Abstract
Although previous studies are beginning to point to the specific types of helix–helix interactions that stabilize the folds of membrane-bound helical proteins, quantitative thermodynamic data on natural membrane proteins has been very limited. Here the database is expanded substantially by adding thermodynamic data for a series of sequence variants of M2 protein from influenza A virus. The M2 protein has a single transmembrane helix that homotetramerizes to form proton-selective channels that are essential to virus function. To determine the contributions of specific residues to the folding of this protein, a series of transmembrane peptides with single-site changes near the core of the protein were studied by using sedimentation equilibrium analytical ultracentrifugation. Remarkably, a large number of the mutations increased the stability of the protein. The free energies of tetramerization of the variants can be understood in terms of current models for the structure of the protein. In general, the energetic consequences of the mutations are smaller than those observed for similar mutations in water-soluble proteins. This observation is consistent with previous studies and hence may represent a general phenomenon.
The increasing number of experimentally determined membrane protein structures and genome-wide sequence analyses are beginning to hint at emerging themes regarding the features that stabilize the structures of α-helical membrane proteins (1, 2). For example, it has become possible to identify sequence motifs and patterns of interactions that seem important for the folding of membrane proteins (3, 4). Furthermore, systematic mutagenesis experiments of a few natural helical membrane proteins (5, 6) as well as model transmembrane helices (7, 8) have contributed insight into interactions important for the association of transmembrane helices. Thus, van der Waals interactions, hydrogen bonding, and electrostatics all seem to contribute to the thermodynamics of folding of membrane proteins. However, because of the lack of thermodynamic data for mutants of membrane proteins, our understanding of the relative importance of these various interactions remains at a primitive level.
The collection of quantitative energetic information on membrane protein folding has been complicated by difficulties associated with establishing conditions for reversible folding, and only a handful of membrane proteins have been demonstrated to fold in a thermodynamically reversible manner (9, 10). Furthermore, membrane proteins fold in a number of distinct steps that involve association of the protein with the surface of the membrane, vertical insertion of helices into the bilayer, and the lateral association of inserted helices to give the fully folded protein (11, 12). Although the processes of surface binding and insertion have been studied extensively, the subsequent association into a native folded protein remains poorly understood.
The self-association of homooligomerizing transmembrane helices is a particularly convenient way to study this latter folding process. For a simple two-state monomer–oligomer system, the “unfolded” state is the isolated, noninteracting monomer, whereas the oligomeric helical bundle represents the native folded form. Analytical ultracentrifugation has been used to probe the energetics of folding of the dimeric, transmembrane two-helix bundle found in glycophorin (13, 14). However, until now data have been entirely lacking for bundles with larger numbers of helices, which might be more representative of the overall structures of more complex membrane proteins. Here we examine the energetics of tetramerization of a series of variants of a tetrameric proton channel, M2 protein from influenza A virus.
The M2 protein from influenza A virus is a 97-aa protein with a single transmembrane helix that forms proton-selective channels that are essential to virus function (15). The hydrophobic transmembrane domain of the M2 protein contains a sequence motif that mediates the formation of functional tetramers in membrane environments (16, 17). The structure of this transmembrane tetramer has been modeled extensively by using unconstrained and experimentally constrained molecular dynamics and energy-minimization techniques (18–29). In particular, Cys-scanning mutagenesis has revealed that the side chains that are most sensitive to mutation show approximate seven-residue periodicity as observed in coiled coils or bundles of straight helices with a left-handed helical crossing angle (18). The most sensitive positions presumably define the central core of the structure and are labeled “a” and “d” in Fig. 1. To determine which residues contribute to the stability and specificity of the transmembrane helix interactions, a series of peptides with sequences that include the transmembrane portion of M2 were synthesized in which the central “a” or “d” residues were changed individually to either Ala or Phe (Fig. 1). The effects of these changes on the free energy of tetramerization were determined and provide data concerning how specific side chains stabilize the energetics of assembly of a functional transmembrane channel protein.
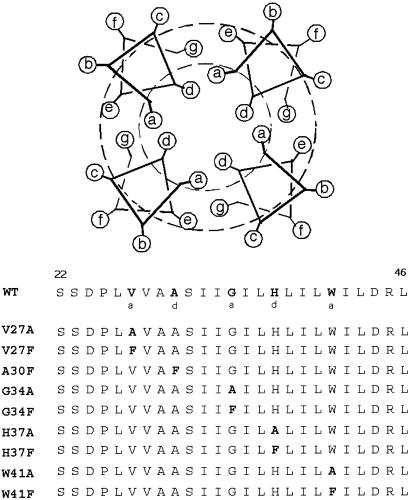
Figure 1.
Sequence of 25-residue synthetic peptide corresponding to the transmembrane segment of wild-type (WT) M2 from influenza A virus as well as all the sequences of the single-site variants studied. The five positions mutated in this study are indicated in bold in the WT sequence and labeled with their postulated corresponding positions in the heptad repeat. The helical wheel diagram shows one heptad of a four-helical bundle viewed down the bundle axis. The helix is drawn with seven-residue periodicity to simulate the left-handed helical crossing angle between the helices within the bundle.
Materials and Methods
Peptide Synthesis and Sample Preparation.
A series of single site variants of M2TM peptide (residues 22–46, C-terminally amidated) were synthesized on an Applied Biosystems 433A peptide synthesizer and purified by using reversed-phase HPLC as described (30). The identities of the purified peptides were confirmed with matrix-assisted laser desorption ionization mass spectrometry.
Samples for ultracentrifugation were prepared by dissolving the desired amount of each peptide in methanol in glass vials and removing organic solvent under a stream of nitrogen. The resulting peptide films were placed under high vacuum overnight. A solution of dodecylphosphocholine (DPC) in buffer was added to each vial, and the samples were vortexed until they became clear.
Analytical Ultracentrifugation.
Sedimentation equilibrium experiments were performed at 25°C on peptides solubilized in DPC micelles by using a Beckman XL-I analytical ultracentrifuge. To eliminate the contribution of DPC detergent micelles to the buoyant molecular weight of the peptide–DPC complex, experiments were carried out at a solvent density adjusted with 2H2O to equal that of the DPC. The density-matched buffer used was 50 mM Tris⋅HCl (pH 7.5)/0.1 M NaCl/51.5% 2H2O. A typical data set for each different peptide studied includes two different peptide/DPC ratios (1:150 and 1:250) with each sample being spun at three different speeds (40,000, 45,000, and 48,000 rpm). Data obtained from absorbance at 280 nm (or 230 nm for W41A and W41F peptides) were analyzed by nonlinear least-squares curve fitting of radial concentration profiles as described (30). The monomer molecular weights and partial specific volumes of each peptide were calculated by using the program SEDINTERP (31). The calculated values of these parameters were held constant in fitting the absorbance versus radius profiles.
Circular Dichroism (CD).
CD measurements were taken by using an Aviv 62A CD spectrometer at 25°C with 1-mm path-length cells. Samples were prepared with a peptide/DPC ratio of 1:150 by using the same buffer conditions as those used in the centrifugation experiments.
Results
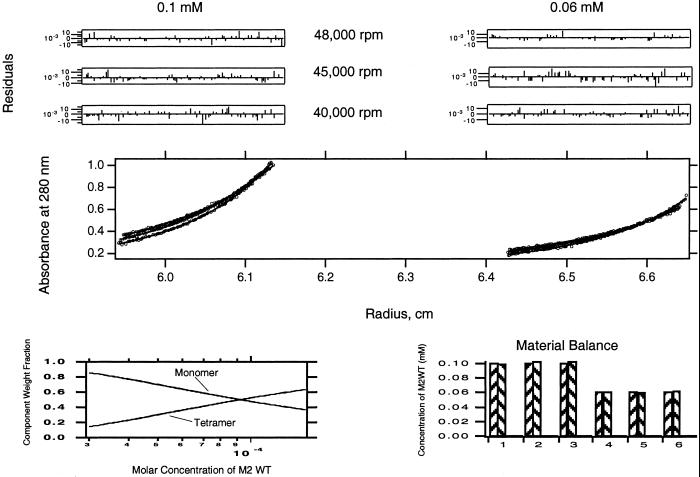
Previous analytical ultracentrifugation and CD studies have demonstrated that M2TM solubilized in DPC micelles exists in a reversible monomer-tetramer equilibrium sensitive to changes in pH and the peptide/detergent ratio (17, 30). Here sedimentation equilibrium analytical ultracentrifugation was conducted on variants of the M2TM peptide to determine the free-energy effect of changing residues that define the conducting core of the protein. Conditions for the centrifugation were chosen (pH 7.5 and peptide/detergent ratios ≈1:150) for which significant amounts of tetramers were present for the parent M2TM peptide. Sedimentation curves were analyzed in terms of the buoyant molecular weight of the peptide alone, because the data were collected under density-matched solvent conditions. Fig. 2 shows typical equilibrium ultracentrifugation data for an M2TM peptide analyzed by using a global fitting procedure and postfitting material balance (32).
Figure 2.
Sedimentation equilibrium of WT M2TM peptide in 15 mM DPC micelles. Six different A280-radius profiles are shown. (Left) The three curves correspond to a peptide/DPC ratio of 1:150 (0.1 mM peptide) collected at three different spinning speeds (40,000, 45,000, and 48,000 rpm). (Right) The set of curves corresponds to a peptide/DPC ratio of 1:250 (0.06 mM peptide) at the same three speeds. Data points were fit to a monomer/tetramer equilibrium, and residuals of the fits are shown in the six upper panels. (Lower Left) The relative contributions (y axis) of tetramers and monomers as a function of total peptide concentration (x axis). (Lower Right) The ideal (left column of each pair) versus calculated amount of total peptide (right column of each pair) in each of the six data sets.
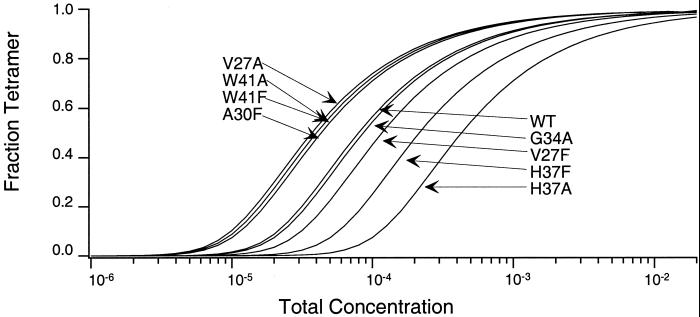
Fig. 3 shows the computed fraction tetramer as a function of total molar concentration for each of the peptides studied. The corresponding differences in the free energy of oligomerization relative to the WT sequence are shown in Table 1 and Fig. 4. Four of the variant peptides form tetramers that are weaker than WT (H37A, H37F, V27F, and G34A) with the remaining variants forming tetramers that associate more strongly (A30F, W41A, W41F, and V27A). The histidine at position 37 is the site at which changes are the most disruptive, with H37A being the peptide most impaired in its ability to form tetramers (ΔΔG = 3.1 kcal⋅mol−1). No data are shown for the G34F peptide, which apparently forms large aggregates that pellet out of solution during the centrifugation experiments. Although the CD spectra of the other M2TM peptides exhibit spectra characteristic of α-helical structure (data not shown), the CD spectrum of G34F (minimum at 209 nm, and a deeper broad minimum at 226 nm) is consistent with β structure and/or formation of higher order light-scattering aggregates.
Figure 3.
The variation in the fraction tetramer as a function of total concentration for each of the M2TM peptides studied. Curves were calculated by using the dissociation constants determined by global fitting of the sedimentation equilibrium data. A curve for the G34F peptide is missing because the sample irreversibly precipitated during the ultracentrifuge runs.
Table 1.
Differences in free energy of association for each of the mutants of M2TM relative to WT
| M2TM mutant | ΔΔG, kcal·mol−1 |
|---|---|
| H37A | 3.1 (± 0.1) |
| H37F | 1.8 (± 0.1) |
| V27F | 0.7 (± 0.1) |
| G34A | 0.2 (± 0.2) |
| A30F | −1.2 (± 0.7) |
| W41A | −1.3 (± 0.1) |
| W41F | −1.3 (± 0.1) |
| V27A | −1.4 (± 0.4) |
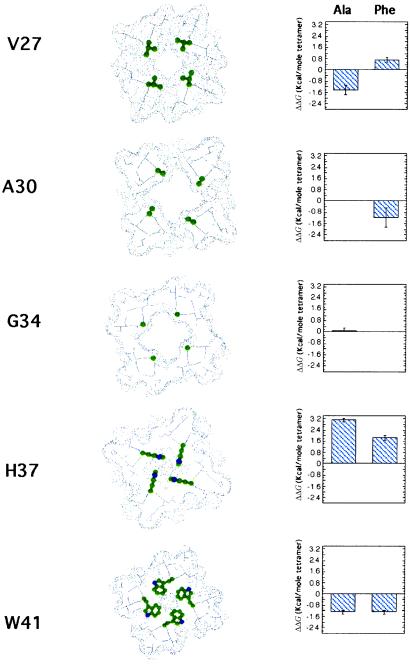
Figure 4.
The differences in the free energies of association (in kcal⋅mol−1) relative to the WT M2TM sequence for peptides, where “a” and “d” positions were changed to either an Ala or Phe. Error bars are calculated by the propagation of errors from the dissociation constants calculated from the centrifugation data for each peptide. Axial slices through the predicted structure of the M2 proton channel (18) are shown to help rationalize observed changes in stability. The sites mutated in this study are shown as ball-and-stick representations in color.
Discussion
Relation of Energetics to the Predicted Structure of M2TM.
It is interesting to consider the energetic consequences of these mutations in light of current models for the structure of M2TM at neutral pH. A variety of models has been proposed based on site-directed mutagenesis in conjunction with computer modeling (18), molecular dynamics calculations (19–23), IR spectroscopy (24), and solid-state NMR (25–29). They vary in the details of the models but are in good agreement with respect to (i) the packing of the helices with a left-handed tilt (ranging from ≈15 to 35°), (ii) the presence of a water-filled pore near the center of the channel, and most importantly, (iii) the identities of the side chains lining the pore (29, 33). Although the lack of a high-resolution structure precludes a detailed discussion of stabilizing interactions, the results of our studies are quite consistent with the more low-resolution features that are common to all the models.
Site-directed mutagenesis indicates that Val-27, Ala-30, Gly-34, His-37, and Trp-41 form a continuous proton-conducting pore. We will consider each of these positions in succession, beginning at the N terminus of the transmembrane helix (Fig. 4). Val-27 lies at the end of the transmembrane helix. Unlike the rest of the sites changed in this study, position 27 exhibits considerable variability in sequence across the range of M2 protein sequences from various natural strains of influenza A virus. A mutation of Val-27 to Ala occurs in naturally occurring variants of the virus and indeed is slightly stabilizing to the structure of the tetramer. By contrast, Phe has not been observed at this position in naturally occurring variants and is destabilizing.
The pore widens substantially at Ala-30 and Gly-34, in part because of the small size of these side chains. Modeling suggests that the pore can accommodate residues as large as a Phe at position 30, and this mutation indeed results in a modest increase in the stability of the tetramer (ΔΔG = −1.2 kcal⋅mol−1). The less drastic mutation of Gly-34 to Ala has no significant effect on stability. Thus, small-to-large mutations in this central cavity of the channel are tolerated, although the Ala-to-Phe mutation had a surprisingly small effect on the stability of the channel.
Progressing through the channel from the outside of the virus toward the interior, one next encounters His-37, which is essential for proton selectivity. In models of the neutral form of the channel, the His side chains are in van der Waals contact, thereby occluding movement of ions through the channel. In the current study, the largest energetic affect is observed after mutating this residue to Ala (ΔΔG = 3.1 kcal⋅mol−1). The energetic penalty associated with mutating this residue is slightly less severe when it is mutated instead to the nearly isosteric but apolar residue, Phe (ΔΔG = 1.8 kcal⋅mol−1). Because His-37 lies near the center of the transmembrane helix, it presumably transitions from being buried in the apolar region of the micelle as a monomer to the core of the tetramer. Presumably, His-37 stabilizes the tetramer because the polar δN and ɛN atoms of its side chain form more favorable interactions within the folded tetramer as compared with the apolar hydrocarbon chains of the detergent micelle. This finding also is consistent with previous studies showing that His is one of a handful of polar side chains that can mediate the association of model hydrophobic helices in micelles and biological membranes (34). Further, the magnitude of ΔΔG for a His-to-Ala mutation is similar to the value observed for mutation of other neutral polar side chains that mediate the association of designed transmembrane helices (8).
The last side chain mutated was Trp-41, which also seems to be essential for the gating and selectivity of the channel (18). It is intriguing that changing Trp-41 to either Ala or Phe is stabilizing. It is not uncommon for changes to functionally essential residues in soluble proteins to be stabilizing, because the structure in these regions is optimized for function rather than stability (35). Furthermore, stability-enhancing mutations in membrane proteins are most prevalent near the edge of the bilayer (36). Also, because this side chain is near the C terminus of the transmembrane helix, in the monomer it should locate to the head-group region of the micelle (or bilayer), which is the most favorable location for this aromatic residue (37). However, after tetramerization, all or some of these favorable atomic contacts are replaced by protein contacts. Thus, tetramerization probably requires the transfer of this side chain from a favorable location within the micelle to a protein interior.
Comparison to Other Transmembrane Proteins.
Data from two other transmembrane proteins in the literature show destabilization for most mutations examined at helical interfaces. Glycophorin A forms homodimers in membrane environments. In a recent study, Ala-scanning mutagenesis and sedimentation equilibrium showed that all mutations to residues at the helix–helix interface of this transmembrane dimer resulted in an unfavorable ΔΔG for dimerization (14). This finding contrasts with the results for M2TM, in which a remarkably large fraction of the mutations at the helix–helix interface actually are stabilizing with respect to WT. These energetic differences almost certainly arise from differences in the structure and the functional roles of the proteins. M2 forms proton channels and hence has a conducting core that must accommodate the demands for function as well as thermodynamic stability. Also, M2TM may have multiple low-energy conformational states that are functionally important for “gating” but also allow more facile rearrangements in response to mutations. By contrast, the transmembrane helices seem to serve a structural role in glycophorin and mediate the formation of very stable dimers.
The glycophorin dimer involves tight packing along a Gly-rich patch, which allows very close approach of the two transmembrane helices (38, 39). A set of reciprocal hydrogen bonds between the Gly Cα–H of one helix and a carbonyl oxygen of a neighboring helix (40) may contribute to a very tight interaction that clamps the dimer within a very steep energy well. Indeed, mutation of one of these Gly residues to Ala within the most tightly packed region of the dimer destabilizes the dimer by 3.2 kcal⋅mol−1 (14). With the exception of this mutant, the range of ΔΔG changes in glycophorin for single site mutations at the dimer interface (ref. 14; ≈0.2–0.8 kcal⋅mol−1 of helix) is approximately the same as the changes seen in M2, when expressed on a per-helix basis.
Phospholamban forms a transmembrane helical homopentamer that has been studied extensively by site-directed mutagenesis. Although the pentameric form of this protein has been reported to form Ca2+ channels in planar bilayers (41), it is believed now that the pentamer serves a more structural role in vivo; the pentamer appears to sequester the active, monomeric form of the protein from interactions with the Ca2+-dependent ATPase of the sarcoplasmic reticulum (42). Phospholamban has a low tolerance for mutations to each of its “a” and “d” positions as revealed by SDS-polyacrylamide gel electrophoresis (43, 44), a finding which again is consistent with this interface serving a structural role to stabilize pentamer formation.
Comparison to Energetics of Mutants of Soluble Proteins.
Mutations in the interior of soluble proteins generally cause larger changes in free energy than the modest effects seen for the M2 protein and glycophorin. For example, a mutation of a buried apolar side chain such as Leu, Val, or Ile to Ala generally destabilizes a protein by 3–5 kcal⋅mol−1 (45). By contrast, much smaller effects are observed in M2TM and glycophorin, in which similar mutations stabilize or destabilize folding by less than a single kcal⋅mol−1 helix. This discrepancy highlights a fundamental difference in the folding energetics of membrane-bound proteins as opposed to soluble proteins. Within the apolar region of a lipid bilayer or detergent micelle, the hydrophobic effect is no longer a major driving force for transmembrane helical association. Thus, the packing of apolar side chains makes a less favorable contribution to the overall free energy of stabilization of membrane proteins. By contrast, the energetic effects of mutating buried neutral polar side chains to alanine is roughly the same in both systems and generally is favorable by 1–2 kcal⋅mol−1 (8, 46). Contributions from buried charged side chains are expected to be even greater, but they occur infrequently in membrane proteins and not at all in some structures. Thus, in net, there appears to be fewer sources for stabilizing membrane proteins versus water-soluble proteins.
How then is it that helical membrane proteins are able to fold and function? As discussed previously (11), the cost in conformational entropy is substantially less in a membrane; the inserted unfolded state is helical already, and thus this penalty does not have to be paid to fold the proteins. A second feature is that membrane proteins might not actually be as stable as water-soluble proteins. In a recent review, Bowie (36) discusses mutagenesis studies on several different membrane proteins and makes the point that stability-enhancing mutations are far from rare, and clearly many stabilizing interactions have gone untapped in membrane proteins. He speculates that the lack of stability optimization in membrane proteins could be necessary for activity, for efficient protein turnover, or simply because the constraints placed on a protein by the surrounding lipid bilayer provide sufficient stability for cell viability. Clearly, more data are needed to test this assumption, but the results of this study, which provide an extensive set of thermodynamic data on a transmembrane helical protein with a well defined function beyond serving as an oligomerization domain, are consistent with this suggestion.
Acknowledgments
K.P.H. thanks David Salom and Holly Gratkowski for helpful discussions. K.P.H. acknowledges funding by the National Institutes of Health, National Science Foundation, and Swarthmore College.
Abbreviations
- WT
wild type
- DPC
dodecylphosphocholine
- CD
circular dichroism
References
- 1.Ubarretxena-Belandia I, Engelman D M. Curr Opin Struct Biol. 2001;11:370–376. doi: 10.1016/s0959-440x(00)00217-7. [DOI] [PubMed] [Google Scholar]
- 2.Adamian L, Liang J. J Mol Biol. 2001;311:891–907. doi: 10.1006/jmbi.2001.4908. [DOI] [PubMed] [Google Scholar]
- 3.Eilers M, Shekar S C, Shieh T, Smith S O, Fleming P J. Proc Natl Acad Sci USA. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senes A, Gerstein M, Engelman D M. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 5.Lemmon M A, Flanagan J M, Treutlein H R, Zhang H R, Engelman D M. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 6.Guan L, Weinglass A B, Kaback H R. J Mol Biol. 2001;312:69–77. doi: 10.1006/jmbi.2001.4933. [DOI] [PubMed] [Google Scholar]
- 7.Zhou F X, Merianos H J, Brunger A T, Engelman D M. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gratkowski H, Lear J D, DeGrado W F. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popot J L, Gerchman S E, Engelman D M. J Mol Biol. 1987;198:655–676. doi: 10.1016/0022-2836(87)90208-7. [DOI] [PubMed] [Google Scholar]
- 10.Lau F W, Bowie J U. Biochemistry. 1997;36:5884–5892. doi: 10.1021/bi963095j. [DOI] [PubMed] [Google Scholar]
- 11.Popot J L, Engelman D M. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 12.White S H, Wimley W C. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 13.Fleming K. Methods Enzymol. 2000;323:63–77. doi: 10.1016/s0076-6879(00)23361-2. [DOI] [PubMed] [Google Scholar]
- 14.Fleming K G, Engelman D M. Proc Natl Acad Sci USA. 2001;98:14340–14344. doi: 10.1073/pnas.251367498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb R A, Holsinger L J, Pinto L H. In: Cellular Receptors and Animal Viruses. Wimmer E, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 303–321. [Google Scholar]
- 16.Duff K C, Ashley R H. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 17.Salom D, Hill B R, Lear J D, DeGrado W F. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto L H, Dieckmann G R, Gandhi C S, Papworth C G, Braman J, Shaughnessy M A, Lear J D, Lamb R A, DeGrado W F. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong Q, Husslein T, Moore P B, Newns D M, Pattnaik P, Klein M L. FEBS Lett. 1998;434:265–271. doi: 10.1016/s0014-5793(98)00988-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhong Q, Newns D M, Pattnaik P, Lear J D, Klein M L. FEBS Lett. 2000;473:195–198. doi: 10.1016/s0014-5793(00)01522-2. [DOI] [PubMed] [Google Scholar]
- 21.Forrest L R, Kukol A, Arkin I T, Tieleman D P, Sansom M S. Biophys J. 2000;78:55–69. doi: 10.1016/s0006-3495(00)76572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerr I D, Sankararamakrishnan R, Smart O S, Sansom M S P. Biophys J. 1994;67:1501–1515. doi: 10.1016/S0006-3495(94)80624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sansom M S P, Kerr I D. Protein Eng. 1993;6:65–74. doi: 10.1093/protein/6.1.65. [DOI] [PubMed] [Google Scholar]
- 24.Torres J, Kukol A, Arkin I T. Biophys J. 2000;79:3139–3143. doi: 10.1016/S0006-3495(00)76547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacs F A, Cross T A. Biophys J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer C M, Pinto L H, Cross T A, Lamb R A. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 27.Song Z, Kovacs F A, Wang J, Denny J K, Shekar S C, Quine J R, Cross T A. Biophys J. 2000;79:767–775. doi: 10.1016/S0006-3495(00)76334-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovacs F A, Denny J K, Song Z, Quine J R, Cross T A. J Mol Biol. 2000;295:117–125. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Kim S, Kovacs F, Cross T A. Protein Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kochendoerfer G G, Salom D, Lear J D, Wilk-Orescan R, Kent S B H, DeGrado W F. Biochemistry. 1999;38:11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 31.Laue T, Shaw B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 32.Arkin M, Lear J D. Anal Biochem. 2001;299:98–107. doi: 10.1006/abio.2001.5396. [DOI] [PubMed] [Google Scholar]
- 33.Forrest L R, DeGrado W F, Dieckmann G R, Sansom M S. Folding Des. 1998;3:443–448. doi: 10.1016/S1359-0278(98)00061-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhou F X, Cocco M J, Russ W P, Brunger A T, Engelman D M. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 35.Soichet B K, Baase W A, Kuroki R, Matthews B W. Proc Natl Acad Sci USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowie J U. Curr Opin Struct Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 37.Yau W M, Wimley W C, Gawrisch K, White S H. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 38.MacKenzie K R, Prestegard J H, Engelman D M. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 39.Smith S O, Song D, Shekar S, Groesbeek M, Ziliox M, Aimoto S. Biochemistry. 2001;40:6553–6558. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- 40.Senes A, Ubarretxena-Belandia I, Engelman D M. Proc Natl Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovacs R J, Nelson M T, Simmerman H K B, Jones L R. J Biol Chem. 1988;263:18364–18368. [PubMed] [Google Scholar]
- 42.Simmerman H K, Jones L R. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 43.Simmerman H K, Kobayashi Y M, Autry J M, Jones L R. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 44.Arkin I T, Adams P D, MacKenzie K R, Lemmon M A, Brünger A T, Engelman D M. EMBO J. 1994;13:4757–4764. doi: 10.1002/j.1460-2075.1994.tb06801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Baase W A, Baldwin E, Matthews B W. Protein Sci. 1998;7:158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pace C N, Shirley B A, McNutt M, Gajiwala K. FASEB J. 1996;10:75–83. doi: 10.1096/fasebj.10.1.8566551. [DOI] [PubMed] [Google Scholar]