Abstract
We present a comprehensive study of the evolutionary origin of the thermodynamic behavior of proteins. With the use of a simplified model, we exhaustively enumerate the space of all sequences and the space of all structures, simulate the evolutionary relationship between sequences and structures, and characterize the steady-state sequence distribution for all structures in terms of several thermodynamic variables. We assess the effects of two major forces of evolution: mutation and recombination. Three simplifications are made. First, a two-dimensional lattice model is used to represent protein sequences and structures. Second, proteins undergo neutral evolution so that the fitness landscape has a flat allowed region inside of which all sequences are equally fit. Third, we ignore otherwise important factors such as finite population size and evolutionary time. Two scenarios emerge from our study. The first occurs when evolution is dominated by mutation events. Even though the prototype sequence that is most mutationally robust is preferred by evolution, the preference is not strong enough to offset the huge size of sequence space. Most native sequences are located near the boundary of the fitness region and are marginally compatible with the native structure. The second scenario occurs when evolution is dominated by recombination events. Now evolutionary preference for prototype sequence is strong enough to overcome the size of sequence space so that most native sequences are located near the center of sequence-structure compatibility. We conclude that the relative frequency of mutation and recombination events is a major determinant of how optimal protein sequences are for their structures.
Understanding the relationship between protein sequences and structures is a central theme in modern molecular biology. Such an understanding can be partially achieved by predicting the folded structure from the amino acid sequence, and by predicting the compatible sequences from a known structure, using physical principles or protein-specific knowledge. However, a better understanding requires a global view of the space of all sequences and the space of all structures and its dynamics as a result of evolution. Because proteins are building blocks of life and a direct result of evolution, it is crucial to understand how evolution shapes the global relationship between protein sequences and structures by simulating the process of evolution in a direct way.
Molecular evolution is often simulated as an adaptive walk over a fitness landscape (1) of connected network of all sequences (2). Simulation of molecular evolution has been used to study RNA sequence-secondary structure relationship (3, 4). Simple tractable models such as lattice and spin-glass models are often used to understand physical principles of protein folding (5–7), because these models are often simple enough to allow for precise statistical mechanical characterization, yet are able to capture the dominant forces in protein folding (8). Recently, simple tractable models are combined with simulation of evolution to study protein sequence–structure relationship on the basis of thermodynamic and kinetic criteria (9–14) and to study recombinatoric exploration of new structures (15). Computational studies can also guide experiments on in vitro evolution (16).
Considerable evidence shows that most accepted molecular mutations have little effect on the fitness of an organism (17). Neutral (and near-neutral) evolution can arise when the fitness landscape is flat (18) or near flat (19), and when the noise in measuring the fitness is comparable with fitness change (20). Neutral evolution has been studied in the context of both RNA secondary structure (21) and model proteins (22). Bornberg-Bauer et al. showed that in neutral evolution of proteins, neutral network topology alone leads to preference for the prototype sequence with maximal mutational stability (23), even though the fitness landscape is flat.
Despite recent advances, a comprehensive view of how evolution shapes the protein sequence–structure relationship is needed. Most previous studies have focused on the sole effects of mutation events, even though evolution involves a combined action of mutation and recombination. In this paper, we perform a detailed study of the effects of both mutation and recombination events on the evolution of protein stability, by using a simplified model for proteins. We assume that to be functional, proteins must assemble and be thermodynamically stable under physiological conditions; once biologically active, the effect of protein stability on protein activity and in general on the organism's fitness is likely to be secondary.
Methods
Protein Model.
We use a 5 × 5 two-dimensional square lattice model for proteins. Protein conformations are represented by self-avoiding compact walks that occupy all lattice vertices, so that all chains are 25 residues long. A total of 1,081 such conformations are not related by symmetry. Each lattice vertex represents a protein residue and is labeled by H (hydrophobic) or P (polar). We use a pairwise contact energy function with eHH = −2.3, eHP = −1, ePP = 0 (24). Despite the limitations of the model (chains are short; all conformations are two-dimensional and maximally compact), this model has protein-like properties. We exhaustively enumerate all possible protein sequences and compute their corresponding native structures and related thermodynamic quantities. A protein sequence is compatible and will fold into a structure if and only if the structure is the unique global minimum of energy among all structures. All sequences that fold into the same structure are assumed to share the same fitness value.
Evolutionary Steady-State with Mutation Events.
We first consider evolution with no recombination events. We construct a neutral network and compute the evolutionary steady-state population in a way similar to Bornberg-Bauer et al. (23). All sequences that fold into the same structure and differ by single mutations are connected to form a neutral network of sequences. A cartoon of a small neutral network is shown in Fig. 1A. Sequence evolution is modeled as a diffusion process over the neutral network in the following way. First, an initial population distribution is established for a pool of N sequences at time 0. At each evolutionary time step, nm mutation events are performed, of which nl are lethal. When N is sufficiently large, this is equivalent to a sequence undergoing single mutation at any position with probability pm = nm/N at any given time, or remain unmutated with probability 1 − pm. The mutated sequence survives if it folds into the same structure, and dies otherwise. After this diffusion step, population distributions for all protein sequences are rescaled so that the population size remains constant. This process is iterated until a steady state is reached and population distribution for all sequences does not change. The steady-state population distribution corresponds to a nonnegative eigenvector of a sparse matrix M determined solely by neutral network topology: Mij is −1 if sequences i and j (i ≠ j) are connected in the neutral network and 0 otherwise. Mii is the number of connections for sequence i in the neutral network. We solve this eigenproblem by transforming M into a nonnegative matrix σ I − M (I is the unit matrix, and parameter σ is determined by a grid search) whose nonnegative eigenvector is determined by a converging power method starting with a nonnegative random vector (25). We perform this computation for every neutral network and locate the prototype sequence that is most populated at steady state.
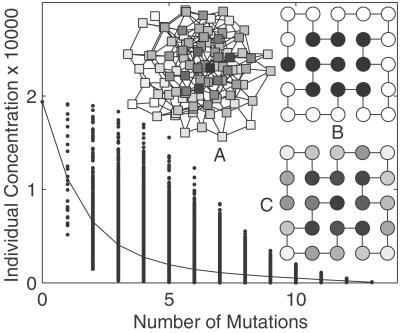
Fig 1.
For evolution with mutation events only, steady-state concentration vs. number of mutations from the prototype sequence for the neutral network with the largest size. Dots mark the concentration for each of the sequences in the neutral network. The solid line connects average concentration for a given number of mutations. A sequence with more mutations from the prototype sequence is less frequent on average, even though the concentration varies widely. (Inset A) A two-dimensional projection of a small neutral network with 99 members. Protein sequences (squares) that differ by a single mutation are connected. Each square is colored in proportion to how frequent it is visited at evolutionary steady state with mutation events only. The darkest square represents the prototype sequence at the center of the sequence cloud. (B and C) Structure with the largest neutral network (also lowest reproduction difficulty). In the long run this structure dominates the population in the absence of additional functional selection. It also has the highest designability. Two neutral networks exist for this structure with 67,614 sequences and 1 sequence, respectively, so the designability for the structure is 67,615. (B) The prototype sequence for this structure. This sequence is most likely to be occupied at steady state. H residues are dark, and P residues are white. (C) Sequence profile constructed from multiple alignments of 1,000 sequences picked at random at steady state. Each vertex is colored in proportion to the probability that it is occupied by an H residue. Even though a typical native sequence is on average more than five mutations away from the prototype sequence, the sequence profile recovers the prototype sequence well.
Evolutionary Steady-State with Mutation and Recombination Events.
For a structure with more than one neutral network, it is possible to move between neutral networks by means of recombination events. Therefore, we determine sequence distributions not for individual neutral networks, but for all sequences that fold into a particular structure. The prototype sequence for a structure is then defined as the prototype sequence for the largest neutral network for the structure. It is not feasible to enumerate all possible recombination events and compute steady-state distribution by matrix computation. Instead, we use a Monte Carlo algorithm to compute steady-state averages of thermodynamic and mutational quantities.
We start with a population of N sequences that fold into a given structure. Two different initial conditions are considered. (i) Every sequence within the population is selected randomly from the set of sequences that fold into the target structure. (ii) One sequence is selected randomly from the set of sequences that fold into the target structure, and all sequences in the population are set to be identical with this sequence. At each evolutionary time step, nm mutation events are performed, followed by nr recombination events. For each mutation event, a random sequence is selected from the population and a random position is mutated. The original sequence is replaced by the mutated sequence if the latter folds into the target structure. Otherwise it is replaced by a copy of a sequence randomly selected from the rest of the population. For each recombination event, two random sequences are selected from the population, and a random sequence position is chosen for a one-point recombination crossover. The two parent sequences are then replaced by the two child sequences. If either child sequence does not fold into the target structure, it is replaced by a copy of a sequence randomly selected from the rest of the population. This process is repeated until convergence.
For evolution with and without recombination events, we compute the distribution of several quantities: d, the number of mutations from the prototype sequence; Tf, the folding temperature, defined as the temperature at which the equilibrium concentration for the native structure is equal to the total concentration for all other conformations, a direct measure of thermodynamic stability of the native state relative to other conformations; ps, the probability that a random mutation produces a more stable sequence, i.e., with a higher Tf. ps is a measure of mutational stability. The steady-state averages only depend on the ratio nr/nm for sufficiently large population size N. At steady state, the quantity nl/nm provides a measure of the reproduction difficulty independent of population size and mutation rate, where at each evolutionary time step, nl is the number of nonviable off-spring produced, and nm is the number of mutation events.
Results
Evolution Favors the Center Sequence of the Neutral Network.
We first used the structure with the largest designability (i.e., number of sequences that fold into the structure) as the target structure to study evolutionary effects on protein thermodynamics. An evolutionary argument for this choice is given later in this paper. Allowing mutation alone, we compute the steady-state population distribution of all sequences in the largest neutral network. Fig. 1B shows the prototype sequence for this structure, which has the highest concentration. This sequence fits the structure well: buried residues are all hydrophobic, and surface residues are all hydrophilic (except for one end). We compute d, the number of mutations from the prototype sequence for all sequences in the neutral network, and plot the corresponding sequence concentration against number of mutations (Fig. 1). For a given number of mutations, concentration varies widely depending on the detailed location of the sequence in the neutral network. The average concentration for sequences with the same number of mutations decreases monotonically as the number of mutations grows. Thus, the prototype sequence located at the center of the neutral network is the most compatible sequence with the target structure in an evolutionary sense.
Mutation Alone Results in Native Sequences That Are Far from Optimal.
Fig. 2 shows population distribution of three quantities (d, Tf, and ps) before and after evolutionary enrichment. In each case evolution by mutation moves the distribution toward the prototype sequence; this shift is most pronounced for Tf. Even though evolution moves the population distribution toward the prototype sequence, the effect is weak. As a result, a sequence is more likely to get lost in the vast sequence space than to find the prototype sequence. At steady state the concentration of the prototype sequence is small. Most sequences are far away from the prototype sequence: a typical sequence at steady state is more than five mutations away from the prototype sequence (Fig. 2A), and no longer has the perfect H-P separation seen in the prototype sequence (Fig. 1B): some surface residues are hydrophobic, and some buried residues are hydrophilic. A typical native sequence is not very stable: its folding temperature, Tf, is just above 0.4, compared with almost 1 for the prototype sequence. It is also easier to engineer a typical native sequence to a more stable one: 20% of the point mutations lead to a more stable sequence, as opposed to just 4% for the prototype sequence.
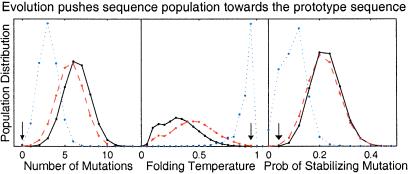
Fig 2.
For the structure shown in Fig. 1B, sequence population distribution before and after selection by neutral evolution with mutation and/or recombination events as measured by three quantities: number of mutations from the prototype sequence, d; folding temperature, Tf; and probability of a random mutation being stabilizing, ps. We set Boltzmann's constant to 1, so the units for temperature match the units for energy. Before evolutionary selection, all sequences are equally populated, and the resulting distribution is shown with a solid line. With mutation events alone, the resulting population by evolutionary selection is shown with a dashed line. With dominant recombination events (nr/nm = 1,000, N = 100,000), the resulting population by evolutionary selection is shown with a dotted line. Arrows mark the prototype sequence. Recombination is much more effective than mutation in pushing the population toward the prototype sequence.
If we imagine a projection of a protein structure onto sequence space as a cloud of sequences folding to that structure, the prototype sequence is at the center of the cloud. Can we recover the prototype sequence from a population of native sequences, most of which are located near the boundary of the cloud? In Fig. 1C, we show the sequence profile constructed by multiple sequence alignments of sequences picked from the steady-state distribution. This profile recovers the prototype sequence almost perfectly; if we transform the sequence profile into an HP sequence, it is only two mutations from the prototype sequence.
Recombination Results in Sequences That Are Close to Optimal.
We now study how evolution shapes protein sequence–structure relationships when recombination dominates. Picking the same target structure, we simulate evolution with a population of N = 100,000 sequences and a 1,000:1 ratio of recombination to mutation rates.
When recombination events are dominant, the steady-state sequence population (Fig. 2) is close to the prototype sequence (just three mutations away), and almost optimal in terms of thermodynamics (Tf > 0.9). Only 10% of the point mutations lead to a more stable sequence. Surprisingly, evolutionary preference for recombinational robustness is much stronger than mutational robustness. It is strong enough to overcome the huge size of sequence space and bring the sequence population close to the prototype sequence.
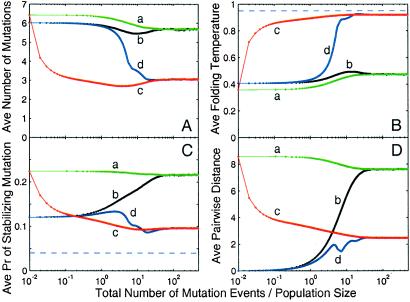
The time evolution of these quantities is more complicated (Fig. 3) and leads to the following observations. (i) Recombination complicates population dynamics. When mutations dominate, the average distance between all pairs of sequences in the population, a measure of the diversity of the population in sequence space, monotonically increases or decreases toward steady state. When recombination dominates and the population starts out as a point in sequence space, the population expands and contracts several times before reaching steady state. This additional complexity occurs as mutation acts on a single sequence and is independent of the population, whereas recombination acts on a pair of sequences and depends on the degree of homogeneity of the population. (ii) Starting from different initial conditions, evolution leads to a converged sequence population. Even though the dynamic behavior is complicated by recombination events, the steady-state distribution is so strong an evolutionary attractor that long-term behavior of the sequence distribution is independent of initial conditions. (iii) Mutation, not recombination, limits the rate of convergence. It takes between 10 and 100 mutation events per sequence to reach convergent steady state, regardless of recombination rate, mutation rate, or population size. Mutation is an irreversible process that progressively reduces the correlation between the current population and the initial population. Convergent steady state is reached when the correlation approaches zero, which occurs when the total number of mutations per sequence is of the same order of magnitude as the diameter of the space of allowed sequences. (iv) In the presence of recombination events, the spread of the sequence population at steady state is smaller. Recombination acts like a spring that holds sequence population together against the diffusion induced by mutation.
Fig 3.
For the structure shown in Fig. 1B, evolutionary dynamics of a sequence population with size N = 100,000 is measured by ensemble averages of four quantities: (A) d; (B) Tf; (C) ps; and (D) average pairwise distance between sequences in the population, a measure of the population spread in sequence space. The x axis is evolutionary time on a logarithmic scale, measured by the total number of mutation events divided by the population size, i.e., total number of mutation events per sequence. Four lines are shown: a, b, c, and d. Lines a and b represent evolutionary dynamics with mutation events only, whereas lines c and d represent evolutionary dynamics with dominant recombination events (nr/nm = 1,000). Lines a and c represent evolutionary dynamics under the initial condition that every member in the population is chosen randomly from the set of sequences compatible with the target structure, whereas lines b and d represent evolutionary dynamics under the initial condition that all members in the population are identical to a sequence chosen randomly from the set of sequences compatible with the target structure. Dashed lines represent the values for the prototype sequence.
Ratio of Recombination to Mutation Rates Determines How Optimal Protein Sequences Are for Their Structure.
In Fig. 4 we show how different steady-state ensemble-averaged properties change with the ratio of recombination to mutation rates. A minimal mutation rate is required to ensure that convergence to steady state is independent of initial conditions. As the ratio of recombination to mutation rates goes up, the spread of population in sequence space decreases gradually, and a typical sequence becomes thermodynamically and mutationally more stable for the target structure. The specific evolutionary history, especially the ratio of recombination to mutation rates, is a major determinant of the optimality of protein sequence–structure relationship.
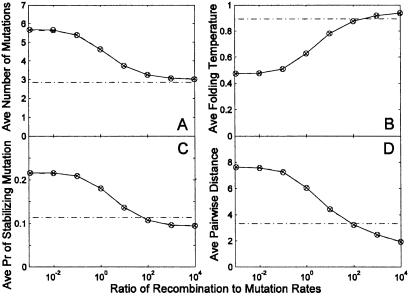
Fig 4.
For the structure shown in Fig. 1B, the dependence of evolutionary converged ensemble-averaged quantities on the ratio of recombination to mutation rates, is calculated using two different initial conditions as outlined in Fig. 3, and represented as circles and x-marks, respectively. The almost complete overlap of the circles and x-marks indicates evolutionary convergence independent of initial conditions. The x axis is the ratio of recombination to mutation rates on a logarithmic scale. The y axis is four different steady-state ensemble-averaged quantities: (A) d; (B) Tf; (C) ps; and (D) average pairwise distance between sequences in the population. The dash-dot line represents these quantities for evolution with recombination but no mutation computed for the initial condition where the population is a random set of sequences compatible with the target structure. The higher ratio of recombination to mutation rate, the more evolution pushes the sequence population toward optimality for the structure. Sequence evolution is closest to optimal when mutation rate is small compared with recombination rate, but not zero.
A Comprehensive Survey of All Neutral Networks and Structures.
To test the generality of our conclusions, we simulate evolutionary behavior with mutation events alone for all 2,977 neutral networks, and with dominant recombination events (nr/nm = 1,000, N = 10,000) for all structures that show convergent evolutionary behavior. Fig. 5 A–C shows the evolution of average thermodynamic stability measured by Tf. Two general trends emerge: first, evolutionary steady state is convergent and does not depend on the initial conditions; second, recombination is much more effective than mutation to push the sequence population toward the prototype sequence. Deviations from these general trends are infrequent and occur mostly for structures with small designability and unusual or competing neutral network topologies. These general trends depend on the special topology that most neutral networks for protein thermodynamics share: an obvious center sequence and a partition between sequences located near the center and those located near the boundary.
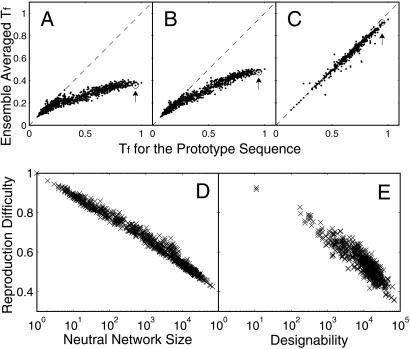
Fig 5.
How does evolution affect protein thermodynamics for all neutral networks and structures. (A and B) Average Tf in sequence population before (A) and after (B) selection by evolution with mutation events only vs. Tf for the prototype sequence, for all 2,977 neutral networks. (C) Average Tf in sequence population after selection by evolution with dominant recombination events (nr/nm = 1,000, N = 10,000) vs. Tf for the prototype sequence, for 638 of a total of 1,081 structures that converge after 100 mutations per individual. Most of the remaining structures that do not converge have small designability (70% of them have designability less than 10,000), and many of them have at least two competing neutral networks of similar size. These structures are not taken into account here. Arrows mark the prototype sequence. (D and E) Reproduction difficulty, nl/nm, is shown to depend primarily on neutral network size/designability for all neutral networks/structures, both in the case with mutation events only (D), and in the case with dominant recombination events (nr/nm = 1,000, N = 10,000) (E). The x axis is neutral network size/designability on a logarithmic scale. Little correlation exists with the detailed neutral network topology. The structure with the highest designability (and also the largest neutral network), depicted in Fig. 1B, has the lowest reproduction difficulty, regardless of the ratio of recombination to mutation rates. In the long run this structure will dominate the population in the absence of additional functional selection.
At evolutionary steady state, the ratio of lethality rate to mutation rate, nl/nm, is a measure of reproduction difficulty. For a sufficiently large sequence pool with arbitrary initial population distributions, the structure with the lowest reproduction difficulty will dominate the population after sufficiently long evolutionary steps. As shown in Fig. 5 D and E, reproduction difficulty correlates with the logarithm of neutral network size in a primarily linear way, regardless of the specific evolutionary mechanism. The structure with the largest designability and the largest neutral network (depicted in Fig. 1B) is also evolutionarily most successful. This observation agrees with the hypothesis that naturally occurring protein folds have high designability (24), and provides a rationale for using the highest designable structure as the target structure to study protein thermodynamics earlier in this paper.
Discussion
What Is the Ratio of Recombination to Mutation Rates Within a Gene?
The ratio, nr/nm, is the most important factor in determining how optimal protein sequences are for their structures. Mutation rates vary widely among species: the rate of spontaneous mutation per genome per replication is ≈0.003 in microbes with DNA chromosomes such as Escherichia coli and Saccharomyces cerevisiae, 0.03–0.43 in retroviruses and retrotransposons, and 0.018–0.49 for higher eukaryotes (26). Spontaneous mutations occur throughout the genome, but are concentrated at hotspots not correlated with gene structures (27).
Recent studies have shown considerable variation in the rates and impact of recombination in different bacteria species (28). Recombination events can occur between genes (intergenic) or within genes (intragenic), but only the latter contribute to protein sequence–structure compatibility. In bacteria, virtually all recombination occurs within genes, and nr/nm is found to be ≈50 in E. coli (29) and 24–100 in Neisseria meningitidis, Streptococcus pneumoniae, and Staphylococcus aureus (28). In yeast, meiotic recombination is initiated by double-strand DNA breaks. In three separate studies, 18 of 18 (30), 20 of the 22 (31), and 70 of 76 (32) mapped double-strand DNA break sites are located within intergenic regions. Because 70% of the yeast genome is covered by genes, this suggests a preference for intergenic versus intragenic recombination of 30:1. Assuming that the recombination rate is 100 per genome per generation, nr/nm is at most 1,000 for yeast. In higher eukaryotes, the genome is considerably longer and the mutation rate per genome is much higher, but the extensive insertion of introns also increases the intragenic recombination rate per genome. The combined result is that nr/nm for higher eukaryotes are probably similar to that for yeast. These rough estimates vary widely over time, between species, and even within a genome, and cannot be directly measured for ancestral species that are long extinct but crucial to the evolution of protein families.
Experimental Results on Protein Stability.
Many experiments suggest that protein sequences are thermodynamically and mutationally close to optimal for their structures, but it is possible to improve native stability by protein design. First, most single mutations are destabilizing. Of 129 single alanine mutations at each of the nonalanine positions of staphylococcal nuclease, 82% decreases stability (33–35). Eighty percent of 51 single alanine mutations at each of the nonalanine positions in the wild-type Arc repressor sequence decreases stability (36). Native proteins have a well-packed stable hydrophobic core, but a fraction of buried residues are hydrophilic, especially for large proteins (37). Second, native proteins are more stable than necessary for proper folding. Among 12 heavily mutated and properly folded variants of protein L, all have lower stability than wild type (38). Third, computational protein design that optimizes native protein stability generally yields a sequence population that is similar, but not identical to that observed in Nature (39, 40). It is also possible to construct a protein that is more stable than the native protein (41).
Linking Protein Sequence–Structure Compatibility to Evolution.
Our simulation connects a protein's thermodynamic and mutational stability with its evolutionary history, in particular the ratio of recombination to mutation rates within the gene. Proteins evolved with a high ratio are close to optimal for their native structures. Proteins evolved with a low ratio are marginally compatible with their native structures. This effect will be most pronounced when the protein fold is highly designable. Our hypothesis can be tested by in vitro protein evolution with controllable mutation and recombination rates, and by studying distributions of sequences for the same protein fold in different organisms. We predict that the spread of the distribution will correlate with the relative rates of recombination and mutation.
A “Levinthal Paradox” for Neutral Evolution of Sequences and Its Resolution Through Recombination, Not Mutation Alone.
The Levinthal paradox for protein folding involves the observation that insufficient time is available to search randomly the entire conformational space available to an unfolded protein (42). Rather, protein folding is more like a directed walk down a folding funnel that explores a small fraction of the conformational space before reaching the native conformational state (43–46). Similarly, a “Levinthal paradox” exists for neutral evolution of sequences: given the exponential size of the sequence space, how does evolution find the optimal sequence in a reasonable amount of time when the fitness landscape is flat? This paradox is also known as the Hoyle Paradox (47). Sometimes evolution fails to solve this problem: when mutation rate is dominant, even though the prototype sequence is preferred by evolution, sequence entropy still dominates, and most steady-state sequences are far away from the prototype sequence. Other times evolution solves this problem effectively: When recombination rate is dominant and mutation rate is minimal but non-zero, sequence population evolves by exploring a small fraction of sequence space before converging to the prototype sequence. The mechanism here is different from that in protein folding; in protein folding, each protein molecule folds up and solves the Levinthal paradox alone, whereas in neutral evolution a sequence that acts alone by means of mutation will almost never find the optimal sequence. Rather, the sequence population needs to act concertedly by recombination to home in to the prototype sequence. A nonzero mutation rate is crucial; without mutations, novel variations cannot be introduced into the sequence population, and evolution is dependent on the initial conditions.
It is tempting to compare the role of the ratio of recombination to mutation rates in sequence evolution to the role of temperature in protein folding. In protein folding, unfolded states are favored at high temperature, and the native state is favored at low temperature. However, when the temperature is too low, protein folding is trapped at a local minimum. In sequence evolution, sequence diversity is favored when the nr/nm ratio is low, and the prototype sequence is favored when it is high. However, when nr/nm approaches infinity (nm is zero), evolution can be trapped at a local minimum. Thus, this ratio is like 1/temperature. Nature is able to adjust the “temperature” of evolution by tuning the relative rates of recombination and mutation.
Limitations of the Model.
Four assumptions are made in our study: (i) many protein sequences share the same fold; (ii) minimal thermodynamic stability is the primary determinant of protein activity; (iii) hydrophobic interactions are the dominant force in protein folding; and (iv) effects of small populations, changing environments, and finite evolutionary time can be ignored. Assumption ii may be problematic for systems in which protein function is strongly correlated with thermodynamic stability. Our model serves as a general framework to understand evolution of protein thermodynamics; system-specific details can be added to provide a more accurate description whenever necessary.
Even though we use a two-dimensional model for protein structures, we believe that our conclusions do not depend on the dimensionality of the model used. First, protein thermodynamic and mutational properties, determined by the sum over all states, are less affected by dimensionality than kinetic properties. Second, our conclusions remain true when other types of protein models are considered, in particular, for a dimensionless model where structure is described by a string of residue states that define the local environment, and the energy only depends on having the right amino acid in the right environment (data not shown).
Implications for Protein Structure Prediction and Sequence Design.
Significant progress has been made in the past several years but much more needs to be done in the field of ab initio protein structure prediction (48–51). Our study explains a way to improve protein structure prediction. The prototype sequence is the most robust representation of the structure in sequence space; when a sequence is far away from the prototype sequence, a small inaccuracy in energy function or move set will fold the sequence into a wrong structure. We see previously that a sequence profile derived from multiple sequence alignments matches the prototype sequence well. This implies that information about multiple homologous sequences can help improve protein structure prediction as shown for both secondary structure prediction (52) and tertiary structure prediction (53–55).
Our study also has implications for sequence design. First, evolution is not a global optimizer but a dynamic process, and the native sequence need not be optimal for the structure. Therefore, care should be taken to calibrate sequence design procedures by comparing designed and native sequences. Second, multiple sequence alignments of remote homologues reveal the sequence distribution for the protein fold as a result of balancing forces in evolution, as compared with sequence population derived by sequence design through global optimization. A comparison of the two will provide deeper insights into protein evolution.
Acknowledgments
We are grateful to the editor and the reviewers for their help with the manuscript. We thank Marcus Feldman, Erik Sandelin, and Patrice Koehl for helpful discussions. This work is supported by National Institutes of Health Grant GM63817 and a Defense University Research Instrumentation Program equipment grant (DAAD- 1901-1-0403).
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wright S. (1932) in Proceedings of the Sixth International Congress on Genetics, ed. Jones, D. F. (Brooklyn Botanic Gardens, New York), Vol. 1, pp. 356–366. [Google Scholar]
- 2.Maynard Smith J. (1970) Nature (London) 225, 563-564.5411867 [Google Scholar]
- 3.Eigen M. (1971) Naturwissenschaften 58, 465-523. [DOI] [PubMed] [Google Scholar]
- 4.Fontana W. & Schuster, P. (1998) Science 280, 1451-1455. [DOI] [PubMed] [Google Scholar]
- 5.Bryngelson J. D. & Wolynes, P. G. (1987) Proc. Natl. Acad. Sci. USA 84, 7524-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dill K. A., Bromberg, S., Yue, K. Z., Fiebig, K. M., Yee, D. P., Thomas, P. D. & Chan, H. S. (1995) Protein Sci. 4, 561-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirny L. & Shakhnovich, E. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 361-396. [DOI] [PubMed] [Google Scholar]
- 8.Dill K. A. (1990) Biochemistry 29, 7133-7155. [DOI] [PubMed] [Google Scholar]
- 9.Govindarajan S. & Goldstein, R. A. (1997) Proteins Struct. Funct. Genet. 29, 461-466. [DOI] [PubMed] [Google Scholar]
- 10.Kaffe-Abramovich T. & Unger, R. (1998) Folding Des. 3, 389-399. [DOI] [PubMed] [Google Scholar]
- 11.Taverna D. M. & Goldstein, R. A. (2000) Biopolymers 53, 1-8. [DOI] [PubMed] [Google Scholar]
- 12.Tiana G., Broglia, R. A. & Shakhnovich, E. I. (2000) Proteins Struct. Funct. Genet. 39, 244-251. [PubMed] [Google Scholar]
- 13.Taverna D. M. & Goldstein, R. A. (2002) J. Mol. Biol. 315, 479-484. [DOI] [PubMed] [Google Scholar]
- 14.Taverna D. M. & Goldstein, R. A. (2002) Proteins Struct. Funct. Genet. 46, 105-109. [DOI] [PubMed] [Google Scholar]
- 15.Cui Y., Wong, W. H., Bornberg-Bauer, E. & Chan, H. S. (2002) Proc. Natl. Acad. Sci. USA 99, 809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voigt C. A., Mayo, S. L., Arnold, F. H. & Wang, Z. G. (2001) Proc. Natl. Acad. Sci. USA 98, 3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura M. (1991) Jpn. J. Genet. 66, 367-386. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M., (1983) The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 19.Ohta T. (1973) Nature (London) 246, 96-98. [DOI] [PubMed] [Google Scholar]
- 20.Levitan B. & Kauffman, S. (1995) Mol. Divers. 1, 53-68. [DOI] [PubMed] [Google Scholar]
- 21.van Nimwegen E., Crutchfield, J. P. & Huynen, M. (1999) Proc. Natl. Acad. Sci. USA 96, 9716-9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bastolla U., Roman, H. E. & Vendruscolo, M. (1999) J. Theor. Biol. 200, 49-64. [DOI] [PubMed] [Google Scholar]
- 23.Bornberg-Bauer E. & Chan, H. S. (1999) Proc. Natl. Acad. Sci. USA 96, 10689-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Helling, R., Tang, C. & Wingreen, N. (1996) Science 273, 666-669. [DOI] [PubMed] [Google Scholar]
- 25.Golub G. H. & Van Loan, C. F., (1989) Matrix Computations (Johns Hopkins Univ. Press, Baltimore).
- 26.Drake J. W., Charlesworth, B., Charlesworth, D. & Crow, J. F. (1998) Genetics 148, 1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewin B., (2000) Genes VII (Oxford Univ. Press, New York).
- 28.Feil E. J. & Spratt, B. G. (2001) Annu. Rev. Microbiol. 55, 561-590. [DOI] [PubMed] [Google Scholar]
- 29.Guttman D. S. & Dykhuizen, D. E. (1994) Science 266, 1380-1383. [DOI] [PubMed] [Google Scholar]
- 30.Wu T.-C. & Lichten, M. (1994) Science 263, 515-518. [DOI] [PubMed] [Google Scholar]
- 31.Gerton J. L., DeRisi, J., Shroff, R., Lichten, M., Brown, P. O. & Petes, T. D. (2000) Proc. Natl. Acad. Sci. USA 97, 11383-11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudat F. & Nicolas, A. (1997) Proc. Natl. Acad. Sci. USA 94, 5213-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shortle D., Stites, W. E. & Meeker, A. K. (1990) Biochemistry 29, 8033-8041. [DOI] [PubMed] [Google Scholar]
- 34.Green S. M., Meeker, A. K. & Shortle, D. (1992) Biochemistry 31, 5717-5728. [DOI] [PubMed] [Google Scholar]
- 35.Meeker A. K., Garcia-Moreno, B. E. & Shortle, D. (1996) Biochemistry 35, 6443-6449. [DOI] [PubMed] [Google Scholar]
- 36.Milla M. E., Brown, B. M. & Sauer, R. T. (1994) Nat. Struct. Biol. 1, 518-523. [DOI] [PubMed] [Google Scholar]
- 37.Kajander T., Kahn, P. C., Passila, S. H., Cohen, D. C., Lehtiö, L., Adolfsen, W., Warwicker, J., Schell, U. & Goldman, A. (2000) Structure (London) 8, 1203-1214. [DOI] [PubMed] [Google Scholar]
- 38.Kim D. E., Gu, H. & Baker, D. (1998) Proc. Natl. Acad. Sci. USA 95, 4982-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koehl P. & Levitt, M. (1999) J. Mol. Biol. 293, 1183-1193. [DOI] [PubMed] [Google Scholar]
- 40.Kuhlman B. & Baker, D. (2000) Proc. Natl. Acad. Sci. USA 97, 10383-10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikolova P. V., Henckel, J., Lane, D. P. & Fersht, A. R. (1998) Proc. Natl. Acad. Sci. USA 95, 14675-14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levinthal C. (1968) J. Chim. Phys. 65, 44-45. [Google Scholar]
- 43.Wolynes P. G., Onuchic, J. N. & Thirumalai, D. (1995) Science 267, 1619-1620. [DOI] [PubMed] [Google Scholar]
- 44.Dill K. A. & Chan, H. S. (1997) Nat. Struct. Biol. 4, 10-19. [DOI] [PubMed] [Google Scholar]
- 45.Pande V. S., Grosberg, A. Y., Tanaka, T. & Rokhsar, D. S. (1998) Curr. Opin. Struct. Biol. 8, 68-79. [DOI] [PubMed] [Google Scholar]
- 46.Dobson C. M. & Karplus, M. (1999) Curr. Opin. Struct. Biol. 1999, 92-101. [DOI] [PubMed] [Google Scholar]
- 47.Wolynes P. G. (1995) in Proceedings of Symposium on Distance-Based Approaches to Protein Structure Determination II, eds. Bohr, H. & Brunak, S. (CRC, Boca Raton, FL), pp. 3–17.
- 48.Simons K. T., Strauss, C. & Baker, D. (2000) J. Mol. Biol. 306, 1191-1199. [DOI] [PubMed] [Google Scholar]
- 49.Xia Y., Huang, E. S., Levitt, M. & Samudrala, R. (2000) J. Mol. Biol. 300, 171-185. [DOI] [PubMed] [Google Scholar]
- 50.Ortiz A. R., Kolinski, A., Rotkiewicz, P., Ilkowski, B. & Skolnick, J. (1999) Proteins Struct. Funct. Genet. S3, 177-185. [DOI] [PubMed] [Google Scholar]
- 51.Liwo A., Lee, J., Ripoll, D. R., Pillardy, J. & Scheraga, H. A. (1999) Proc. Natl. Acad. Sci. USA 96, 5482-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rost B. & Sander, C. (1993) J. Mol. Biol. 232, 584-599. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein R. A., Luthey-Schulten, Z. A. & Wolynes, P. G. (1992) Proc. Natl. Acad. Sci. USA 89, 4918-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keasar C., Tobi, D., Elber, R. & Skolnick, J. (1998) Proc. Natl. Acad. Sci. USA 95, 5880-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonneau R., Strauss, C. E. M. & Baker, D. (2001) Proteins Struct. Funct. Genet. 43, 1-11. [DOI] [PubMed] [Google Scholar]