Abstract
Acute rheumatic fever (ARF), a sequelae of group A Streptococcus (GAS) infection, is the most common cause of preventable childhood heart disease worldwide. The molecular basis of ARF and the subsequent rheumatic heart disease are poorly understood. Serotype M18 GAS strains have been associated for decades with ARF outbreaks in the U.S. As a first step toward gaining new insight into ARF pathogenesis, we sequenced the genome of strain MGAS8232, a serotype M18 organism isolated from a patient with ARF. The genome is a circular chromosome of 1,895,017 bp, and it shares 1.7 Mb of closely related genetic material with strain SF370 (a sequenced serotype M1 strain). Strain MGAS8232 has 178 ORFs absent in SF370. Phages, phage-like elements, and insertion sequences are the major sources of variation between the genomes. The genomes of strain MGAS8232 and SF370 encode many of the same proven or putative virulence factors. Importantly, strain MGAS8232 has genes encoding many additional secreted proteins involved in human–GAS interactions, including streptococcal pyrogenic exotoxin A (scarlet fever toxin) and two uncharacterized pyrogenic exotoxin homologues, all phage-associated. DNA microarray analysis of 36 serotype M18 strains from diverse localities showed that most regions of variation were phages or phage-like elements. Two epidemics of ARF occurring 12 years apart in Salt Lake City, UT, were caused by serotype M18 strains that were genetically identical, or nearly so. Our analysis provides a critical foundation for accelerated research into ARF pathogenesis and a molecular framework to study the plasticity of GAS genomes.
Keywords: Streptococcus pyogenes‖GAS‖complete genome sequence‖ DNA microarray‖genomic diversity
Acute rheumatic fever (ARF), the leading cause of preventable pediatric heart disease worldwide, is a sequelae of group A Streptococcus (GAS) infection. Although the pathogenesis of ARF and rheumatic heart disease has been studied extensively (1), there is not a comprehensive understanding of the contributing pathogen and host factors. GAS are rarely isolated from patients with ARF, in part because symptoms begin well after the antecedent infection. This problem, together with the lack of an animal model, has hindered identification of GAS virulence determinants that contribute to these devastating diseases. Multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and molecular genetic analyses have shown that strains of GAS have extensive chromosomal diversity (2–4). For example, more than 100 M protein serotypes have been described, and allelic variation within a serotype has been identified (4, 5). Epidemiologic and molecular population genetic studies have implicated serotype M18 GAS strains with several ARF outbreaks in the U.S. (6). The genome of a serotype M1 organism was recently sequenced (7), but the strain is genetically distinct from M1 strains commonly responsible for GAS infections (8). Moreover, serotype M1 organisms are rarely associated with contemporary ARF outbreaks. As a first step toward gaining new insight into ARF pathogenesis and developing new therapeutics, we sequenced the genome of strain MGAS8232, a serotype M18 organism isolated from a patient with ARF. DNA microarray analysis of 36 serotype M18 strains collected in the U.S. from a wide range of infection types over 73 years also was conducted.
Materials and Methods
Genome Sequencing, Closure, and Annotation.
A library was made for random shotgun sequencing by cloning sheared chromosomal DNA from strain MGAS8232 in a TOPO-BLUNTII vector (Invitrogen). Nucleotide sequence data from colony-PCR-amplified clones were obtained with Big-Dye terminator chemistry and an ABI3700 automated capillary electrophoresis sequencer (Applied Biosystems). Sequence data storage and assembly were conducted with the FINCH data management system (Geospiza, Seattle), sps-phrap (Southwest Parallel Software, Albuquerque, NM), and an E450 microcomputer (Sun Microsystems, Mountain View, CA). Sequence and physical gaps were determined by aligning contiguous sequence fragments to the GAS strain SF370 genome sequence (http://www.genome.ou.edu) with cross_match (P. Green, unpublished data) and SEQUENCHER4.1 (Gene Codes, Ann Arbor, MI). Primers (Sigma–Genosys, The Woodlands, TX) for gap closure were designed with PRIMER3 (http://www-genome.wi.mit.edu). Sequence and physical gaps located in unique regions of the MGAS8232 genome were closed with a combination of directed PCR, multiplex PCR, and sequencing suggested by the “autofinish” module of consed (9). The genome sequence was completed to a minimum Q40 consensus-base quality. Manual verification of the final assembly after initial subassembly of problematic regions (e.g., insertion elements and rrn operons) was performed with PCR amplification of overlapping 5–15-kb fragments and comparison of predicted to observed SmaI restriction fragments.
ORFs ≥99 bp were identified with GLIMMER2 (10) (www.tigr.org). The model training set consisted of ≥500-bp ORFs from GAS strain SF370 obtained from the WIT2 analysis (www.genome.ou.edu/strep.html). ORFs were identified with blastp [BLAST2.0.11 (11); maximum E = 10−6]. Secretion signals were identified with signalp (www.cbs.dtu.dk/services/ SignalP-2.0), cell wall-attachment motifs (12) were identified with WORD98 (13), and transmembrane domains were identified with dnastar (Lasergene, Madison, WI) and assigned to functional categories based on cluster of orthologous gene analysis (14). Alignments between M18 and M1 ORF sequences (blastn; maximum E = 10−6; ≥75% ORF coverage) were done to identify ORFs shared between the M18 and M1 genomes.
Serotype M18 GAS Strains and DNA Microarray Analysis.
Serotype M18 GAS strains associated with several ARF outbreaks were used for DNA microarray analysis (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). Strains identified as “ARF-associated” or “RF” were collected from patients with ARF, and strains from Texas and Utah identified as pharyngeal were collected from patients with pharyngitis during ARF outbreaks (Table 2). Additional serotype M18 strains from invasive infections and asymptomatic carriers were also analyzed. Growth of isolates and purification of chromosomal DNA were done as described elsewhere (15). Contaminating RNA was removed by RNase digestion, mild alkaline hydrolysis, and filtration with Multiscreen-PCR plates (Millipore).
The DNA microarray consisted of PCR products representing 1,705 ORFs in GAS strain SF370 (7, 16) and was supplemented with PCR products representing M18-specific ORFs and small ORFs shared by SF370 and MGAS8232 ORFs identified in this study. PCR products ranged in size from 100 to 500 bp. Polyamine-coated slides (Corning) were spotted in quadruplicate with a Chipwriter Robotic Arrayer (Virtek, Waterloo, ON, Canada). DNA was crosslinked with UV light and blocked (17). DNA microarray hybridization of each test strain vs. control strain MGAS8232 was done in triplicate. Chromosomal DNA from each test strain and strain MGAS8232 was digested with RsaI for 3 h. Postdigest purification, direct incorporation of Cy3- and Cy5-labeled dCTP (NEN), and hybridization were performed as described (18) except that HybSolution3 (Ambion, Austin, TX) was used. Hybridization was detected with a ScanArray 5000 instrument (Packard). During scanning, fluorescence intensity between channels was normalized to a serial dilution of MGAS8232 chromosomal DNA by adjusting laser power and/or photomultiplier gain. Images were analyzed with quantarray (Packard). For each slide, spot intensities were adjusted for background and normalized to the median intensity of the slide. The median ratio of normalized test strain fluorescence to normalized control strain (MGAS8232) fluorescence (n = 12 spots) was used to identify gene differences. Genes were considered lacking or highly variable in the test vs. control strain below a 0.7 test/control ratio and considered present in greater copy number in the test vs. control strain above a 1.5 test/control ratio. Genes were considered to be different when 10 of 12 spots exceeded the threshold ratio. These threshold values are conservative metrics based on cross hybridization of related genes. In the aggregate, ORFs exceeding these threshold ratios were considered “gene differences.” Gene differences for each ORF in each strain were plotted in a three-color scheme with excel98 (Microsoft). In a hybridization of MGAS8232–MGAS8232 DNA, these analysis parameters produced a type-1 error rate for the microarray that was less than 0.0005.
Phage Nomenclature.
Two nomenclatures have been used in reference to the 4 phages and phage-like elements present in the genome of strain SF370. The strain SF370 genome sequence deposited in GenBank (accession no. AE004092) refers to these elements as 370.1, 370.3, 370.2, and 370.4 in clockwise order around the chromosome. This nomenclature is at variance with information presented in refs. 7 and 19, which refer to these 4 elements as 370.1, 370.2, 370.3, and 370.4. We will use the nomenclature used in refs. 7 and 19.
Results
Overview of the Genome of Strain MGAS8232 and Comparison with the Genome of GAS Strain SF370 (Serotype M1).
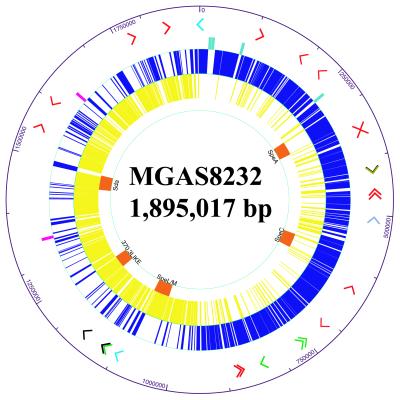
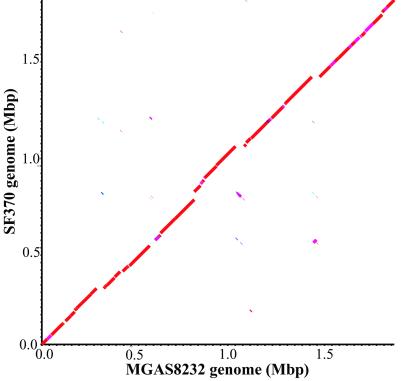
The genome of strain MGAS8232 is a single, circular chromosome of 1,895,017 bp (Fig. 1) containing 1,889 ORFs. The G + C content is 38.6%, a value virtually identical to the genome of GAS strain SF370 (38.5%). The genomes of strains MGAS8232 and SF370 each have six ribosomal RNA operons (rrn) and share 1,532 of 1,696 ORFs originally identified in strain SF370 and 179 small ORFs identified in this study (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). The nucleotide sequence identity between the 1,532 shared genes ranged from 83 to 100%. Each strain has many unique genes. Strain MGAS8232 has 178 unique ORFs compared with strain SF370 (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org), and strain SF370 has 112 unique ORFs compared with strain MGAS8232 (Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). blastp analysis of the unique ORFs of each strain with the current protein database from the human genome project matched regions of many GAS proteins with human proteins (Tables 3 and 4). Comparison of the genomes of MGAS8232 and SF370 with cross_match and MUMMER (20) found that 1.7 Mb of chromosome is shared between these organisms (Fig. 2). There are 24 regions of difference between the genomes that are greater than 2,000 bp in size. Five of these areas represent insertion of lysogenic prophage or phage-related elements, and three large regions of variation represent sequence divergence of a phage region that is shared by the two strains. The additional 16 regions of difference are 2–15-kb islands of DNA that are unique to the individual genomes, are in different locations in the genomes, or are highly variable regions of DNA sequence. In addition, the two genomes differ by 7,161 single nucleotide polymorphisms, 2,650 small regions of variation (<2,000 bp), and 271 small sequence insertion/deletions (<2,000 bp) as identified with MUMmer (Table 1).
Figure 1.
Atlas of the chromosome of serotype M18 strain MGAS8232. Arrowheads in the outermost ring depict the position and orientation of all transposase genes or gene fragments identified in the genome. Transposase genes are color coded to represent the transposase family with the closest BLASTP match (red, IS1239; black, IS904A; olive, IS1562; gray blue, tnpA; green, IS 861; light blue, Streptococcus salvarius transposase). The middle three rings show the position and orientation of the six RNA operons (light blue, clockwise; pink, counterclockwise) and ORFs (blue, clockwise; yellow, counterclockwise) identified in the genome. The innermost ring shows the location and name of the phage sequences (orange) identified in the genome.
Figure 2.
Dot plot of a cross_match comparison of strains MGAS8232 and SF370 genome sequences. cross_match was run with default parameters except the minimum match was set to 100. Deflections of segments along either axis indicate insertions of DNA sequence. Segments not aligning on the diagonal line represent sequences that are similar but located in different portions of the genomes. The color of the segments shows the similarity between the nucleotide sequences (green, 72–76%; forest green, 76–80%; light blue, 80–84%; dark blue, 84–88%; maroon, 88–92%; pink, 92–96%; red, 96–100%).
Table 1.
Summary of MGAS8232 and SF370 genome comparison
| Types of genome variation* | Strain
|
|
|---|---|---|
| MGAS8232 | SF370 | |
| Genome size, bp | 1,895,017 | 1,852,442 |
| No. of ORFs† | 1,889 | 1,696 |
| Large insertion | 8 | 10 |
| Small insertion | 64 | 56 |
| Single nucleotide insertion | 71 | 80 |
| Large region of variation | 6 | |
| Small region of variation | ||
| 2–50 bp | 2,591 | |
| 51–500 bp | 48 | |
| 501–1,999 bp | 11 | |
| Single nucleotide polymorphism | 7,161 | |
Large insertions and regions of variation are greater than 2 kb, and small insertions and regions of variation are less than 2 kb. Single nucleotide polymorphisms were identified on the basis of a minimum reported MUM of 10 with MUMmer analysis.
The number of ORFs deposited in GenBank is reported. The minimum, median, and maximum ORF lengths were 99, 741, and 4,941 bp in strain MGAS8232 and 81, 789, and 6,135 bp in strain SF370.
Mobile Genetic Elements in the Genome of Strain MGAS8232.
There are many apparently mobile genetic elements (e.g., phages and insertion elements) located in the genome of strain MGAS8232 (Fig. 1). Strains MGAS8232 and SF370 share homologues of TnpA, IS861, and ORFs with similarity to transposases from other Gram-positive cocci (e.g., SpyM18_0534, SpyM18_1318, and SpyM18_1393). Variants of insertion sequence IS1239 (21) are present in the genome of strain MGAS8232 but not the genome of strain SF370. Strain MGAS8232 has 11 copies of an intact IS1239 transposase gene and 4 copies of IS1239 transposase variants (Fig. 1). These transposase variants had either a 72-bp in-frame deletion at position 762 of the transposase gene (three copies) or a deletion of 600 nucleotides at the 3′ terminus (one copy). IS1239 and the IS1239-like elements do not disrupt ORFs but they are frequently located upstream of ORFs in putative regulatory sequences (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Among the ORFs that are in the same orientation as their upstream IS1239 elements, two have putative functions [thiophene degradation protein (thdF) and dipeptidase (pepQ)]. It is unknown whether expression of these genes or the other ORFs encoding hypothetical proteins is influenced by the IS1239 elements.
The genome of strain MGAS8232 has a 13.8-kb island of DNA that is absent from the strain SF370 genome. Insertion sequence IS1562, located in the Mga regulon in the genome of strain SF370 and other serotype M1 GAS strains (22), is located between spyM18_0535 and spyM18_0537 in the genome of strain MGAS8232. The IS1562 transposase gene in MGAS8232 seems to have inserted in a transposase of another insertion sequence and forms the center of this 13.8-kb island. The remainder of the island is composed of ORFs that are genes and pseudogenes homologous to ABC transporter and two-component regulatory system genes (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Proteins encoded by spyM18_543, spyM18_542, and spyM18_541 have homology to ABC transporters of Streptococcus pneumoniae. SpyM18_542 and SpyM18_541 have homology to the N-terminal 162-aa residues and C-terminal 248-aa residues of the S. pneumoniae bacteriocin-like protein transporter, BlpB (23). An ORF downstream of spyM18_541 is predicted to encode a 41-aa residue peptide that has a GlyGly cleavage site common to peptides processed and transported by ABC transporter systems (24). The predicted mature 17-aa peptide has an acidic residue at position +1, and the C-terminal residues RKK are similar to streptococcal competence pheromones, including CSP2 in S. pneumoniae and ComC in Streptococcus oralis (25, 26). In addition, SpyM18_0538 and SpyM18_0539, a two-component regulatory system, are encoded by the reverse strand, downstream from this ABC transporter system (Fig. 6). SpyM18_0538 and SpyM18_0539 have homology to a peptide-inducible response regulator and histidine kinase in S. pneumoniae [51%/72% and 34%/57% (identity/similarity), respectively] and seem to be a classical two-component regulatory system.
Five regions of the strain MGAS8232 genome are composed of phages or phage-like elements, each approximately 34–47 kb in length (Fig. 1). Each element has a copy of a described hyaluronidase gene, hylP (27, 28). The sequences of two phage regions (ΦspeC and Φ370.3-like) are similar to 370.1 and 370.3, respectively (phage regions in the strain SF370 genome), and they are inserted in the same location in each genome. The three phage regions unique to the genome of strain MGAS8232 are inserted between the murC and snf genes (ΦspeA); in the tmRNA coding region between spy1289 and spy1290, hypothetical genes (ΦspeL/M); and between spy1736 and spy1737, encoding xanthine/uracil permease and conserved hypothetical protein homologues, respectively (Φsda). The genome of strain MGAS8232 lacks phage regions present in the genome of strain SF370 areas designated 370.2 and 370.4, although some sequence of 370.2 is similar to sequence in ΦSpeL/M (Fig. 2). Four phage regions present in strain MGAS8232 (ΦspeA, ΦspeC, Φsda, and Φ370.3-like) have genes encoding characterized phage-associated proven and putative virulence factors, including SpeA, SpeC, MF2, Sda, and MF3 (7, 29). In addition, the T12-like phage region (ΦspeL/M) is inserted at the predicted T12att site (30), lacks a speA gene, and has two contiguous ORFs (designated speL and speM) encoding inferred newly identified superantigens (Fig. 4).
Genes Encoding Proven and Putative Virulence Factors Present in the Genome of Strain MGAS8232.
Many proven and putative virulence factors are encoded by the genome of strain MGAS8232, including a potent cysteine protease (SpeB), streptolysin O (SLO), streptokinase (Ska), and two streptococcal collagen-like proteins (Scl1 and Scl2). Genes encoding streptodornase (Sda), streptococcal protective antigen (Spa), and streptococcal pyrogenic exotoxin A (SpeA1 variant) are present in strain MGAS8232 but not strain SF370 (Table 3). Genes encoding collagen-binding protein (Cpa), LepA, and protein F2 (PrtF2) homologues are located near the negative regulator of virulence factors locus (nra) in strain MGAS8232. Several ORFs in this region (e.g., cpa and lepA) are highly variable between strains MGAS8232 and SF370 (Tables 3 and 4). The streptolysin S (sag) and hyaluronic acid capsule synthase (has) operons are also present in the genome of strain MGAS8232. The two strains share 21 genes predicted to encode extracellular proteins that are anchored to the cell surface by LPXTG and related motifs (12). In addition, secretion signal and cell wall-binding motif (LPXTG and related motifs) analyses predict that a cell surface-anchored amidase (SpyM18_2055) and a conserved hypothetic protein (SpyM18_0130) are encoded by MGAS8232 but not SF370. signalp analysis and Kyte–Doolittle hydrophilicity plot analysis (DNAstar) predict that the genome of strain MGAS8232 encodes eight freely diffusible extracellular proteins and one membrane-anchored extracellular protein not encoded by SF370 (Table 3). Given the importance of many characterized extracellular products in GAS pathogenesis (5), these proteins may contribute to host–pathogen interactions as well.
Expression of many GAS virulence factors is controlled by two-component regulatory systems and transcription factors (e.g., Mga, CovR-CovS, Rgg, Nra, and SagA). The genomes of strains MGAS8232 and SF370 share 11 two-component regulatory systems and more than 36 transcription factors. ORFs encoding two additional putative histidine kinases (SpyM18_0538 and SpyM18_1569) are present in the genome of strain MGAS8232. SpyM18_0538 has homology to a peptide-inducible histidine kinase in S. pneumoniae, and SpyM18_1569 is encoded by an ORF that has 91% sequence identity with a pseudogene in strain SF370. The pseudogene contains a frame-shift mutation and is located downstream of spy1556. SpyM18_1569 completes an uncharacterized two-component regulatory system with SpyM18_1570 in strain MGAS8232 (Spy1556 in SF370). The transcription regulators RofA and Mga are present in regions of chromosomal plasticity among GAS strains of diverse M protein serotype (31, 32). The gene encoding Nra (originally described in serotype M49 GAS) is present in the genome of strain MGAS8232 at the location where RofA is located in the genome of strain SF370, in agreement with Podbielski et al. (31). However, the nra gene in strain MGAS8232 (and 12 of 12 additional M18 strains studied) has a single nucleotide change that creates a stop codon and results in truncation of Nra. The molecular architecture of the mga regulon in strain MGAS8232 is similar to the gene arrangement present in several serum opacity factor-negative strains (32).
Microarray Analysis of Serotype M18 Strains Associated with ARF.
The availability of a chromosome sequence for a serotype M18 strain permits genome-scale analysis of the molecular population genetics of strains expressing this M protein type. To assess the nature and extent of genome variation among serotype M18 strains and gain additional insight into the molecular population genetics of ARF outbreaks, we conducted DNA microarray analysis of 36 M18 strains cultured from diverse localities. Few gene differences were detected among strains, and the overall greatest difference between a test strain (62654-134) and MGAS8232 was 3.0% of genes. ORFs with a difference between at least one test strain and MGAS8232 were plotted in MGAS8232 genomic order (Fig. 3). Phages and phage-like elements were the primary source of variation in gene content among strains. Indeed, of the regions of difference detected in the comparison between strains MGAS8232 and SF370, only phage regions varied among the M18 strains. Variation in phage regions among the M18 strains ranged from the absence of an entire phage region (MGAS1585) to minor differences in the gene composition of the regions.
Figure 3.
Subset of 223 strain-specific ORFs identified by DNA microarray analysis of 36 serotype M18 strains of GAS. All ORFs had a test/control ratio that either was below the threshold value of 0.7 (yellow) or above the threshold of 1.5 (blue) in at least 1 of 36 strains (black, ORFs with test/control values between the threshold ratios). ORFs are sorted in MGAS8232 genomic order (clockwise from oriC). Strains are numbered 1–36 as they are listed in Table 2. Phage-associated ORFs are identified with a black vertical bar at the phage region. Other ORFs that showed a “gene difference” represent potential transposase genes, transcription regulators, carbohydrate and metal transport genes, and hypothetical genes.
Both temporal and geographic patterns in phage-related genes emerged from the microarray analysis. Twenty-four of the 36 strains had the same or similar (<10 gene differences in the genome) phage gene content as strain MGAS8232. All of these strains were collected since 1963 and include an ARF strain and all analyzed pharyngeal isolates collected from Salt Lake City, UT, during both recent ARF outbreaks (Fig. 3). In addition, ARF strains collected from Ohio and Colorado, invasive isolates from Nebraska and Washington, and pharyngeal isolates collected during an ARF event at Lackland Air Force Base, San Antonio, TX (Fig. 3), had virtually identical phage gene patterns observed in the Utah strains. Strains with substantially different phage-gene patterns were collected before 1963, during outbreaks of invasive disease in Texas and Illinois, or from an asymptomatic Air Force recruit. Thus, it is clear from these microarray data that horizontal gene transfer events involving phage-related genes are important sources of genomic diversity among serotype M18 strains.
Discussion
Comparison of the Genomes of Strains MGAS8232 and SF370.
The genomes of strains MGAS8232 and SF370 have a colinear 1.7-Mb backbone of DNA interspersed with large insertions and deletions but no major rearrangements. Both genomes contain genes necessary for vegetative growth, including genes required for replication, protein synthesis, metabolic processes, and nutrient uptake. The genome of MGAS8232, like SF370, lacks tricarboxylic acid cycle and electron transport genes, and both organisms have few amino acid biosynthetic genes. As with the specialized genomes of obligate parasites and symbionts (33–35), the loss of biosynthetic genes and accumulation of scavenging systems may also reflect the human specialization of GAS.
Sources of MGAS8232 Genomic Diversity.
Phages and phage-related sequences are the primary sources of diversity in gene content between strains SF370 and MGAS8232 and among the 36 serotype M18 strains analyzed. Seven distinct phage regions and phage-like elements present in strains SF370 and MGAS8232 encode 10 proven or putative GAS toxins and virulence factors (SpeA, SpeC, SpeH, SpeI, SpeL, SpeM, MF2, MF3, Sda, and HylP). Variation in the phage gene content among GAS genomes and allelic variation in phage-encoded genes (e.g., speA and hylP) suggest that phages are important contributors to differences in GAS virulence (2). The variation in content of phage genes among serotype M18 strains collected at different times and in distinct localities throughout the U.S. demonstrates the complexity and plasticity of lysogenic phages in GAS and indicates that horizontal gene transfer events have contributed extensively to shaping genetic variation among strains of this serotype.
Insertion sequences are common features of bacterial genomes that can alter genotype and phenotype by mediating recombination events such as insertions and deletions and influencing transcription of linked genes. IS1239 is in multiple copies in the genome of strain MGAS8232 and all serotype M18 strains studied by DNA microarray. Kapur et al. (21) reported that IS1239 is present in 26 of 78 strains representing a variety of M serotypes. IS1239 is not present in the genome of the sequenced M1 strain but is present upstream of the gene (ssa) encoding streptococcal superantigen in an M15 strain (36), and the has operon in strain Vaughn (37), but the element is not present at these sites in strain MGAS8232. It is not known whether IS1239 influences gene expression or gene mobility in strain MGAS8232 or other GAS. However, we note that insertion elements are known to alter expression of genes in many bacteria (38) and may form composite transposons in β-hemolytic Gram-positive bacteria as suggested by Franken et al. (39).
Microarray Analysis of Serotype M18 Strains Associated with Acute Rheumatic Fever.
The focus of our DNA microarray study was the genetic diversity within a rheumatogenic serotype of GAS. Overall, the 36 serotype M18 strains analyzed had little or no variation in gene content. These results are in contrast to data reported for microarray analysis of Staphylococcus aureus (18) and Helicobacter pylori (40) in studies that analyzed random strains. The microarray data clearly indicate that M18 strains recovered during two ARF outbreaks in Salt Lake City, occurring 12 years apart, were genetically identical or nearly so. These results suggest that the increase of ARF cases in 1998–99 was associated with a resurgence of an M18 clone common in 1986–87 in the same area. This hypothesis is supported by comparative sequencing of genes in 500 serotype M18 pharyngeal isolates collected in Salt Lake City during the two ARF outbreaks (41). Indeed, the lack of variation in gene content among the M18 strains collected in the west-central U.S. between 1985 and 1998 suggests that these closely related organisms were the dominant serotype M18 strains in this geographic area. We cannot exclude the possibility that variation exists in genes not present in the MGAS8232 and SF370 genomes, and therefore not represented on the microarray. However, the consistent presence of phage-related genes among these strains suggests that undetected variation is unlikely to be mediated by phages.
Concluding Comment
This study provides an expanded understanding of GAS genetics and biology, and the results have relevance to studies of host–pathogen interactions. Coupled with classic genetic methods and newly formulated experimental approaches, the availability of genomic data creates extensive opportunities for research designed to gain new insight into the molecular pathogenesis of GAS infections, including ARF and rheumatic heart disease. Discovery of new putative virulence determinants and analysis of their distribution among M18 GAS strains provide a more complete view of the molecular armament present in members of this species. Importantly, our research shows that two distinct epidemics of ARF in Salt Lake City were caused by the same clone of GAS as assessed by the apparent identity in gene content. The sequenced M1 and M18 GAS genomes provide insight into the genetic elements responsible for diversity in the species, and thus assist development of new therapeutic and surveillance methods. Completion of genome sequences for additional GAS strains that have biomedically relevant correlates will permit identification of the metagenome and provide an expanded framework for GAS pathogenesis research.
Supplementary Material
Acknowledgments
We are indebted to L. Actis, Y. Liu, J. Slagel, and R. Cole for assistance and helpful discussions. This article is dedicated to Dr. Richard M. Krause for 50 years of contributions to GAS research.
Abbreviations
- GAS
Group A Streptococcus
- ARF
acute rheumatic fever
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE009949).
References
- 1.Cunningham M W. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaussee M S, Liu J, Stevens D L, Ferretti J J. J Infect Dis. 1996;173:901–908. doi: 10.1093/infdis/173.4.901. [DOI] [PubMed] [Google Scholar]
- 4.Facklam R, Beall B, Efstratiou A, Fischetti V, Johnson D, Kaplan E, Kriz P, Lovgren M, Martin D, Schwartz B, et al. Emerg Infect Dis. 1999;5:247–253. doi: 10.3201/eid0502.990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoe N P, Nakashima K, Lukomski S, Grigby D, Liu M, Kordari P, Dou S, Pan X, Vuopio-Varkila J, Salmelinna S, et al. Nat Med. 1999;5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- 6.Musser J M, Krause R M. In: Emerging Infections. Krause R M, editor. New York: Academic; 1998. pp. 185–218. [Google Scholar]
- 7.Ferretti J J, McShan W M, Ajdic D, Savic D J, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov A N, Kenton S, et al. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoe N, Nakashima K, Grigsby D, Pan X, Dou S J, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, Musser J M. Emerg Infect Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon D, Desmarais C, Green P. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delcher A L, Harmon D, Kasif S, White O, Salzberg S L. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janulczyk R, Rasmussen M. Infect Immun. 2001;69:4019–4026. doi: 10.1128/IAI.69.6.4019-4026.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horsburgh M J, Ingham E, Foster S J. J Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov R L, Natale D A, Garkavtsev I V, Tarusova T A, Shankavaram U T, Rao B S, Kiryutin B, Galperin M Y, Fedorova N D, Koonin E V. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smoot L M, Smoot J C, Graham M R, Somerville G A, Sturdevant D E, Migliaccio C A, Sylva G L, Musser J M. Proc Natl Acad Sci USA. 2001;98:10416–10421. doi: 10.1073/pnas.191267598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald J R, Sturdevant D E, Mackie S M, Gill S R, Musser J M. Proc Natl Acad Sci USA. 2001;98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desiere F, McShan W M, van Sinderen D, Ferreti J J, Brussow H. Virology. 2001;288:325–341. doi: 10.1006/viro.2001.1085. [DOI] [PubMed] [Google Scholar]
- 20.Delcher A L, Kasif S, Fleischmann R D, Peterson J, White O, Salzberg S L. Nucleic Acids Res. 1999;27:2369–2376. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapur V, Reda K B, Li L L, Ho L J, Rich R R, Musser J M. Gene. 1994;150:135–140. doi: 10.1016/0378-1119(94)90872-9. [DOI] [PubMed] [Google Scholar]
- 22.Berge A, Rasmussen M, Björck L. Infect Immun. 1998;66:3449–3453. doi: 10.1128/iai.66.7.3449-3453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins J, Alborn W E, Arnold J, Blaszczak L C, Burgett S, DeHoff B S, Estrem S T, Fritz L, Fu D J, Fuller W, et al. J Bacteriol. 2001;183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Havarstein L S, Diep D B, Nes I F. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 25.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein L S, Piccoli L, Simon D, Morrison D A. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Havarstein L S, Hakenbeck R, Gaustad P. J Bacteriol. 1997;179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hynes W L, Ferretti J J. Infect Immun. 1989;57:533–539. doi: 10.1128/iai.57.2.533-539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hynes W L, Hancock L, Ferretti J J. Infect Immun. 1995;63:3015–3020. doi: 10.1128/iai.63.8.3015-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Podbielski A, Zarges I, Flosdorff A, Weber-Heynemann J. Infect Immun. 1996;64:5349–5356. doi: 10.1128/iai.64.12.5349-5356.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu C, Ferretti J J. Mol Gen Genet. 1991;231:161–168. doi: 10.1007/BF00293833. [DOI] [PubMed] [Google Scholar]
- 31.Podbielski A, Woischnik M, Leonard B A, Schmidt K H. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 32.Whatmore A M, Kehoe M A. Mol Microbiol. 1994;11:363–374. doi: 10.1111/j.1365-2958.1994.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 33.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 34.Andersson S G E, Zomorodipour A, Andersson J O, Sichertitz-Ponten T, Alsmark U C M, Podowski R M, Naslund A K, Eriksson A, Winkler H H. Nature (London) 1998;396:133–143. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 35.Read T D, Brunham R C, Shen C, Gill S R, Heidelberg J F, White O, Hickey E K, Peterson J, Utterback T, Berry K, et al. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reda K B, Kapur V, Mollick J A, Lamphear J G, Musser J M, Rich R R. Infect Immun. 1994;62:1867–1874. doi: 10.1128/iai.62.5.1867-1874.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashbaugh C D, Alberti S, Wessels M R. J Bacteriol. 1998;180:4955–4959. doi: 10.1128/jb.180.18.4955-4959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galas D J, Chandler M. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 109–162. [Google Scholar]
- 39.Franken C, Haase G, Brandt C, Weber-Heynemann J, Martin S, Lammler C, Podbielski A, Lutticken R, Spellerberg B. Mol Microbiol. 2001;41:925–935. doi: 10.1046/j.1365-2958.2001.02563.x. [DOI] [PubMed] [Google Scholar]
- 40.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smoot, J. C., Korgenski, E. K., Daly, J. A., Veasy, L. G. & Musser, J. M. (2002) J. Clin. Microbiol., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.