar]
- 44.Schroppel, K., M. Rotman, R. Galask, K. Mac, and D. R. Soll. 1994. Evolution and replacement of Candida albicans strains during recurrent vaginitis demonstrated by DNA fingerprinting. J. Clin. Microbiol. 32:2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shemer, R., Z. Weissman, N. Hashman, and D. Kornitzer. 2001. A highly polymorphic degenerate microsatellite for molecular strain typing of Candida krusei. Microbiology 147:2021-2028. [DOI] [PubMed] [Google Scholar]
- 46.Sternberg, S. 1994. The emerging fungal threat. Science 266:1632-1634. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennett, and D. C. Coleman. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterisation of a novel species associated with oral candidiasis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]
- 48.Tautz, D. 1998. Evolutionary biology. Debatable homologies. Nature 395:17-19. [DOI] [PubMed] [Google Scholar]
- 49.Tavanti, A., N. A. Gow, S. Senesi, M. C. Maiden, and F. C. Odds. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J. Clin. Microbiol. 41:3765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tavanti, A., N. A. Gow, M. C. Maiden, F. C. Odds, and D. J.Shaw. 2004. Genetic evidence for recombination in Candida albicans based on haplotype analysis. Fungal Genet. Biol. 41:553-562. [DOI] [PubMed] [Google Scholar]
- 51.Thanos, M., G. Schonian, W. Meyer, C. Schweynoch, Y. Graser, T. G. Mitchell, W. Presber, and H. J. Tietz. 1996. Rapid identification of Candida species by DNA fingerprinting with PCR. J. Clin. Microbiol. 34:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsang, P. W., B. Cao, P. Y. Siu, and J. Wang. 1999. Loss of heterozygosity, by mitotic gene conversion and crossing over, causes strain-specific adenine mutants in constitutive diploid Candida albicans. Microbiology 145:1623-1629. [DOI] [PubMed] [Google Scholar]
- 53.Vázquez, J. F., T. Pérez, J. Albornoz, and A. Domínguez. 2000. Estimation of microsatellite mutation rates in Drosophila melanogaster. Genet. Res. 76:323-326. [DOI] [PubMed] [Google Scholar]
- 54.Weber, J. L., and C. Wong. 1993. Mutation of human short tandem repeats. Hum. Mol. Genet. 2:1123-1128. [DOI] [PubMed] [Google Scholar]
- 55.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with the increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wickes, B. L., and R. Petter. 1996. Genomic variation in C. albicans. Curr. Top. Med. Mycol. 7:71-86. [PubMed] [Google Scholar]