Abstract
DNA from fossil human bones could provide invaluable information about population migrations, genetic relations between different groups and the spread of diseases. The use of ancient DNA from bones to study the genetics of past populations is, however, very often compromised by the altered and degraded state of preservation of the extracted material. The universally observed postmortem degradation, together with the real possibility of contamination with modern human DNA, makes the acquisition of reliable data, from humans in particular, very difficult. We demonstrate that relatively well preserved DNA is occluded within clusters of intergrown bone crystals that are resistant to disaggregation by the strong oxidant NaOCl. We obtained reproducible authentic sequences from both modern and ancient animal bones, including humans, from DNA extracts of crystal aggregates. The treatment with NaOCl also minimizes the possibility of modern DNA contamination. We thus demonstrate the presence of a privileged niche within fossil bone, which contains DNA in a better state of preservation than the DNA present in the total bone. This counterintuitive approach to extracting relatively well preserved DNA from bones significantly improves the chances of obtaining authentic ancient DNA sequences, especially from human bones.
Keywords: ancient DNA, preservation
Remnants of DNA preserved in fossil bones are potentially invaluable sources of information on the evolution, population dynamics, migrations, diets, and diseases of vertebrates, including humans. Despite almost two decades of work in the ancient DNA (aDNA) field, it has proven to be difficult to obtain reliable results, especially from human bones.
The enormous power of the PCR to amplify even a few copies of a particular DNA sequence makes working with partially degraded DNA possible (1, 2). However, for PCR to produce reliable results, the segment of DNA of interest has to be well preserved. aDNA samples are, however, often damaged and/or contaminated (3–7). Damaged DNA may produce different cloned sequences, making it difficult to identify the authentic sequence (5–9). Furthermore, it has recently been found that the preferred sites for naturally occurring mutations are often also the preferred sites where postmortem damage occurs (10–12), although this observation has been challenged (13). The majority of postmortem mutations arise from the deamination of cytosine (C) to uracil, an analogue of thymine (T) or the deamination of adenine (A) to hypoxanthine, an analogue of G. Because both transitions occur on both strands of the DNA, they can appear in the sequenced strand as A to G and T to C (type 1) or as C to T and G to A (type 2) transitions. In addition to deamination, a small proportion of miscoding lesions may also be the result of oxidatively derived transversions (9).
Another serious problem is the contamination of ancient DNA by contemporary DNA (2). Contemporary DNA contaminants are common in almost all ancient remains (6, 14) and are more likely to be amplified by PCR and to provide reproducible cloned sequences compared with damaged DNA (15). It is thus very difficult to obtain reliable genetic information from aDNA, and it is essential to optimize access to the best preserved DNA available in a fossil specimen.
Here, we show that relatively well preserved DNA can be preserved in intergrown crystal aggregates within fossil bones. The approach used also alleviates a major source of modern contamination. The existence of crystal aggregates in bone was reported by Weiner and Price (16). They showed that a significant portion of the crystals in modern bones exists as intergrown crystal aggregates that cannot be disaggregated when the collagenous matrix is removed by oxidation with sodium hypochlorite (NaOCl), even if the bone is ground into a fine powder. Sodium hypochlorite will readily oxidize almost all organic matter that is accessible in the bone powder. This organic matter includes external DNA contaminants and degraded organic matrix molecules that are exposed to water from the surroundings. DeNiro and Weiner (17) showed that remnants of collagen and probably other proteins are preserved within these aggregates in fossil bones and are not destroyed by the NaOCl.
We initially determined that DNA is present in aggregates first in modern bones and then in fossil bones of different species. We then evaluated the state of DNA preservation in fossil bone aggregates compared with untreated bone powder from the same bone. Larger and better preserved DNA fragments were obtained from the aggregates.
Materials and Methods
Samples. Compact bone from six fossil and two modern animal bones were analyzed. The fossil bones, the periods from which they were derived, and their radiocarbon ages are listed in Table 1. Five of the six fossil bones were excavated in archaeological sites in the Levant, whereas the penguin (Pygoscelis adeliae) bone was excavated in King George Island, Antarctica. The five samples from the Levant include a human (Homo sapiens) femur from the site of Wadi Makuch, which is a closed dry cave in the Judean desert, a Bos primigenius tibia from the site of Motza, near Jerusalem and humeri from a cat (Felis catus) and a chicken (Gallus gallus) from the site of Akko. The modern bones are femora from a pig (Sus scrofa) and a bovid (Bos taurus). Aggregates were also extracted from a modern human (H. sapiens) femur. No special precautions were taken to prevent DNA contamination during excavation and handling before DNA analysis.
Table 1. HCl-insoluble collagen contents of five fossil bones of different ages.
| Bone sample | Calibrated age | 14C age B.P. uncal. | Period | Collagen content, wt % |
|---|---|---|---|---|
| G. gallus | 2nd—3rd century A.D.* | Roman | 6.8 | |
| F. catus | 2nd—3rd century A.D.* | Roman | 20 | |
| H. sapiens | 6,540-6,170 B.P.† | 5,560 ± 80‡ | Chalcolithic | 12.6 |
| P. adeliae | 8,600-8,390 B.P.† | 7,715 ± 60‡ | 15 | |
| B. primigenius | 10,100-10,600 B.P.§ | Early PPNB | 2.5 |
Age is based on the archaeological context (G. Bar Oz, personal communication).
Calibration is performed for ±2 SD using oxcal 3.9 (34).
Radiocarbon dating for samples RTT-4434 and RTT-4388 was performed at the Radiocarbon Laboratory of The Weizmann Institute.
Calibrated age is based on several 14C ages from charcoal and bone samples from the same stratigraphic unit (22). A.D., anno Domini; PPNB, pre-pottery Neolithic B.
Precautions Regarding Contamination During DNA Analysis. Following Hofreiter et al. (18), Cooper and Poinar (19), and Pääbo et al. (20), we paid close attention to contamination in the laboratory. All pre-PCR procedures were performed in a dedicated aDNA laboratory with high efficiency particulate air (HEPA) filters. Amplifications were carried out in separated rooms, and post-PCR procedures were performed in another building. All reagents and equipment were bleached and UV irradiated (1.0 J/cm2, 254 nm for 45 min) before use. Molecular biology grade reagents were used (obtained from Sigma unless a different company is mentioned). No modern DNA was ever introduced into the aDNA laboratory, and positive PCR controls were never performed. A blank reaction was performed in parallel with each extraction, and several negative control PCRs were made. Note that the hypervariable region 1 (HV-1) sequence of M.S., the person who conducted the DNA analyses, is the Cambridge Reference Sequence.
Determining Weight Percent Insoluble Collagen in Bone. Powdered bone (150 mg, particle size <500 μm) was dissolved in 4 ml of 1 M HCl for 10 min with gentle shaking and centrifuged (2 min, 12,000 × g), and the supernatant was discarded. The insoluble fraction was resuspended twice with distilled water and centrifuged. It was then dried at 58°C overnight. The dried material was weighed, and its content was calculated as the weight percent (wt%) of total bone powder. We verified that this dried material is collagen and assessed its quality by means of Fourier transform infrared (FTIR) spectrometry. FTIR spectra (MIDAC, Costa Mesa, CA) were obtained by mixing ≈0.1 mg of powdered sample with ≈80 mg of KBr. Spectra were collected at 4 cm–1 resolution. For more details, see Weiner et al. (21).
Quantitative Separation Between Crystal Aggregates and Single Crystals. This procedure followed Weiner and Price (16) with several modifications. Ground compact bone powder (100 mg, particle size <500 μm) was mixed continuously with 10 ml of 2.5% NaOCl for 2 h, at room temperature. The sample was centrifuged (2 min, 3,000 × g), and the insoluble fraction was reground and resuspended in 10 ml 2.5% NaOCl. This procedure was repeated two more times, each time for 1 h using a fresh 2.5% NaOCl solution. The pellet was then washed twice with 10 ml of distilled water (saturated with hydroxyl apatite and filtered through a 0.45-μm Corning filter) and twice with 10 ml of 95% ethanol (saturated with hydroxyl apatite). The sample was centrifuged between washes to remove the supernatant. The pellet was resuspended in 2 ml of absolute ethanol and dried at 55°C overnight. Fifteen milligrams of the dried material was placed in a 12 × 75-mm test tube (Kimble, Toledo, OH) with 3 ml of 95% ethanol (saturated with respect to hydroxyl apatite) and sonicated (55,000 cycles per s) for 2 min. The sample was then vortexed and allowed to settle for 5 min. The heaviest fraction that settles within 5 min is defined as the aggregate fraction. The lightest fraction that did not settle even after 2 h contains mainly single crystals. These fractions were dried and weighed. An aliquot was examined in FTIR (see above) and in a scanning electron microscope (Leo Gemini, Oberkochen, Germany) after coating with chromium.
Preparation of Aggregates for DNA Extraction. We followed the above procedure for treating the bone with NaOCl with some modifications. Two hundred milligrams of NaOCl-treated bone powder was placed in a 15-ml sterile tube with 3 ml of 95% ethanol and sonicated for 1 min. The sample was then vortexed and allowed to settle for 1 min. The precipitant was washed for 1 min once with 2.5% NaOCl and twice with double distilled water (irradiated for 20 min at 254 nm) before DNA extraction, to assure no contamination of foreign DNA during aggregate handling with ethanol. No solutions saturated with hydroxyl apatite were used.
Preparation of Whole Bone Powder for DNA Extraction. A fragment of compact bone (2 × 2 cm2) was briefly rinsed with 2.5% bleach solution, and 2 mm of the outer layer was removed in one piece by using an electric drill (Dremel, Racine, WI). The bone was crushed in a mortar and pestle and then sieved to obtain particles of <500 μm.
DNA Extraction. Two hundred milligrams of whole bone powder or aggregates were decalcified with 10 ml of EDTA solution (0.5 M, pH 8.0). This solution was incubated at room temperature with continuous rotation. EDTA solution was exchanged every 1–2 h followed by centrifugation using a swing-out rotor (5 min, 2,000 × g), until decalcification was complete. Decalcified bone was incubated in proteinase K buffer (10 mM Tris base, pH 8.0/1 mM DTT/50 mM EDTA/100 mM NaCl/0.5% SDS/2 mg of proteinase K) (Roche Applied Science) for 10–45 min at 60°C (to convert the collagen into gelatin) and continued overnight at 37°C. The DNA was then extracted with 1 vol of 25:24:1 phenol:chloroform:isoamyl alcohol saturated with 10 mM Tris base (pH 8.0) and 1 mM EDTA and 2 vol of 1-butanol. The extracted DNA was purified and concentrated by using a YM30 unit (Millipore) to a final volume of 250 μl. To avoid cross-contamination between the whole bone extract and the aggregates extract, DNA extraction from whole bone powder was always performed after the extraction from the aggregates.
DNA Analysis. To avoid false negative results, we first analyzed the DNA extracts for the presence of PCR inhibitors by spiking a lambda PCR containing 0.5 pg of λDNA with variable amounts of aDNA extract (0.1–5 μl). The magnitude of inhibition was determined by a comparison of PCR products with and without aDNA extract. Second, we analyzed all extracts for modern human DNA contaminants. Non-human samples were analyzed for a 132-bp fragment from the human HV-1 region and human samples for a fragment of 520 bp, which is greater than the length we expect from ancient human samples. The state of DNA preservation was then assessed by amplifying DNA fragments of increasing size by using the same 5′ primer. These fragments are integrated within HV-1, cytochrome b, or 12S of the mtDNA genome. Primers were targeted to conserved regions within these regions (see supporting information, which is published on the PNAS web site). Amplifications were performed by using PTC-200 Thermal Cycler (MJ Research, Cambridge, MA) or iCycler iQ Real-Time PCR Detection System (Bio-Rad). The former reaction of a 50-μl volume contains 0.1–2.5 μl of DNA, 1.5 units of AmpliTaq Gold DNA Polymerase (PE Applied Biosystems), 1× reaction buffer including 1.5 mM MgCl2, 8 μg of BSA, 0.2 mM of each dNTP and 10 pmol of each primer. The second reaction mix of a 25-μl volume contains iQ SYBR Green Supermix (Bio-Rad) and 10 pmol of each primer. Primers were synthesized by using the high purity salt free (HPSF) method and analyzed by MALDI-TOF mass spectroscopy (MWG Biotech, Ebersberg, Germany). A touchdown protocol was applied to increase the yield of a specific product. Cycling conditions were 95°C for 3–10 min, followed by 30–50 cycles of 94°C for 30 s, 30 s at the relevant annealing temperature (a set point annealing temperature of 51°C was lowered to 48°C throughout the first 10 cycles) and 72°C for 30 s, with a final extension cycle at 72°C for 5 min. In the quantitative PCR using the iCycler 3, replicates were produced for each standard or sample and the primer-dimer signal was excluded from analysis. The set point temperature for melt curve data collection was 55°C, and it increased by 0.5°C per cycle for 80 cycles total. Only samples amplified in parallel with all clean PCR negative controls were analyzed by electrophoresis in 3% agarose gels, cloned, and sequenced. Sequences were compared with GenBank sequences by using blastn (National Institutes of Health database).
Cloning. The PCR product was inserted into a pGEM-T Easy vector (Promega), which was then inserted into XL1-Blue competent cells (Stratagene). Competent cells were grown on LB/Amp medium containing IPTG and X-Gal (Sigma). The inserts were amplified directly from the white colonies and were sequenced on an Applied Biosystems 3700 DNA analyzer machine.
Results
Insoluble collagen contents of the fossil bones are listed in Table 1 according to increasing age. All of the bones used in this study do contain insoluble collagen, which we regard as an indicator of a relatively good preservational state (unpublished data). Furthermore, there is no correlation between collagen content and age of the sample.
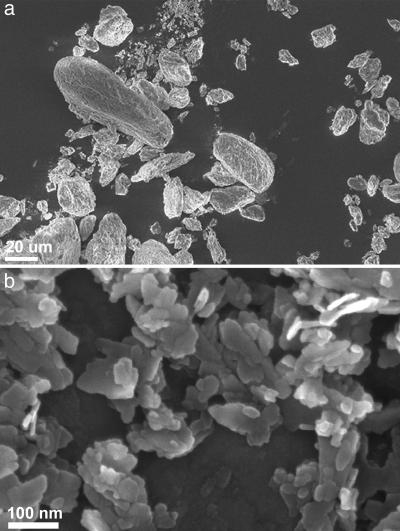
We extracted bone aggregates from these five fossil bones and three modern bones. Fig. 1a shows an SEM image of some aggregates from a fossil human bone from Wadi Makuch, and Fig. 1b shows individual bone crystallites extracted from the same bone. The crystal shapes and sizes are similar to modern bone crystals, namely flat thin plates (16, 23). Analysis of the structural organization of the crystals in the aggregates shows that it, too, is similar to modern bone lamellar structure. This finding implies that the aggregates are in essence regions within normal bone where the crystals have intergrown such that the oxidizing agent and mild sonication does not disaggregate them.
Fig. 1.
SEM of aggregate fraction from ancient human bone (a) and single crystals (b) from the same bone. Note that the aggregates are in the μm scale range, whereas the crystals are in the nanometer scale range.
The modern B. taurus femur produced a yield of ≈50 wt% aggregates (of the total bone mineral phase), which is similar to that reported by Weiner and Price (16). The modern H. sapiens femur aggregate content was higher than the value reported by Weiner and Price (16), 48% compared with 22%. The aggregate contents of four of the five fossil bones were around 50 wt% aggregates, whereas the fossil H. sapiens sample contained 24 wt% aggregates. It is not known whether the differences are due to age differences among the individuals and/or diagenetic effects.
Aggregates from the modern bovine (B. taurus) and pig (S. scrofa) bones were analyzed for the presence of DNA. After extracting DNA from these aggregates, as well as from the untreated whole bone powder, species-specific DNA fragments were amplified and sequenced. The sequences obtained from the aggregates were identical to those from the whole bone powder (accession nos. DQ020118 and DQ020119, respectively), and they aligned perfectly with published sequences from individuals from the same species (Fig. 2). It is thus clear that some DNA is occluded inside the aggregates and that the treatment with the aggressive oxidant does not affect its primary structure.
Fig. 2.
DNA sequences from crystal aggregates (A) and whole bone powder (W) of modern bone samples (1) B. taurus HV-1 sequence, positions 15943–16302. (2) S. scrofa CYTB sequence, positions 14285–14505. Published sequence of B. taurus (24, 25) and S. scrofa (26, 27) are shown in the top line. Bases identical to the reference sequence are indicated by dots.
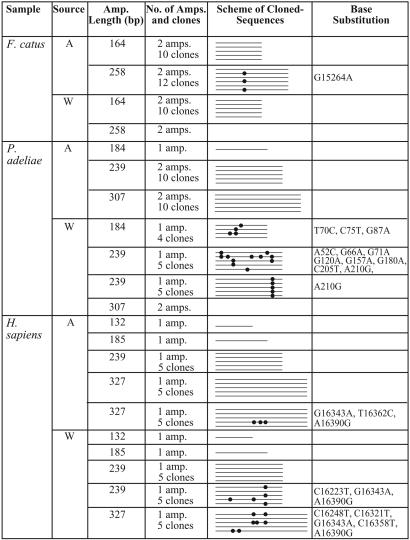
DNA was then extracted from five fossil bones by using aggregates as well as untreated whole bone powders. Although the primers were designed to express conserved sequences, no amplification was obtained from either the aggregates or the whole bone powders of two of the five fossil bones, namely B. primigenius and G. gallus. The fragments that failed to amplify were the shortest chosen for this study. Thus, these samples were not analyzed further. Using the other three fossil samples, P. adeliae, F. catus, and H. sapiens, we could amplify the shortest fragment defined for each species and therefore continued to analyze longer fragments. A comparison of the PCR products obtained from aggregate and whole bone powder showed the advantages of the aggregates over whole bone powder: larger and better preserved fragments were obtained by using aggregates compared with those from whole bone powder. Table 2 summarizes these results (for full sequences and comments on the sequence variations, see the supporting information). In all three samples, we defined a consensus sequence (authentic sequence) as the reproducible sequence among amplifications of different fragment lengths from different extracts (accession nos. DQ020115–DQ020117). The spectrum of variant bases within these ancient cloned sequences is consistent with previous studies of aDNA; 41.5% type 1 transition, 53.7% type 2 transition, and 4.8% transversion events (9, 28). Both transition events occurred as either singletons or in multiple clones. The percentage of these variants in each sequence analyzed was calculated (Table 3) and found to be significantly more frequent in sequences from whole bone powder. As expected from ancient sequences, increasing fragment length increases the number of variants detected. The variants percentage obtained is higher than the expected error of the polymerases used here, 1 × 10–5 to 1 × 10–6.
Table 2. Species-specific sequences of increasing length used in this study to compare the state of DNA preservation in aggregates (A) and whole bone powder (W).
Partial sequences from the HV-1 region and the CYTB and 12S genes were analyzed in the human, cat, and penguin bone samples, respectively. Total number of amplifications (amp.) and cloned sequences are shown. The scheme portrays sequences aligned with each other within each species, and the dots represent variations from the consensus sequence found here. The position and type of each variation are listed. Most of the sequences obtained from amplifications of different fragment lengths and from different extracts are identical. Two independent amplifications with identical cloned sequences were combined into one line, and only sequences from one are shown. An empty box means no PCR product.
Table 3. Comparison between variant bases per total sequence obtained using DNA extracts from aggregates and whole bone powder of five fossil bones.
| Variant bases per total sequence amplified
|
|||
|---|---|---|---|
| Fossil sample | Fragment length, bp | Whole bone powder | Aggregates |
| B. Primigenius | 121 | N.O. | N.O. |
| G. gallus | 168 | N.O. | N.O. |
| F. catus | 164 | 0% (0/1270) | 0% (0/1270) |
| 258 | N.O. | 0.2% (6/2664) | |
| P. adeliae | 184 | 0.7% (4/580) | 0% (0/145) |
| 239 | 0.8% (16/2000) | 0% (0/2000) | |
| 307 | N.O. | 0% (0/2670) | |
| H. sapiens | 132 | 0% (0/90) | 0% (0/90) |
| 185 | 0% (0/145) | 0% (0/145) | |
| 239 | 0.25% (5/1990) | 0% (0/995) | |
| 327 | 0.5% (7/1375) | 0.1% (3/2750) | |
Amplifications of increasing length were performed for each species. When the short fragment could not be obtained by PCR, analysis of longer fragments was not performed (N.O.). Primers were excluded from the calculation.
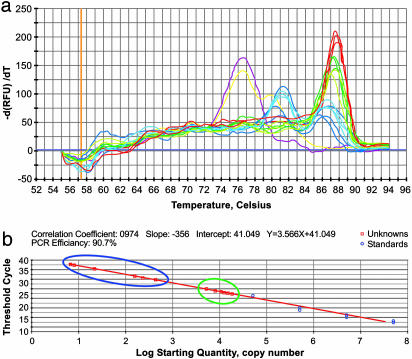
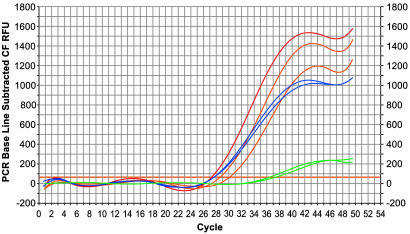
Quantitative PCRs performed on the P. adeliae bone sample using 0.1 μl, 2.5 μl, and 5 μl of DNA extract from aggregates produced a product of 239 bp, with means of 80 ± 125, 200 ± 135, and 807 ± 500 template molecules, respectively. In contrast, 0.1 μl and 2.5 μl of DNA extract from whole bone powder produced means of 15,000 ± 2,300 and 8,000 ± 2,200 molecules (Fig. 3), with no product using 5 μl of extract. Thus, DNA extracted from whole bone powder contained ≈2 orders of magnitude more DNA templates as compared with DNA extracted from aggregates. We also note that identical sequences of 239 bp were obtained from the aggregates extract using the three different volumes. However, the whole bone extract showed multiple “miscoding lesions” (Table 2 and supporting information). Based on the λ inhibition test, we found that whole bone powder extract inhibits the PCR by 5 orders of magnitude more than DNA extracts from aggregates (Fig. 4). The same inhibition pattern was obtained when using all other fossil bone samples (data not shown).
Fig. 3.
qPCR for 239 bp within the 12S rRNA of the fossil P. adeliae bone. DNA template [0.1 μl (dark color) or 2.5 μl (light color)] from aggregates (blue) and whole bone powder (green) were amplified in parallel with blanks of both DNA extraction procedures (yellow) and blanks composed of water for controlling the amplification procedure (pink). (a) Melting curve showing a real product with a melting temperature of 87°C using both extracts and standard (red) but not in the blanks. (b) Comparison between copy numbers of DNA template in each extract (□) with respect to standards (○). The amounts of available DNA template in 0.1 and 2.5 μl of aggregates extract are marked blue whereas those from whole bone powder are marked green.
Fig. 4.
The λ inhibition test using the fossil P. adeliae bone. Amplification plot displays the relative fluorescence for each sample at every cycle; red, λDNA (control); blue, λDNA spiked with 0.1 or 2.5 μl of DNA extract from aggregates; green, λDNA spiked with 0.1 or 2.5 μl of DNA extract from whole bone powder; orange, λDNA spiked with 2.5 μl of DNA extraction blanks. No inhibition is seen in reactions with DNA extract from aggregates, whereas a strong inhibition is seen in the reactions with the whole bone powder extract.
Human DNA contamination was not found in any of the fossil non-human animal bones analyzed in this study in both the aggregates and the whole bone powder extracts. Furthermore, no large human mitochondrial sequences (520 bp) were obtained using the fossil human bone. However, three contaminant cloned sequences of unknown origin were obtained using the whole bone powder of P. adeliae. All extraction and PCR blanks were consistently negative throughout the study, indicating that the results are unlikely to be derived from contaminants in the extraction or PCR processes.
Discussion
We demonstrate the presence of DNA in crystal aggregates in modern and fossil bones, and showed that in some fossil bones these aggregates contain better preserved DNA than untreated whole bone powder, based on frequently obtained reproducible sequences from the aggregates. We regard reproducibility in powdered samples treated with NaOCl as an important criterion for authenticity.
The counterintuitive idea of treating finely ground bone powder with a strong oxidant to obtain well preserved DNA is based on the hypothesis that the intergrown crystal aggregates will protect the DNA from the environment. Others have used the oxidant NaOCl to clean the surfaces of macroscopic pieces of bone (6, 29, 30). In our approach, not only is the hypothesis shown to be correct for some fossil bones, but the treatment has the enormous advantage of also removing modern contaminants as well as many of the inhibitors that interfere with the PCR amplification process.
Our study also shows that in all modern bones well preserved DNA is occluded within aggregates. However, not all aggregate fractions from fossil bones have preserved DNA, and as can be expected from fossil material in different states of preservation, the benefits of using the aggregate fraction for DNA extraction vary between bones. Good examples are the fossil cat and chicken bones examined here. Both are from the same archaeological site and from the same period and have similar proportions of aggregates. Nevertheless, the cat bone produced amplifiable DNA from the aggregate fraction whereas in the chicken bone no amplifiable DNA could be obtained. This difference could not be explained by the nature of the mineral, because the mineral in the aggregates of both bones, and in fact all of the bones in this study, did not show any systematic differences in infrared splitting factor. Furthermore, similar splitting factors were found in their whole bone powders. The splitting factor reflects the crystal size and extent of imperfections (31). Thus, the more important parameter for DNA preservation may be the quality of the aggregates and not their quantity. Much more work needs to be done to understand the nature of the protective environment in the aggregates of modern and fossil bones.
The major benefits of using the aggregate fraction as opposed to whole bone powder are as follows. (i) Longer and better preserved DNA molecules were found in aggregates compared with those obtained by using whole bone powder. Most of the short sequences analyzed here were identical in both extracts (aggregates and whole bone powder extracts). However, as the fragment length analyzed increased, more “miscoding lesions” appeared, mainly in whole bone extracts. The presence of multiple miscoding lesions at a single position in a longer fragment can be explained by jumping PCR, namely obtaining long amplification products even if all of the templates are too short (3, 11), especially when no product could be obtained while trying to increase the fragment analyzed further on. This problem was seen in whole bone extracts in shorter fragments compared with the extracts from aggregates. We therefore defined authenticity based on better preserved DNA extracts with reproducibility in sequences of increasing length. (ii) Fewer PCR inhibitors were coextracted with the DNA from aggregates. PCR inhibitors are well known in fossil samples (6), where they prevent DNA fragments from being amplified. The approach currently used to minimize this problem is to reduce the amount of these inhibitors by dilutions of the extract 1:10–1:1,000,000 (6, 12). These dilutions, however, dramatically reduce the number of DNA molecules available for the PCR, sometimes down to a single molecule. By using the aggregates, no dilutions were needed for the polymerase activity. On the contrary, increasing volumes produced more PCR product, with identical DNA sequences. Finding more DNA damage in whole bone extracts containing more PCR inhibitors is in agreement with refs. 6 and 12. (iii) The use of NaOCl-treated bone powder greatly reduces the possibility of contamination by extraneous sequences of the same species and from sample handling before extraction. Because modern DNA is more likely to be amplified by PCR, we expect to see such a sequence in all fragment lengths analyzed. However, there is no such sequence except for the one we regard as authentic (see above). Moreover, because the samples analyzed in this study are the first samples per species to be analyzed in our laboratory and because the variations in the sequences are not present in the blstn database, their presence as a laboratory contaminant therefore seems unlikely. Contaminating modern human DNA has been noted in many studies despite the use of rigorous protocols (6, 15, 18, 32, 33). Because we could not detect such a contaminant in the samples treated with NaOCl, we propose that this treatment may be particularly beneficial when studying fossil human bones.
The detailed study of the fossil penguin bone provided a better understanding of the DNA preserved in the aggregates. The penguin whole bone powder extract contained tens of thousands of DNA molecules per PCR that produced many different cloned sequences. Among the different cloned sequences, most contained A to G substitutions that may have been considered authentic if a comparison with the aggregate extract results had not been made. Our results therefore suggest that nonrandomness of changes occurring during diagenesis can result in artifacts due to damaged template DNA being mistaken for genuine sequence features. This observation is in agreement with several previous studies (6, 10–12, 28).
The aggregate extract resulted in reproducible cloned sequences that enabled us to determine which of the different cloned sequences from the penguin whole bone powder is authentic. Pääbo et al. (20) noted that specific errors stemming from postmortem changes are not expected to predominate in an amplified population of molecules and that these errors should be minimized when amplifications start from >1,000 molecules. They also noted that in such samples a single amplification is sufficient. On the contrary, in this study, better preserved cloned sequences were found in amplifications using aggregates extracts containing tens to hundreds of template molecules. Highly damaged cloned sequences were obtained in amplifications using whole bone extracts, despite the presence of thousands of template molecules. Thus, clarifying the quality of the template DNA more than its quantity may be a more important criterion for authenticity.
In conclusion, to increase the probability that fossil bones will produce authentic sequences, we propose the use of crystal aggregates as a substrate that may contain well preserved DNA. Using NaOCl-treated aggregates improves our ability to differentiate between contaminant DNA, highly damaged DNA, and better preserved DNA. Reproducible sequences of well preserved DNA from a NaOCl-treated sample can be used as a reliable criterion for authenticity.
Supplementary Material
Acknowledgments
We thank J. Beckman for his help and advice. We also thank Guy Bar Oz (Haifa University, Haifa, Israel), Hamudi Khalally (Israel Antiquities Authority, Jerusalem), Moshe Inbar (Haifa University), and Josef Zias (Science and Antiquity Group, Hebrew University, Jerusalem) for supplying us the bones. We thank E. Boaretto (The Weizmann Institute) for her help with the radiocarbon dating. We thank the Laboratories of Analytical Biology of the Smithsonian Institution for their support. This study was funded by the Kimmel Center for Archaeological Science at The Weizmann Institute, by Mr. George Schwartzmann (Sarasota, FL), as well as by a Burch Fellowship to S.W. from the Smithsonian Institution. S.W. is the incumbent of the Dr. Walter and Dr. Trude Borchardt Professorial Chair in Structural Biology. M.S. received a Dan David scholarship.
Author contributions: M.S., N.T., and S.W. designed research; M.S. performed research; M.S. and S.W. contributed new reagents/analytic tools; M.S., S.W., and N.T. analyzed data; N.T. and S.W. supervised ancient biomolecules; B.A. supervised anatomy; S.W. supervised bone structure; and M.S., N.T., and S.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: aDNA, ancient DNA; HV-1, hypervariable region 1.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ020115–DQ020119).
References
- 1.Hagelberg, E., Sykes, B. & Hedges, R. (1989) Nature 342, 485. [DOI] [PubMed] [Google Scholar]
- 2.Pääbo, S., Higuchi, R. & Wilson, A. (1989) J. Biol. Chem. 264, 9709–9712. [PubMed] [Google Scholar]
- 3.Pääbo, S., Irwin, D. & Wilson, A. (1990) J. Biol. Chem. 265, 4718–4721. [PubMed] [Google Scholar]
- 4.Lindahl, T. (1997) Cell 90, 1–3. [DOI] [PubMed] [Google Scholar]
- 5.Hofreiter, M., Jaenicke, V., Serre, D., Heaseler, A. & Pääbo, S. (2001) Nucleic Acids Res. 29, 4793–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolman, C. & Tuross, N. (2000) Am. J. Phys. Anthropol. 111, 5–23. [DOI] [PubMed] [Google Scholar]
- 7.Hoss, M., Jaruga, P., Zastawny, T. H., Dizdaroglu, M. & Pääbo, S. (1996) Nucleic Acids Res. 24, 1304–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen, A., Willerslev, E., Wiuf, C., Mourier, T. & Arctander, P. (2001) Mol. Biol. Evol. 18, 262–265. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert, M. T. P., Hansen, A. J., Willerslev, E., Rudbeck, L., Barnes, I., Lynnerup, N. & Cooper, A. (2003) Am. J. Hum. Genet. 72, 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, M. T. P., Willerslev, E., Hansen, A. J., Barnes, I., Rudbeck, L., Lynnerup, N. & Cooper, A. (2003) Am. J. Hum. Genet. 72, 32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee, M. & Brown, T. A. (2004) J. Archaeol. Sci. 31, 59–63. [Google Scholar]
- 12.Pusch, C. & Bachmann, L. (2004) Mol. Biol. Evol. 21, 957–964. [DOI] [PubMed] [Google Scholar]
- 13.Serre, D., Hofreiter, M. & Pääbo, S. Mol. Biol. Evol. 21, 1463–1467. [DOI] [PubMed]
- 14.Cooper, A., Rambaut, A., Macaulay, V., Willerslev, E., Hansen, A. J. & Stringer, C. (2001) Science 292, 1655–1656. [DOI] [PubMed] [Google Scholar]
- 15.Richards, M., Sykes, B. & Hedges, R. (1995) J. Archaeol. Sci. 22, 291–299. [Google Scholar]
- 16.Weiner, S. & Price, P. (1986) Calcif. Tissue Int. 39, 365–375. [DOI] [PubMed] [Google Scholar]
- 17.DeNiro, M. & Weiner, S. (1988) Geochim. Cosmochim. Acta 52, 2197–2206. [Google Scholar]
- 18.Hofreiter, M., Serre, D., Poinar, H., Kuch, M. & Pääbo, S. (2001) Nat. Rev. Genet. 2, 353–359. [DOI] [PubMed] [Google Scholar]
- 19.Cooper, A. & Poinar, H. (2000) Science 289, 1139. [DOI] [PubMed] [Google Scholar]
- 20.Pääbo, S., Poinar, H., Serre, D., Jaenicke-Després, V., Hebler, J., Rohland, N., Kuch, M., Krause J., Vigilant, L. & Hofreiter, M. (2004) Annu. Rev. Genet. 38, 645–679. [DOI] [PubMed] [Google Scholar]
- 21.Weiner, S., Goldberg, P. & Bar-Yosef, O. (1993) J. Archaeol. Sci. 20, 613–627. [Google Scholar]
- 22.Yizhaq, M., Mintz, G., Cohen, I., Khalally, H., Weiner, S. & Boaretto, E. (2005) Radiocarbon 47, 193–206. [Google Scholar]
- 23.Robinson, R. A. (1952) J. Bone Joint Surg. 34, 389–434. [PubMed] [Google Scholar]
- 24.Anderson, S., de Bruijn, M. H. L., Coulson, A. R., Eperon, I. C., Sanger, F. & Young, I. G. (1982) J. Mol. Biol. 156, 683–717. [DOI] [PubMed] [Google Scholar]
- 25.Mannen, H., Kohno, M., Nagata, Y., Tsuji, S., Bradley, D. G., Yeo, J. S., Nyamsamba, D., Zagdsuren, Y., Yokohama, M., Nomura, K., et al. (2004) Mol. Phylogenet. Evol. 32, 539–544. [DOI] [PubMed] [Google Scholar]
- 26.Kijas, J. M. & Andersson, L. A. (2001) J. Mol. Evol. 52, 302–308. [DOI] [PubMed] [Google Scholar]
- 27.Yang, J., Wang, J., Kijas, J., Liu, B., Han, H., Yu, M., Yang, H., Zhao, S. & Li, K. (2003) J. Hered. 94, 381–385. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert, M., Shapiro, B., Drummond, A. & Cooper A. (2005) J. Archaeol. Sci. 32, 1053–1060. [Google Scholar]
- 29.Yang, Y., Eng, B. & Saunders, S. (2003) Hum. Biol. 75, 355–364. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert, M. T. P., Rudbeck, L., Willerslev, E., Hansen, A. J., Smith, C. I., Penkman, K. E. H., Prangenberg, K., Nielsen-Marsh C. M., Jans, M. E., Arthur, P., et al. (2005) J. Arch. Sci. 32, 785–793. [Google Scholar]
- 31.Weiner, S. & Bar-Yosef, O. (1990) J. Archaeol. Sci. 17, 187–196. [Google Scholar]
- 32.Handt, O., Krings, M., Ward, R. & Pääbo, S. (1996) Am. J. Hum. Genet. 59, 368–376. [PMC free article] [PubMed] [Google Scholar]
- 33.Krings, M., Stone, A., Schmitz, R. W., Krainitzki, H., Stoneking, M. & Pääbo, S. (1997) Cell 90, 19–30. [DOI] [PubMed] [Google Scholar]
- 34.Bronk Ramsey, C. (1995) Radiocarbon 37, 425. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.