Abstract
Low temperature regulates gene expression in bacteria, yeast, and animals as well as in plants. However, the signal transduction cascades mediating the low temperature responses are not well understood in any organism. To identify components in low temperature signaling genetically, we isolated Arabidopsis thaliana mutants in which cold-responsive genes are no longer induced by low temperatures. One of these mutations, los1–1, specifically blocks low temperature-induced transcription of cold-responsive genes. Surprisingly, cold-induced expression of the early response transcriptional activators, C-repeat/dehydration responsive element binding factors (CBF/DREB1s), is enhanced by the los1–1 mutation. The los1–1 mutation also reduces the capacity of plants to develop freezing tolerance but does not impair the vernalization response. Genetic analysis indicated that los1–1 is a recessive mutation in a single nuclear gene. The LOS1 gene encodes a translation elongation factor 2-like protein. Protein labeling studies show that new protein synthesis is blocked in los1–1 mutant plants specifically in the cold. These results reveal a critical role of new protein synthesis in the proper transduction of low temperature signals. Our results also suggest that cold-induced transcription of CBF/DREB1s is feedback inhibited by their gene products or by products of their downstream target genes.
Low temperature is an important environmental factor influencing the growth and survival of all organisms. Exposure to low temperatures has been shown to induce the expression of specific sets of genes in both prokaryotes and eukaryotes. In higher plants, a number of low temperature-induced genes have been identified (1, 2). Many of these genes and gene products were termed CAPs for cold acclimation proteins (3), CORs for cold responsive (4), or LTIs for low temperature induced (5). Some of the cold-induced proteins in plants share sequence similarity with proteins of known function (1, 2). It remains a challenge to determine the functions of cold-induced proteins that do not share sequence homology with proteins of known function. It is encouraging to note that chloroplasts and protoplasts taken from plants overexpressing COR15A (a nuclear-encoded, chloroplast-located protein) show increased freezing tolerance compared with those from control plants (6). More recently, it was demonstrated that COR15A deters the formation of deleterious membrane structures during freezing (7).
A major gap in the understanding of cellular responses to cold is that of the signal transduction pathways that lead to the transcription of cold-induced genes. Significant progress has been made in the identification of the cis element and trans factors that control cold-induced gene transcription in plant cells. The CCGAC core sequence that has been termed dehydration responsive element (DRE), or C-repeat, is necessary and sufficient for gene transcription under cold stress (8, 9). The element also mediates osmotic stress regulation of gene transcription (8, 9). A small group of highly homologous transcription activators [C-repeat binding factor (CBF)/DRE binding factor (DREB1)] that bind the DRE/C-repeat sequence and induce cold-regulated gene expression were identified (10, 11). CBF/DREB1 genes are rapidly and transiently induced by cold treatments (11). The important role of CBF/DREB1s in plant cold acclimation was underlined by the observation that their ectopic expression activated COR genes and enhanced freezing as well as osmotic stress tolerance of nonacclimated Arabidopsis plants (10, 11).
To dissect low temperature signal transduction genetically, we developed a screen for mutations in Arabidopsis thaliana that affect cold-responsive gene expression (12). The screening procedure uses transgenic Arabidopsis plants that express the firefly luciferase coding sequence under control of the RD29A promoter. The RD29A promoter contains, among other things, the DRE/C-repeat sequence and thus confers cold-inducible bioluminescence to the transgenic plants. We isolated a group of mutants that appear to be impaired in cold sensing or signal transduction because the bioluminescence is no longer induced by cold stress. Characterization of one of these mutants identified a genetic locus, LOS1, as essential for cold induction of gene expression and critical for the development of freezing tolerance. The LOS1 gene was cloned based on its map location. It encodes a translation elongation factor 2-like protein. Protein synthesis is normal in los1–1 mutant plants at warm temperatures but is blocked in the cold. These results point to a vital role of new protein synthesis in low temperature signal transduction during cold acclimation.
Materials and Methods
Plant Materials.
Transgenic A. thaliana in the C24 background containing homozygous RD29A-LUC transgene (referred to as wild type) were mutagenized with ethyl methanesulfonate (12). M2 seedlings grown on 0.8% agar plates containing MS salts (Murashige and Skoog salt base, JRH Biosciences, Lenex, KS) were screened for altered RD29A-LUC expression in response to low temperature, exogenous abscisic acid (ABA), or osmotic stress by using a video-imaging system as described by Ishitani et al. (12). Plants used for luciferase imaging were 1-week-old seedlings grown on MS agar plates under constant white fluorescent light at room temperature (22 ± 2°C). Plants for genetic, vernalization, or freezing tolerance analysis were grown in pot soil in growth chambers with 16 h light at 22°C, 8 h dark at 18°C, and 70% relative humidity.
Genetic Analysis and Map-Based Cloning of LOS1.
Mutants were crossed with the wild type by rubbing stamens from the mutants onto the stigma of emasculated wild-type flowers. The cold response of F1 and F2 seedlings arising from the crosses was determined by luminescence imaging. For mapping of the LOS1 locus, homozygous los1–1 plants in the C24 background were crossed to wild-type plants of the Columbia background. From the segregating F2 generation, a total of 885 homozygous los1–1 mutant plants based on the luminescence phenotype were selected for mapping with molecular markers that are polymorphic between C24 and Columbia. Genetic mapping was performed as described (13, 14). Simple sequence length polymorphism markers were developed by searching for simple repeat sequences with the repeatmasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker).
Primer pairs flanking the identified simple repeats that generate PCR products with size polymorphisms on 4% agarose gels between the C24 and Columbia ecotypes were used as molecular markers for mapping. Genomic DNA corresponding to candidate genes were amplified by PCR from wild-type and los1–1 mutant plants and sequenced to identify the los1–1 mutation.
For los1–1 mutant complementation, a PCR fragment containing the hypothetical T6H22#13 gene, including 1,391 bp upstream of the initiation codon and 231 bp downstream of the stop codon, was amplified from bacterial artificial chromosome (BAC) clone T6H22. The fragment was cloned into the binary vector pCAMBIA 1200. The insert was sequenced completely to verify that there were no PCR or cloning errors. The binary construct was then transformed into Agrobacterium strain GV3101 by electroporation and then introduced into los1–1 mutant plants by vacuum infiltration of the inflorescence. Hygromycin-resistant transgenic T2 and T3 plants were tested for RD29A-LUC expression in response to cold, ABA, and NaCl by luminescence imaging.
To obtain the cDNA sequence of LOS1, reverse transcription–PCR was performed with mRNA extracted from wild-type C24 plants by using the following primers: 5′-GGGATCCATGGT GAAGTTTACA GCTGATGAGC TTC-3′ and 5′-GGAATTCTTAAAGC TTGTCTTCGA ACTCAGATAG-3′. The reverse transcription–PCR product was cloned into pBluescript II KS(+) vector and sequenced.
Low Temperature and Other Stress Treatments.
For screening of mutants, the low temperature treatment was 0°C for 48 h in the dark. For detailed characterization of RD29A-LUC expression in response to low temperature treatment, agar plates with 1-week-old seedlings of mutant and wild-type plants were placed in an incubator set at the designated temperatures (±0.1°C) for 24 h. At the completion of the treatments, agar plates were removed from the incubator, and luminescence images were taken immediately (12). Treatment at subfreezing temperature (−5°C) lasted for 3 h and the plates were then placed at room temperature for 2 h to thaw before luminescence imaging. For ABA treatment, 100 μM ABA (mixed isomers) in water was sprayed on leaves of the seedlings and luminescence images were taken 3 h later. NaCl treatment was conducted on filter paper saturated with MS salt solution supplemented with 300 mM NaCl, and the luminescence images were taken 5 h after the seedlings were transferred to the filter paper.
For vernalization treatment, mutant and wild-type seeds were sown in pot medium and the pots were kept at 4°C for different periods (days) as indicated in the text. After vernalization, the pots were placed in growth chamber until flowering. At the emergence of the first flower, the number of rosette and cauline leaves was counted.
Freezing Tolerance Assay.
Plants for freezing tolerance assays were grown in pot medium in growth chambers. For cold acclimation, seedlings at the rosette stage (3 weeks old) were incubated at 4°C under white fluorescent light for 48 h before sampling the leaves for the freezing assay.
Fully developed rosette leaves were used to determine electrolyte leakage essentially as described by Ristic and Ashworth (15). Briefly, for each temperature treatment, an excised leaflet was placed in a test tube containing 100 μl deionized H2O, and the tube was incubated in a refrigerated circulator (freezing bath) (model 1187, VWR Scientific) with temperature set at 0°C. The temperature of the bath was programmed to decrease to −12°C with 1°C decrements in 30 min. The tubes were removed from the bath when the designated temperature was reached and placed immediately on ice to allow gradual thawing. The sample was then transferred to another tube containing 25 ml deionized water and shaken overnight, and the conductivity of the solution was measured. The tubes with the leaflets were then autoclaved, and after cooling to room temperature, conductivities of the solutions were measured again. Percent electrolyte leakage was calculated as the conductivity before autoclaving as a percentage of that after autoclaving.
For the whole-plant freezing test, 3-week-old plants in soil were cold-acclimated at 4°C in the light for 4 days, and then subjected to freezing at −7°C for 4 h. After the freezing treatment, the plants were moved to 4°C and incubated for 12 h before being returned to 22°C in a growth chamber. Pictures were taken 1 week later.
RNA Analysis.
Ten-day-old seedlings grown on MS agar plates were treated with either low temperature, ABA, NaCl, or polyethylene glycol (PEG), respectively (see text for details). Total RNA from control or stressed plants was extracted as described by Liu and Zhu (16). The RD29A gene-specific probe was from the 3′ noncoding region. COR15A and COR47 cDNAs (17, 18) were kindly provided by M. F. Thomashow (Michigan State University, East Lansing). DNA probes for RD19, RD22, and RD29B (19) were cloned from genomic DNA of wild-type Columbia plants by PCR. The ADH probe was a 1.6-kb SalI–NotI fragment of the Arabidopsis expressed sequence tag clone 199P20T7. The DREB2A probe was amplified by using the following primers: 5′-CAAAACAATATGAAGCTTTTTGG-3′ and 5′-AGTGTGTATTATTCATTCCTG-3′. CBF-specific probes were amplified by using the primer pairs as described (13).
Labeling with 35S Amino Acids.
Wild-type and los1–1 seeds were germinated and seedlings were allowed to grow on a MS medium containing 2% sucrose and 1.2% agar. One-week-old plants were pretreated at 0°C for 4 days and labeled with 250 μCi of >1,000 Ci (37.0 TBq)/mmol [35S]Met and [35S]Cys (EXPRE35S35S Labeling Mix, NEN) that was diluted to 200 μl in 0.1% (vol/vol) Tween 20 (20). The labeling mix was painted onto leaves and roots. The plants were then incubated at 0°C or 22°C for 36 h. After the treatment, plants were harvested and rinsed briefly in water, and total proteins were extracted by homogenization in a protein-loading buffer (67 mM Tris⋅HCl, pH 6.8/2.7% SDS/10% glycerol/0.5% β-mercaptoethanol/0.1% Bromophenol blue). The protein samples were denatured by boiling for 5 min and separated in a 10% SDS/PAGE gel. The gel was stained, dried, and exposed to an x-ray film.
Results
Identification of the LOS1 Locus.
Although the function of the RD29A gene product is unclear, its promoter has been extensively studied as a paradigm of transcriptional activation by cold, osmotic stress, or ABA (8, 9). To facilitate genetic analysis of cold and osmotic stress signaling, we previously generated transgenic Arabidopsis plants with stress-inducible bioluminescence by introducing a chimeric gene consisting of the firefly luciferase coding sequence under control of the RD29A promoter. Construction of the RD29A-LUC plants, mutagenesis, and mutant screening were described in Ishitani et al. (12). From a large collection of mutants with defective regulation of bioluminescence caused by cold, ABA, and/or high salt, several lines were identified as specifically blocked in regulation of gene expression by low temperature treatment. One of these lines, designated los1–1, was chosen for detailed characterization.
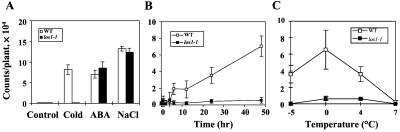
As shown in Fig. 1A, the los1–1 mutant exhibited significantly lower levels of luciferase expression than the wild type (the unmutagenized RD29A-LUC parental line) when treated with low temperature stress. However, its responses to ABA or high salt were not substantially different from those of wild-type plants. Fig. 1B illustrates the time course of RD29A-LUC response in both wild type and los1–1 during 0°C treatment. The mutant showed a much lower response throughout the cold treatment. The defect in los1–1 is also evident after treatments at temperatures below or above 0°C (Fig. 1C).
Figure 1.
RD29A-LUC expression in los1–1 and wild-type (WT) plants. (A) Luminescence intensity (RD29A-LUC expression) in wild type and los1–1 with different stress treatments. Control, 22 ± 2°C without treatment; Cold, 0°C for 24 h; ABA, 100 μM ABA, 3 h; NaCl, 300 mM NaCl, 5 h. Error bars represent sd (n = 20). (B) Time course of RD29A-LUC expression at 0°C. One-week-old los1–1 and wild-type seedlings grown in the same agar plates were placed in an incubator at 0°C. Plates were removed from the incubator at different time periods, and luminescence images were taken immediately and the intensities were determined. (C) RD29A-LUC expression in los1–1 and wild-type plants under different temperature treatments. los1–1 and wild-type plants were planted in the same agar plates and allowed to grow for 1 week under constant light at 22 ± 2°C. The plates were then treated at indicated temperatures (±0.1°C) for either 3 h (−5°C treatment) or 24 h (all other temperature treatments) before taking luminescence images. Error bars represent SD (n = 20).
los1–1 was backcrossed to the wild type. The resulting F1 seedlings were examined for RD29A-LUC (luminescence) expression at 0°C, and all were found to exhibit the wild-type phenotype. Selfed F2 populations from the backcross segregated ≈3:1 of wild type/mutant (data not shown). The results suggest that the mutant is caused by a recessive mutation in a single nuclear gene. All subsequent molecular and physiological experiments were carried out on the mutant that had been backcrossed at least three times with the wild type.
Transcript Levels of Cold-Regulated Genes in los1–1.
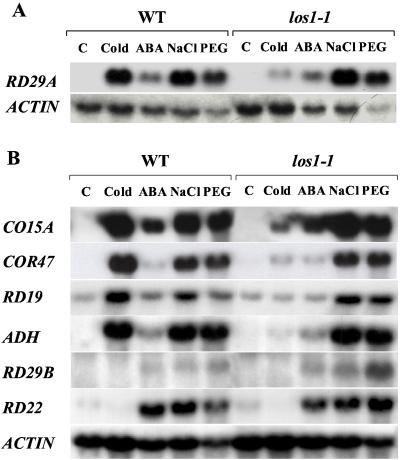
To determine whether the los1–1 mutation affects endogenous gene expression, total RNA was extracted from los1–1 and wild-type seedlings that were treated with low temperature, ABA, NaCl, or PEG. Cold induction of the endogenous RD29A gene was blocked in the los1–1 mutant (Fig. 2A). However, RD29A induction by ABA, NaCl, or PEG was not impaired by los1–1 (Fig. 2A). Cold-induced expression of several other genes was also greatly reduced in los1–1 compared with that seen in the wild type (Fig. 2B). The COR15A gene encodes a cryoprotective protein (6, 17). The results in Fig. 2 indicate that COR15A expression under cold stress was blocked by the los1–1 mutation. Similar to RD29A, the COR47 gene has no established function but has sequence similarity with genes encoding group II LEA proteins (18). Cold induction of COR47 was also greatly reduced in the los1–1 mutant (Fig. 2B). The mutation eliminated cold induction of RD19 that has sequence similarity with cysteine proteases (21). Cold induction of the alcohol dehydrogenase gene (ADH) was nearly abolished by the los1–1 mutation. The los1–1 mutation did not influence the expression of RD29A (12) (Fig. 2A), COR15A, COR47, RD19, or ADH in response to ABA or osmotic stress (Fig. 2B). The RD29B gene, which has high sequence similarity with RD29A, is not induced by low temperature stress (Fig. 2B). However, it is interesting to note that RD29B induction by PEG was higher in the los1–1 mutant (Fig. 2B). The los1–1 mutation does not affect the expression of the constitutive actin gene under cold or other stress treatments (Fig. 2).
Figure 2.
The steady-state transcript levels of stress-responsive genes in los1–1 and wild-type (WT) plants. Plants were treated with: low temperature, 0°C for 24 h; ABA, 100 μM ABA for 3 h; NaCl, 300 mM NaCl for 5 h; and 30% PEG (average molecular weight 6,000) for 5 h.
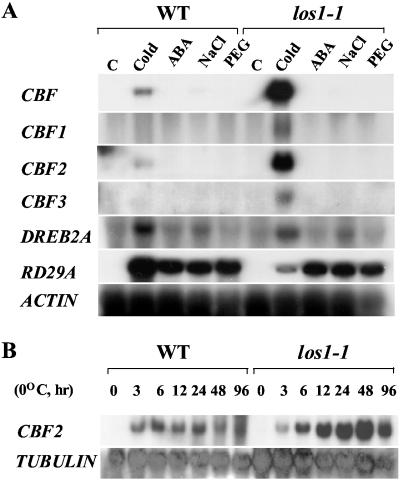
It was of interest to know whether the expression of cold-specific transcription factors that activate the expression of the above cold-regulated genes is altered in los1–1 mutant plants. RNA blot analysis using the CBF1 coding sequence as a probe revealed that CBF transcripts were superinduced by cold in the mutant (Fig. 3). Subsequent experiments using CBF1-, CBF2-, and CBF3-specific probes found that cold induction of all three transcription factor gene transcripts was enhanced by the los1–1 mutation (Fig. 3A). A related transcription factor gene DREB2A was induced by cold as well as by NaCl treatments, under our conditions (Fig. 3A), although this gene was previously reported not to be induced by cold (10). However, the cold induction of DREB2A was not enhanced in the los1–1 mutant (Fig. 3). Although not much different at the 3-h time point, the transcript levels for CBF2 (Fig. 3B) and other CBFs (data not shown) were consistently higher in los1–1 than in wild type at all other time points during the cold treatment.
Figure 3.
Transcript levels of CBF and DREB2A genes in wild-type (WT) and los1–1 plants. The probe for CBF is CBF1 coding sequence. CBF1, CBF2, CBF3, and DREB2A probes are gene-specific and are from the 3′ untranslated regions of the respective genes. (A) Transcript levels after stress treatments. C, control, no stress treatment; Cold, 0°C for 24 h; ABA, 100 μM ABA for 3 h; NaCl, 300 mM NaCl for 5 h; PEG, 30% PEG for 5 h. (B) CBF2 expression after treatments at 0°C for the indicated times. An actin or tubulin gene was used as loading control.
los1–1 Mutation Impairs Cold Acclimation but Not Vernalization.
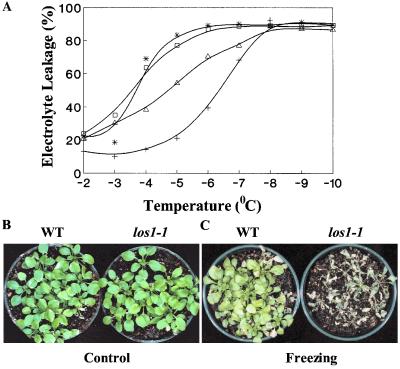
An electrolyte leakage test was performed to evaluate the effect of the los1–1 mutation on plant freezing tolerance (Fig. 4A). There was little difference between los1–1 and the wild-type plants without acclimation. Temperatures at 50% ion leakage (LT50) were estimated to be −3.4°C and −3.6°C for nonacclimated los1–1 and wild-type plants, respectively. After 2 days of acclimation at 4°C, wild-type plants became considerably more tolerant. The LT50 value of acclimated wild-type plants dropped to −6.4°C (Fig. 4A). The los1–1 mutant also increased in freezing tolerance after acclimation; however, the increase was substantially less than that of the wild type. The LT50 value of acclimated los1–1 plants was estimated to be −4.6°C (Fig. 4A). The ion leakage experiments were repeated three times with different batches of plants, and similar results were obtained in these experiments, which showed that acclimated los1–1 plants are less freezing tolerant.
Figure 4.
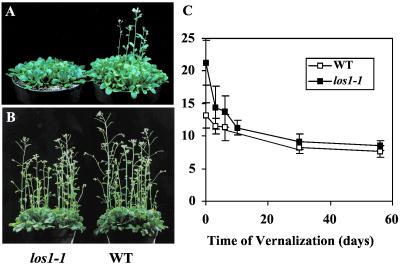
Freezing sensitivity of los1 mutant. (A) Freezing induced leakage of electrolytes from los1–1 and wild-type leaves. For cold acclimation, plants were incubated at 4°C for 48 h under white fluorescent light. □, Nonacclimated wild type; *, nonacclimated los1–1; +, acclimated wild type; and ▵, acclimated los1–1. (B) Three-week-old wild-type (WT) and los1–1 plants without freezing treatment (Control). (C) los1–1 and wild-type plants after freezing treatment. Three-week-old wild-type and mutant plants were first incubated at 4°C for 4 days, and then subjected to freezing treatment (−7°C, 4 h). The pictures were taken 1 week after the treatment.
Intact wild-type and los1–1 rosette plants growing in soil were also subjected to the freezing test. The plants were treated at 4°C for 4 days for cold acclimation and then subjected to freezing at −7°C for 4 h. Fig. 4C shows that most of the wild-type plants survived the freezing treatment, whereas virtually all los1–1 plants were killed. This result further illustrates that los1–1 mutant plants are defective in cold acclimation.
Under normal growth conditions, los1–1 plants grow and develop just like wild-type plants, except that los1–1 plants flower a little late (Fig. 5 A and B). Under long day conditions (16 h light, 8 h dark), los1–1 plants flowered ≈5 weeks after imbibition, whereas the wild type flowered within 4 weeks after imbibition. The total leaf number (LN) at first flower opening may be a more accurate measurement of flowering time (22). The LN values were 21.3 ± 3.4 and 13.2 ± 1.9 for los1–1 and wild-type plants, respectively. Because flowering time in Arabidopsis can be influenced by long periods of low temperature treatment (i.e., vernalization), we determined the vernalization responses of los1–1 and wild-type plants. As shown in Fig. 5C, both los1–1 and wild-type plants responded to vernalization. After 8 weeks of vernalization at 4°C, the LN value for wild-type plants was decreased to 7.7 ± 0.9, and for los1–1, it was decreased to 8.5 ± 0.7. The levels of reduction in LN values in response to 8 weeks of vernalization are 42% and 60%, respectively, for wild type and los1–1. Thus, the los1–1 mutation does not impair the vernalization response, rather it appears that the mutation actually enhances vernalization (Fig. 5C).
Figure 5.
Development and vernalization response of los1–1 and wild-type (WT) plants. (A) Four-week-old wild-type and los1–1 plants under growth chamber conditions. (B) Six-week-old wild-type and los1–1plants under growth chamber conditions. (C) Vernalization response. Seeds of los1–1 and the wild type were sown in pot medium and incubated in a cold room (4°C) for the indicated time periods before being placed in a growth chamber. At emergence of the first flower, 20 plants were counted for total LNs.
Positional Cloning of LOS1.
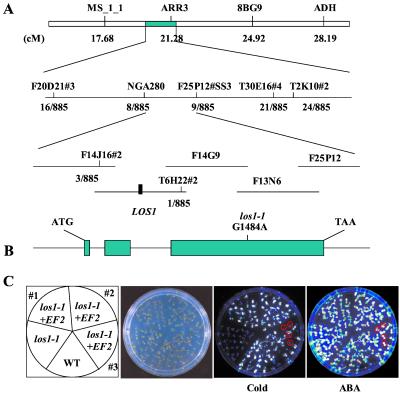
To map the LOS1 gene genetically, los1–1 mutant plants in the C24 ecotype were crossed with wild-type plants of the Columbia ecotype. From the segregating F2 population, los1–1 mutant seedlings were identified by their low luminescence under cold treatment but normal luminescence after ABA application. The chromosomal map position of los1–1 was determined by using PCR-based molecular markers (23, 24). los1–1 showed linkage to nga111 on chromosome I and no linkage to markers on other chromosomes. Segregation of the microsatellite marker nga280 on chromosome I was then determined. The results show that the LOS1 locus is tightly linked to nga280. Several simple sequence length polymorphism markers were developed based on the genomic sequences of BAC clones in the nga280 region (Fig. 6A). Fine mapping with these markers delimited LOS1 to an approximately 80-kb region, between the markers F14J16#2 and T6H22#2. Further mapping became very difficult because of a lack of recombination events. The ORFs of candidate genes in this region were sequenced from wild-type as well as los1–1 mutant plants. The sequence analysis revealed a single nucleotide substitution in the los1–1 mutant in the hypothetical T6H22#13 gene, i.e., a G-to-A change at position 1484 (Fig. 6B). No mutation was found in los1–1 mutant plants in any other candidate genes sequenced.
Figure 6.
Positional cloning of the LOS1 gene. (A) Genetic mapping delimited LOS1 to BAC clones F14J16 and T6H22. The los1–1 mutation was identified by sequencing and comparing all predicted genes on these BAC clones from los1–1 mutant and wild-type plants. (B) Structure of LOS1 and the position of the los1–1 mutation. Positions are relative to the translation initiation codon. Filled boxes indicate the ORF, and lines between boxes indicate introns. The G1484A mutation in the los1–1 mutant is indicated. (C) Complementation of los1–1 mutant by the wild-type (WT) LOS1 gene. Ten-day-old wild-type, los1–1, and two homozygous T3 progenies and one T2 progeny of the los1–1 plant containing the wild-type LOS1 (indicated as los1–1 + EF2) were assayed for complementation. From left, picture indicates the position of wild-type, los1–1, and the T3 (nos. 1 and 2) and T2 (no. 3) progenies; picture of the plants in an agar plate; luminescence image of the plants incubated at 0°C for 48 h; and luminescence image of the plants treated with 100 μM ABA for 3 h. los1–1 plants in the segregating T2 progeny (no. 3) were circled, which showed low luminescence in the cold but normal luminescence under ABA treatment.
To confirm that T6H22#13 is LOS1, a wild-type genomic fragment corresponding to this gene was cloned from BAC clone T6H22 and then introduced into los1–1 mutant plants. Several T2 and T3 transformants were tested for RD29A-LUC expression by luminescence imaging. All transformants containing the introduced wild-type T6H22#13 gene showed a wild-type phenotype. Fig. 6C shows the luminescence responses from two independent homozygous T3 lines and one segregating T2 line. Plants from the homozygous lines all exhibited a wild-type RD29A-LUC expression in response to cold. The T2 line segregated for wild-type and los1–1 phenotypes in cold-induced luminescence. As expected, those showing a mutant cold response reacted normally to ABA (Fig. 6C). These results demonstrated that los1–1 mutant phenotypes were complemented by the wild-type T6H22#13 gene, thus confirming that T6H22#13 is indeed LOS1.
LOS1 Encodes a Translation Elongation Factor 2-Like Protein.
LOS1 cDNA was obtained by reverse transcription–PCR. Comparison of the cDNA and genomic sequences showed that the LOS1 gene contains three exons and two introns (Fig. 6B). The LOS1 cDNA (GenBank accession no. AY054461) predicts an 843-aa protein with an estimated molecular mass of 94 kDa and pI of 5.9. The los1–1 mutation occurs in the third exon and changes cysteine495 to tyrosine.
Database searches revealed that the predicted LOS1 protein is similar to eukaryotic translation elongation factor 2 (eEF-2) proteins. LOS1 shares more than 60% amino acid identity with eEF-2 proteins from yeast, nematode, and fruitfly. This high sequence homology with eEF-2 proteins extends throughout the entire length of LOS1. LOS1 contains all five functional domains (not shown) shared by eEF-2s and prokaryotic elongation factors G (25). LOS1 appears to be the only cytoplasmic eEF-2 in Arabidopsis. Two other proteins (GenBank accession nos. At1g06220 and At5g25230) encoded by the Arabidopsis genome share only 36–37% amino acid identity with LOS1, but are highly similar to the human U5–116kD and yeast Snu114p that are part of the spliceosomal small nuclear ribonucleoprotein (26). Cys495 in LOS1 is not conserved between plant and nonplant eEF-2 proteins (see Fig. 8A, which is published as supporting information on the PNAS web site, www.pnas.org). However, this residue is absolutely conserved in all plant eEF-2 proteins we have examined (Fig. 8B).
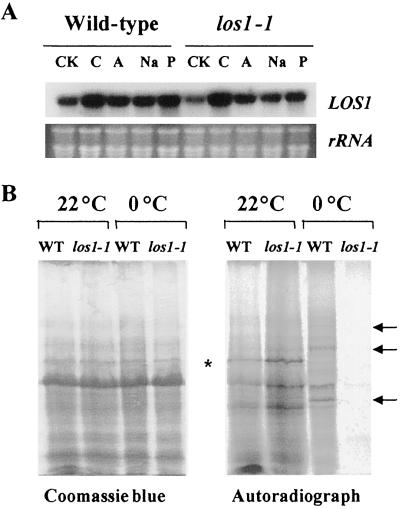
The LOS1 gene is expressed in every plant part (i.e., root, stem, leaves, flowers, and siliques) examined (data not shown). Although constitutively expressed, the level of LOS1 transcript was slightly enhanced by cold treatment but was not obviously affected by other stress or ABA (Fig. 7A). Additionally, the los1-1 mutation does not appear to affect LOS1 transcript level (Fig. 7A).
Figure 7.
The expression of LOS1 gene and new protein synthesis in los1 plants. (A) Transcript levels of LOS1 in wild-type and los1–1 plants under control (CK), cold (C) (0°C, 24 h), ABA (A) (100 μM, 3 h), NaCl (300 mM, 3 h), or PEG (30%, 5 h) treatment. (B) In vivo labeling with [35S]Met and [35S]Cys. One-week-old wild-type (WT) and los1–1 plants were incubated at 0°C for 4 days and then labeled with [35S]Met and [35S]Cys at either 0°C or 22°C for 36 h. Total proteins were extracted and resolved on a 10% SDS/PAGE gel. Arrows point to cold up-regulated proteins in the wild type. * indicates a cold down-regulated protein in the wild type.
los1–1 Mutant Plants Are Defective in Protein Synthesis in the Cold.
The LOS1 sequence suggests that it functions in protein synthesis. To determine whether los1–1 mutant plants are impaired in protein synthesis at normal growth temperatures as well as under cold treatments, we carried out in vivo labeling studies by feeding 1-week-old seedlings a mixture of [35S]Met and [35S]Cys. The 35S precursors were applied to the leaf surfaces and roots of intact seedlings of wild type and los1–1. The feeding was carried out for 36 h at either 22°C or 0°C. Fig. 7B shows that the incorporation of [35S]Met and [35S]Cys into new proteins was not different between wild-type and los1–1 mutant plants at 22°C. At 0°C, new protein synthesis took place in the wild-type seedlings (Fig. 7B). Several cold-induced protein bands were evident in the wild-type sample (Fig. 7B, indicated by arrows). However, there was very little new protein synthesis in los1–1 seedlings at 0°C. At 4°C, los1–1 mutant seedlings also had reduced protein synthesis compared with the wild type, but the reduction was not as dramatic as at 0°C (data not shown). These results suggest that the los1–1 mutation impairs new protein synthesis specifically in the cold.
Discussion
RD29A expression in wild type is induced by treatment with cold, osmotic stress, or ABA. In the los1–1 mutant there is a loss of induction by cold, but normal induction by ABA and NaCl. In addition to RD29A, the expression of all other downstream cold-regulated genes analyzed is reduced substantially in los1–1 plants after cold treatment (Fig. 2). Surprisingly, cold induction of CBF/DREB1 genes is higher in los1–1 than in the wild type (Fig. 3). This intriguing result points to the possibility that the expression of CBF/DREB1 genes is down-regulated by their own gene products or by their target genes. The los1–1 mutation appears to block the events downstream of the induction of CBF/DREB1 transcripts and thus the feedback regulation of CBF/DREB1 genes does not occur, which would then lead to the overaccumulation of CBF/DREB1 transcripts.
Results from LOS1 cloning support this hypothesis. LOS1 is predicted to encode a translation elongation factor 2. eEF-2 is essential for protein synthesis by mediating the translocation step in peptide chain elongation. eEF-2 is a GTPase that promotes the transfer of peptidyl-tRNA from the A to the P site of the ribosome (27). In the los1–1 mutant, Cys495 was changed to Tyr. This Cys residue is conserved in all plant eEF-2 proteins. The Cys495 to Tyr change in los1–1 may affect the functionality of this elongation factor specifically at low temperatures but not at warm temperatures. Indeed, in vivo labeling studies (Fig. 7B) showed that protein synthesis in los1–1 mutant plants is normal at 22°C but is impaired at 0°C. These results suggest that los1–1 is a cold-sensitive allele.
These results demonstrate that cold induction of CBF/DREB1 genes does not require new protein synthesis. An obvious implication of this finding is that all signaling components required for CBF/DREB1 induction are constitutively present. Because los1–1 mutant plants are defective in protein synthesis in the cold, the induced CBF/DREB1 transcripts cannot be translated. Without the CBF/DREB1 proteins, the downstream cold-responsive genes (i.e., RD/COR/LTI/KIN) cannot be activated. This scenario is reminiscent of a number of transcriptional cascades in mammalian cells where “early response” transcription factor genes are induced without the need for new protein synthesis but these transcription factor proteins need to be made to activate their downstream “delayed response” genes (28). The CBF/DREB1 genes are rapidly and transiently induced by cold (29). In comparison, the downstream cold-responsive genes are induced later by cold, but the induction is not transient (29). The transient nature of CBF/DREB1 induction suggests that CBF/DREB1 proteins may feedback suppress their own transcription. Alternatively, some of their downstream gene products may suppress the transcription of CBF/DREB1 genes. The fact that the CBF/DREB1 genes are superinduced by cold in the los1–1 mutant is consistent with this notion of feedback suppression of CBF/DREB1 expression in the wild type.
The los1–1 mutation not only impairs cold induction of RD/COR/LTI/KIN gene transcripts, it also reduces the capacity of the mutant plant to cold-acclimate. los1–1 plants were less tolerant to freezing stress than were the wild type after both had been acclimated at 0°C (Fig. 4). However, the los1–1 mutation did not abolish the ability to cold acclimate totally, because freezing tolerance in the mutant still increased after acclimation, although the increase was less than in the wild type (Fig. 4A). The decline in cold acclimation in the mutant is probably a direct consequence of reduced cold induction of genes. COR15A, for example, has been demonstrated to contribute to freezing tolerance (6). Other gene products such as RD29A and COR47 presumably also have cryoprotective functions. The remaining cold acclimation response in los1–1 suggests that part of the cold acclimation responses is independent of new protein synthesis and independent of the CBF/DREB1 regulon.
Besides the activation of gene expression and acclimation, another important plant response to low temperature is vernalization (22). Vernalization refers to the acceleration effect of low temperature on flowering time. Unlike cold-induced gene expression and acclimation, vernalization results from relatively long term (weeks to months) exposure to cold. The los1–1 mutation clearly does not impair the response to vernalization at 4°C (Fig. 5). The fact that los1–1 mutant plants are impaired in protein synthesis in the cold but can still be vernalized suggests that new protein synthesis may not be critical for vernalization. It is possible that the residual protein synthesis at 4°C is sufficient to allow normal vernalization of los1–1 mutant plants.
Supplementary Material
Acknowledgments
We thank Dr. Andre Jagendorf for critical reading of the manuscript and Becky Stevenson and Hojoung Lee for excellent technical assistance. This work was supported by U.S. Department of Agriculture National Research Initiative Grant 2000-0064 to (J.-K.Z.).
Abbreviations
- DREB
dehydration responsive element binding factor
- CBF
C-repeat binding factor
- ABA
abscisic acid
- MS
Murashige and Skoog
- BAC
bacterial artificial chromosome
- PEG
polyethylene glycol
- LN
leaf number
- eEF-2
eukaryotic translation elongation factor 2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Guy C L, Anderson J V, Haskell D W, Li Q-B. In: Biochemical and Cellular Mechanisms of Stress Tolerance in Plants. Cherry J H, editor. Berlin: Springer; 1994. pp. 479–499. [Google Scholar]
- 2.Thomashow M F. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 3.Guy G L, Haskell D. Plant Physiol. 1987;84:872–878. doi: 10.1104/pp.84.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajela R K, Horvath D P, Gilmour S J, Thomashow M F. Plant Physiol. 1990;93:1246–1252. doi: 10.1104/pp.93.3.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordin K, Heino P L, Palva E T. Plant Mol Biol. 1991;16:1061–1071. doi: 10.1007/BF00016077. [DOI] [PubMed] [Google Scholar]
- 6.Artus N N, Uemura M, Steponkus P L, Gilmour S J, Lin G, Thomashow M F. Proc Natl Acad Sci USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steponkus P L, Uemura M, Joseph R A, Gilmour S J, Thomashow M F. Proc Natl Acad Sci USA. 1998;95:14570–14575. doi: 10.1073/pnas.95.24.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockinger E J, Gilmour S J, Thomashow M F. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Kasuge M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaglo-Ottosen K R, Gilmour S J, Zarka D G, Schabenberger O, Thomashow M F. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 12.Ishitani M, Xiong L, Stevenson B, Zhu J-K. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu J K. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong L, Ishitani M, Lee H, Zhu J K. Plant Cell. 2001;9:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ristic Z, Ashworth E N. Protoplasma. 1993;172:111–123. [Google Scholar]
- 16.Liu J, Zhu J-K. Plant Physiol. 1997;114:591–596. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C, Thomashow M F. Plant Physiol. 1992;99:519–525. doi: 10.1104/pp.99.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmour S J, Artus N N, Thomashow M F. Plant Mol Biol. 1992;18:13–32. doi: 10.1007/BF00018452. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K. Plant Cell Physiol. 1992;33:217–224. [Google Scholar]
- 20.Hansen E R, Petracek M E, Dickey L F, Thompson W F. Plant Physiol. 2001;125:770–778. doi: 10.1104/pp.125.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. Gene. 1993;129:175–182. doi: 10.1016/0378-1119(93)90266-6. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Zapater J M, Coupland G, Dean C, Koornneef M. In: Arabidopsis. Clark S E, Meyerowitz E M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 403–433. [Google Scholar]
- 23.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 24.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 25.Czworkowski J, Wang J, Steitz T A, Moore P B. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabrizio P, Laggerbauer B, Lauber J, Lane W S, Luhrmann R. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proud C G. Mol Biol Rep. 1994;19:161–170. doi: 10.1007/BF00986958. [DOI] [PubMed] [Google Scholar]
- 28.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D, editors. Molecular Biology of the Cell. New York: Garland; 1994. pp. 729–731. [Google Scholar]
- 29.Gilmour S J, Zarka D G, Stockinger E J, Salazar M P, Houghton J M, Thomashow M F. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.