Abstract
Aquatic photosynthetic organisms live in quite variable conditions of CO2 availability. To survive in limiting CO2 conditions, Chlamydomonas reinhardtii and other microalgae show adaptive changes, such as induction of a CO2-concentrating mechanism, changes in cell organization, increased photorespiratory enzyme activity, induction of periplasmic carbonic anhydrase and specific polypeptides (mitochondrial carbonic anhydrases and putative chloroplast carrier proteins), and transient down-regulation in the synthesis of Rubisco. The signal for acclimation to limiting CO2 in C. reinhardtii is unidentified, and it is not known how they sense a change of CO2 level. The limiting CO2 signals must be transduced into the changes in gene expression observed during acclimation, so mutational analyses should be helpful for investigating the signal transduction pathway for low CO2 acclimation. Eight independently isolated mutants of C. reinhardtii that require high CO2 for photoautotrophic growth were tested by complementation group analysis. These mutants are likely to be defective in some aspects of the acclimation to low CO2 because they differ from wild type in their growth and in the expression patterns of five low CO2-inducible genes (Cah1, Mca1, Mca2, Ccp1, and Ccp2). Two of the new mutants formed a single complementation group along with the previously described mutant cia-5, which appears to be defective in the signal transduction pathway for low CO2 acclimation. The other mutations represent six additional, independent complementation groups.
Acclimation to changed environmental conditions is a key to survival for all organisms. In response to perceived environmental signals, organisms may exhibit specific adaptive changes, such as changes in the expression of key genes to survive specific environmental changes. Because CO2 can vary substantially in aquatic habitats and represents the major substrate for photosynthetic CO2 fixation via the enzyme Rubisco, CO2 concentration is an important environmental signal in aquatic photosynthetic organisms including cyanobacteria and Chlamydomonas reinhardtii.
Unlike terrestrial higher plants, aquatic photosynthetic organisms can face difficulties in acquiring CO2. Because the CO2 diffusion rate in water is much slower than that in air (Badger and Spalding, 2000), the CO2 supply to Rubisco in these aquatic photosynthetic organisms can become limited. C. reinhardtii and other aquatic photosynthetic organisms have a genetic program to allow them to acclimate to low CO2. This acclimation includes induction of a CO2-concentrating mechanism (CCM) that allows the cells to acquire CO2 efficiently by increasing the CO2 concentration around Rubisco under limiting CO2 conditions (Badger et al., 1980; for review, see Spalding, 1998; Kaplan and Reinhold, 1999).
Along with the induction of the CCM, C. reinhardtii shows adaptive changes to limiting CO2 conditions, such as changes in cell organization (Geraghty and Spalding, 1996), increased photorespiratory enzyme activity (Marek and Spalding, 1991), induction of periplasmic carbonic anhydrase (CA) (pCA1, encoded by the Cah1 gene; Fujiwara et al., 1990; Fukuzawa et al., 1990; Ishida et al., 1993), mitochondrial CA (mtCA, encoded by the Mca1 and Mca2 genes; Eriksson et al., 1996; Geraghty and Spalding, 1996), and putative chloroplast carrier protein (Ccp, encoded by the Ccp1 and Ccp2 genes; Geraghty et al., 1990; Ramazanov et al., 1993; Chen et al., 1997), and transient down-regulation in the synthesis of Rubisco (Coleman and Grossman, 1984; Winder et al., 1992).
The signal for acclimation to limiting CO2 in C. reinhardtii is unidentified. It is not known how they sense a change of CO2 availability, whether by CO2 concentration directly or indirectly via a cellular process such as carbohydrate metabolism. Whatever the limiting-CO2 signal, it must be transduced into the changes in gene expression observed during acclimation, such as expression of Cah1. A powerful way to identify components of the CCM and of the signal transduction pathway for low CO2 acclimation is through the analysis and characterization of mutants specifically defective in growth in limiting CO2, like the ca-1, pmp-1, and cia-5 mutants (Spalding et al., 1983a; 1983b; Moroney et al., 1989). Using advances in nuclear transformation of C. reinhardtii (Kindle, 1990), a collection of insertionally generated high CO2-requiring (HCR) mutants unable to grow in limiting CO2 was obtained and is described here.
RESULTS
Generation and Isolation of Mutants
Using glass bead transformation (Kindle, 1990; Davies et al., 1994), CC425 (Table I) was complemented by transformation with p-Arg7.8 (Debuchy et al., 1989) to generate a pool of insertional mutants on CO2-minimal medium. Cells from each of more than 7,000 transformant colonies were suspended in air-minimal medium and grown on plates in high CO2 (5% [v/v] CO2 in air), normal air, and low CO2 (50–100 μL L−1 CO2). HCR mutants, defined as those showing little or no growth either in normal air or in low CO2, should include mutants, like cia-5, that are defective in acclimation to limiting CO2, as well as those with functional defects in the CCM. Sixteen putative HCR mutants were identified, and eight of those are described here (Table II).
Table I.
Strains of C. reinhardtii used in the study
| Strain | Genotype | Description | Reference |
|---|---|---|---|
| ars301 | cw15 sr-u-2-60 mt+ | Generated by transformation of CC425 with Arg7; used as wild type in liquid growth experiments | Provided by John P. Davies (Exelixis, Inc., South San Francisco) |
| CC124 | mt− | Wild type (137C) | Harris (1989) |
| CC425 | arg2 cw15 sr-u-2-60 mt+ | Cell wall-less, Arg-requiring, and streptomycin-resistant mutant | Harris (1989) |
| CC849 | cw10 mt− | Cell wall-less mutant used as wild type in RNA analyses | Harris (1989) |
| CC1068 | arg2 nr-u-2-1 mt− | Arg-requiring and kanamycin-resistant mutant | Harris (1989) |
| CC2702 | cia-5 mt+ | No acclimation to limiting CO2 | Moroney et al. (1989); Spalding et al. (1991) |
| cia-5 mt− | Generated by CC2702 × CC124 | This report | |
| CC1219 | ca1-1 mt+ | Defective in Cah3, thylakoid lumen CA | Spalding et al. (1983a); Funke et al. (1997); Karlsson et al. (1998) |
| ca1-1 mt− | Generated by CC1219 × CC801 | Provided by Kensaku Suzuki (Tohoku National Agri Research, Morioka, Japan) | |
| CC1860 | pmp1-1 mt+ | Deficient in Ci transport | Spalding et al. (1983b) |
| pmp1-1 mt− | Generated by CC1860 × CC124 | This report | |
| CC2648 | pgp1-1 mt+ | Deficient in phosphoglycolate phosphatase | Suzuki et al. (1990) |
| pgp1-1 mt− | Generated by CC2648 × CC124 | This report | |
| HCRP34 mt− | Generated by HCRP34 × CC1068 | This report | |
| HCR209 mt− | Generated by HCR209 × CC1068 | This report | |
| HCR3510 mt− | Generated by HCR3510 × CC1068 | This report | |
| HCR86 mt− | Generated by HCR86 × CC1068 | This report | |
| HCR89 mt− | Generated by HCR89 × CC1068 | This report | |
| HCR90 mt− | Generated by HCR90 × CC1068 | This report |
Table II.
Characteristics of HCR mutants
| Mutants | High CO2Requirementa
|
|||||
|---|---|---|---|---|---|---|
| Air | Low CO2 | Arg7 Insertb | Vector Sequence Present | Cosegregation with Arg+ Phenotype and Arg7 Insert | Diploid Analysis | |
| cia-5 | Leakyc | Stringentc | NAd | NA | NA | Recessive |
| HCRP34 | Leaky | Stringent | 1 | +e | Yes | Recessive |
| HCR209 | Leaky | Stringent | 2 | + | NDf | Recessive |
| HCR3510 | Wild type | Stringent | 1 | + | Yes | Recessive |
| HCR86 | Stringent | Stringent | 1 | + | Yes | Recessive |
| HCR89 | Leaky | Leaky | 1 | − | Yes | Recessive |
| HCR90 | Leaky | Stringent | 1 | + | Yes | Recessive |
| HCR95 | Leaky | Leaky | 1 | + | No | Recessive |
| HCR105 | Stringent | Stringent | 1 | ND | No | Recessive |
Growth phenotype in low CO2 was determined by spot test on agar in high CO2 (5% [v/v] CO2 in air) versus either air (300–500 μL L−1 CO2) or low CO2 (50–100 μL L−1 CO2).
Arg7 insert detected by Southern analysis using 1.3-kb SalI fragment of Arg7 as probe.
Leaky, Cells grow very slowly; stringent, cells did not grow at all.
NA, Not applicable.
+/− Indicate the presence (+) or absence (−) of vector sequence.
ND, Not determined.
General Characteristics of HCR Mutants
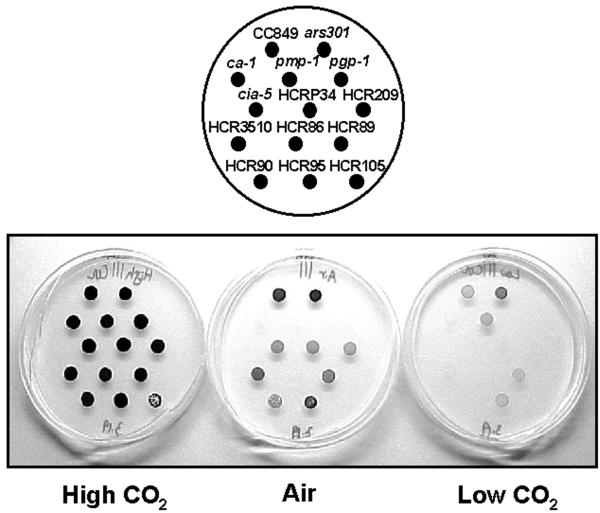
The eight HCR mutants and their general characteristics are shown in Table II and Figure 1. When grown in high CO2 on agar, all HCR mutants except HCR105 were indistinguishable from the wild type (Fig. 1). The eight HCR mutants could be divided into four groups based on their apparent high CO2 requirement for photoautotrophic growth. The first group, including HCRP34, HCR209, and HCR90, showed a leaky HCR phenotype in air but a stringent phenotype in low CO2. The second group, including HCR86 and HCR105, showed a stringent HCR phenotype both in air and in low CO2. HCR89 and HCR95, comprising the third group, had a leaky HCR phenotype both in air and in low CO2. HCR3510 lacked a significant growth phenotype in air but had a stringent phenotype in low CO2.
Figure 1.
Spot tests for growth response to different CO2 concentrations for wild-type strains (CC849 and ars301), four previously described HCR mutants (cia-5, ca-1, pmp-1, and pgp-1), and eight new HCR mutants. Plates were kept either at high CO2 (5% [v/v] CO2), at air level of CO2, or at low CO2 (50–100 μL L−1) for 10 d.
Genetic Characteristics of HCR Mutants
Seven of the eight HCR mutants were found by Southern analysis (data not shown) to contain only one copy of the Arg7 insert, and the presence of vector sequences was confirmed in six mutants (Table II). The presence of vector sequences provides an opportunity for the cloning of sequences flanking the insert by plasmid rescue (Quarmby and Hartzell, 1994).
Selected random progeny and/or tetrads from HCR mutants were tested in crosses with another arg2 mutant (CC1068, Table I) for linkage of the Arg insert with Arg+ and HCR phenotypes (Table II). Five of the eight mutants showed cosegregation of the single Arg insert with the HCR phenotype, suggesting that the Arg insert is responsible for the HCR phenotype in these five mutants. In two of the mutants, HCR95 and HCR105, the inserts did not cosegregate with the HCR phenotype, indicating that insertion of the Arg plasmid was not directly responsible for the HCR phenotype in these two mutants. In HCR209, which has two inserts, cosegregation crosses were not conclusive, but other evidence (see below) suggests the two inserts are tandemly arranged and are responsible for the phenotype.
Heterozygous vegetative diploids, generated in crosses with CC1068 and selected by their resistance to both kanamycin and streptomycin, were used to determine the dominant/recessive nature of the HCR phenotype of each mutant. Based on growth tests of the heterozygous diploids, the mutant phenotype of all eight HCR mutants was judged to be recessive.
Complementation Group Analysis
Crossing with the various known mutants such as cia-5, ca-1, pmp-1, and pgp-1 should help identify new alleles of previously characterized mutants. If any wild-type colonies appear under low CO2 conditions (50–100 μL L−1 CO2) after mating with HCR mutants, this indicates they are not allelic to each other, because these known mutants also show HCR phenotypes.
Rapid allelism tests were used to place the various HCR mutants into different complementation groups. Complementation analysis was tested with the eight HCR mutants (Table II) along with cia-5, ca-1, pmp-1, and pgp-1 (Table I). Only crosses between cia-5 × HCRP34, cia-5 × HCR209, and HCRP34 × HCR209 failed to generate wild type colonies. Thus, HCR3510, HCR86, HCR89, HCR90, HCR95, and HCR105 each define a new HCR locus. HCRP34 and HCR209 have been confirmed as defective in the same locus as cia-5 by comparison of the sequence of the DNA flanking the inserts with a cloned cia-5 gene (Xiang et al., 2001) and by complementation with a cloned cia-5 gene (data not shown).
Liquid Growth Experiments
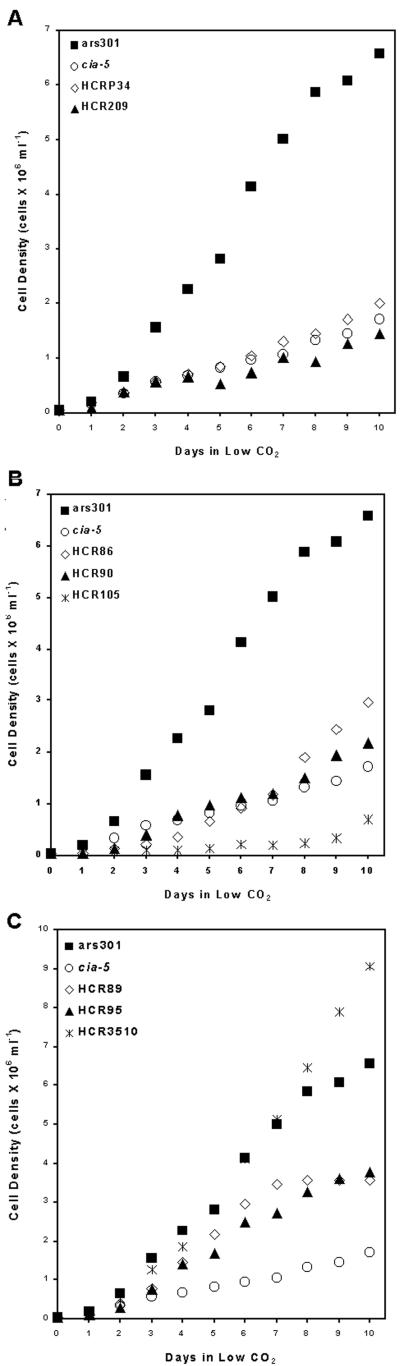
Growth experiments showed patterns of high CO2 requirement for photoautotrophic growth consistent with those seen in spot tests (Fig. 1). Active, 1-d-old air-adapted cells were inoculated into liquid minimal medium with similar starting cell densities (5 × 104 cells ml−1), grown with no aeration, and the cell densities measured daily at the same time of day for 10 d. HCRP34 and HCR209, judged to be allelic to cia-5, grew very similar to cia-5 in air (Fig. 2A). The growth rates of HCR86 and HCR90 also were only slightly better than that of cia-5 in air (Fig. 2B), but the growth rates of HCR89 and HCR95 were intermediate between wild type (ars301; see Table I) and cia-5 (Fig. 2C). HCR105 was able to grow slightly in air but bleached within a few days (Fig. 2B). HCR3510, which showed a wild-type phenotype in air on agar, also grew as well as wild type (ars301) in air in liquid culture (Fig. 2C). Chlorophyll content also was measured in these cultures along with cell density, and the growth curves based on chlorophyll content showed the same pattern as those of cell density (data not shown).
Figure 2.
Liquid cell growth curves for wild type (ars301), cia-5, and HCR mutants grown at pH 7 on an orbital shaker without aeration. A, HCRP34 and HCR209. B, HCR86, HCR90, and HCR105. C, HCR89, HCR95, and HCR3510. The growth curves shown are averages of three independent growth experiments.
Accumulation of Low CO2-Inducible Transcripts
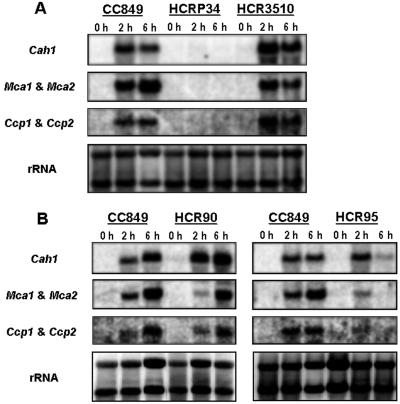
Because the expression of low CO2-inducible polypeptides (pCA1, mtCA1, mtCA2, Ccp1, and Ccp2) has been reported to change differentially during acclimation to limiting CO2 (Villarejo et al., 1996, 1997; Eriksson et al., 1998), accumulation of these three transcripts also was analyzed. The cia-5-like mutants, HCRP34 and HCR209, showed no detectable Cah1 mRNA, Mca1 and Mca2 mRNA, and Ccp1 and Ccp2 mRNA (Fig. 3A; data shown only for HCRP34). HCR90, which showed a leaky HCR phenotype in air but a stringent phenotype in low CO2, had reduced expression of only Mca1 and Mca2 mRNA (Fig. 3B). In separate, long-term experiments, the expression of the other genes was somewhat variable, but only Mca1 and Mca2 showed reproducibly decreased mRNA abundance (data not shown). HCR3510, which showed a wild-type phenotype in air but a stringent HCR phenotype in low CO2, had normal expression of these genes compared with wild type (CC849; see Table I; Fig. 3A). However, HCR95 showed a much different pattern of expression for these three genes. From cells exposed for 2 h to air, Cah1 mRNA of HCR95 was detected at normal levels, whereas much-reduced levels of Mca1 and Mca2 mRNA and Ccp1 and Ccp2 mRNA were detected relative to wild type (Fig. 3B). After 6 h, wild type showed the same or increased levels of these three mRNAs, but expression of all three mRNA in HCR95 was dramatically reduced (Fig. 3B), suggesting only a transient induction of their expression in this mutant. In separate, long-term experiments, this apparent transient induction in HCR95 also was confirmed up to 24 h (data not shown). The other HCR mutants (HCR86, HCR89, and HCR105) did not show reproducibly different patterns of expression for the three low CO2-inducible transcripts relative to wild type (data not shown).
Figure 3.
Northern-blot analyses for wild type (CC849) and HCR mutants. A, HCRP34 and HCR3510. B, HCR90 and HCR95. Total RNA (10 μg per lane) was isolated 2 h to 6 h after transfer of cells to air levels of CO2 from high CO2. Cah1 mRNA was probed with the 1.4-kb BglII and NcoI fragment of Cah1 cDNA (Van and Spalding, 1999). Mca1 and Mca2 mRNA was probed with the full-length Mca2 cDNA (Eriksson et al., 1996, 1998). Ccp1 and Ccp2 mRNA was probed with the 1.2-kb EcoRI and HindIII fragment of Ccp1 G1 (Chen et al., 1997). The rRNA was probed with 25S and 5.8S rDNA (Marco and Rochaix, 1980).
DISCUSSION
HCR mutants have been useful for investigation of various processes, both in algae and in higher plants. HCR mutants with defects in several of the enzymes of the photorespiratory pathway have been isolated in the C3 plants Arabidopsis (Somerville and Ogren, 1982) and barley (Hordeum vulgare) (Joy et al., 1992; Leegood et al., 1996; Wingler et al., 1999). These photorespiratory mutants exhibited lethality (HCR phenotype) in air levels of CO2 for various reasons, including accumulation of toxic intermediates during photorespiration and depletion of exchangeable nitrogen in photorespiratory intermediates. In C. reinhardtii, the photorespiratory mutant pgp-1 (lacks PGPase) has a HCR phenotype, indicating that the oxygenase activity of Rubisco was not completely suppressed by operation of the CCM and that photorespiratory mutants in C. reinhardtii also are lethal in air levels of CO2 (Suzuki et al., 1990; Spalding, 1998).
Mutants defective in functional components of the CCM also exhibit an HCR phenotype in C. reinhardtii (Spalding et al., 1983a, 1983b; Moroney et al., 1986; Suzuki and Spalding, 1989; Funke et al., 1997; Karlsson et al., 1998) and cyanobacteria (Price and Badger, 1989; Ogawa, 1991, 1992; Marco et al., 1993; Ohkawa et al., 1998; Price et al., 1998). Isolation and characterization of the C. reinhardtii mutants, ca-1 and pmp-1, demonstrated the requirement for active transport and accumulation of Ci (Badger et al., 1980; Spalding et al., 1983b) and for a thylakoid lumen CA (Spalding et al., 1983a; Funke et al., 1997; Karlsson et al., 1998) for function of the CCM. Another C. reinhardtii HCR mutant, cia-5, exhibits no apparent low-CO2 acclimation responses, such as induction of CCM, up-regulation of low CO2-inducible polypeptides, up-regulation of photorespiratory enzymes, or down-regulation of Rubisco biosynthesis (Moroney et al., 1989; Marek and Spalding, 1991; Spalding et al., 1991; Burow et al., 1996). This mutant is thought to be defective in the signal transduction pathway for acclimation to limiting CO2. The gene responsible for this mutation (Cia5) has been cloned recently (Fukuzawa et al., 2001; Xiang et al., 2001), and its characterization suggests it may encode a transcription factor. The identification of this important gene opens the way for more rapid progress in delineation of the signal transduction pathway for acclimation to limiting CO2.
Because many changes involved in acclimation to limiting CO2 conditions appear to be controlled at different gene expression levels, it is possible that mutations in several different loci might yield signal transduction mutants like cia-5 with HCR phenotypes. Thus, the HCR phenotype should be a good indicator of nonacclimation to low CO2 as well as for a dysfunctional CCM, so isolation of HCR mutants should be helpful for identification of loci required for either function of the CCM or for signal transduction leading to low CO2 acclimation.
Among the eight new HCR mutants described here, six represent new complementation groups and the other two represent new alleles of the previously described cia-5 locus. The patterns of growth and of low CO2-inducible transcript accumulation for HCRP34 and HCR209 were similar to those of cia-5, and complementation group analyses confirmed that the three are allelic. As new alleles of cia-5, HCRP34 and HCR209 may prove valuable in understanding the function of the gene product from this important locus.
Other than for HCRP34 and HCR209, the growth responses to air and low CO2 varied among these new HCR mutants, as did the pattern of accumulation of limiting-CO2-inducible genes. HCR90, which showed a stringent HCR phenotype in low CO2 and grew only slightly better than cia-5 in air (Fig. 2B), had reproducibly reduced expression of only one pair of the limiting-CO2-inducible transcripts, Mca1 and Mca2. No disruption of the structural gene for either Mca1 or Mca2 was found in genomic Southern blots probed with the Mca1 and Mca2 promoter region (data not shown), so HCR90 may be defective in a regulatory component that preferentially affects expression of Mca1 and Mca2. HCR86, which has a growth phenotype very similar to HCR90, showed limiting-CO2-inducible transcripts accumulations that were not reproducibly different from those of wild type (data not shown). The leaky phenotype in low CO2 of HCR89 and HCR95 was supported by their growth patterns (Fig. 2C), but only HCR95 reproducibly showed reduced level of low CO2-inducible transcripts (Fig. 3B).
HCR3510 showed no significant differences from wild type in terms of low CO2-inducible transcript accumulation, suggesting it is unlikely to be defective in the limiting-CO2-responsive signal transduction pathway. The growth phenotype of this mutant, near wild-type growth in normal air but a stringent phenotype in low CO2, suggests a defect in a functional component of the CCM (or another pathway required for acclimation to limiting CO2) that is essential in very low CO2 but not in air levels of CO2.
The advantage of using insertional mutagenesis to generate mutants lies in the use of the inserted DNA as a “tag” to clone the disrupted gene, but of course this only works if the insert cosegregates with the mutant phenotype, i.e. if the insert is responsible for the mutation. As judged by the Arg+ phenotype, the Arg7 inserts in mutants HCRP34, HCR3510, HCR86, HCR89, and HCR90 cosegregate with the HCR phenotype (Table II), suggesting the insert caused the mutation in each of these strains. As indicated above, both HCRP34 and HCR209 are allelic to cia-5 and the insert in each has been confirmed to disrupt the cia-5 gene. Thus, we know the defect in both these mutants, even though cosegregation of the Arg+ and HCR phenotypes has not been demonstrated for HCR209.
It is unfortunate that the inserts in mutants HCR95 and HCR105 do not cosegregate with the HCR phenotype, so identification of the disrupted gene responsible for the HCR phenotype in these mutants will have to be accomplished without the aid of an insertional tag. The three remaining tagged mutants (HCR3510, HCR86, and HCR 90) remain as viable candidates for identification of novel genes essential for acclimation of C. reinhardtii to limiting CO2. Cloning of the disrupted genes in these three HCR mutants is in progress.
MATERIALS AND METHODS
Cell Strains and Culture Conditions
All Chlamydomonas reinhardtii strains (Table I) were grown as previously described (Geraghty et al., 1990). Cells were cultured on an orbital shaker under aeration with 5% (v/v) CO2 in air (high CO2-grown cells) or no aeration (air-adapted cells). For experiments monitoring the accumulation of low CO2 inducible transcripts, cell cultures were switched from aeration with 5% (v/v) CO2 to aeration with normal air for 2 h to 6 h. For growth on solid media, cells were maintained under 5% (v/v) CO2 in air (high CO2), normal air, or 50 to 100 μL L−1 CO2 (low CO2).
Generation and Isolation of Mutants
Glass bead transformations were performed as described previously (Van and Spalding, 1999). To generate a pool of insertional mutants on CO2-minimal medium, CC425 (Table I) was transformed with linearized p-Arg7.8 (Debuchy et al., 1989) containing the structural gene (Arg7) for argininosuccinate lyase to complement the arg2 mutation. Each of more than 7,000 colonies was screened by spot tests to identify HCR mutants. After replica plates with transformants were made, each plate was placed in high CO2 and air or high CO2 and low CO2. Mutants identified in this primary screen as having HCR phenotypes were screened again by western immunoblots of extracellular protein to identify mutants in which pCA1 expression was decreased or absent (Van and Spalding, 1999).
Spot Growth Tests and Growth Experiments
For spot growth tests, actively growing cells were suspended to similar cell densities in minimal medium, spotted (10 μL) onto minimal agar plates, and grown in different concentrations of CO2 for 10 d (Harris, 1989).
For liquid growth experiments, active, 1-d-old air-adapted cells were inoculated into liquid minimal medium at similar cell densities (5 × 104 cells ml−1). The cultures were grown on an orbital shaker without aeration for the next 10 d. The cell density was determined using a hemacytometer (Reichert Scientific Instruments, Buffalo, NY; Harris, 1989). Chlorophyll content was estimated after extraction with 96% (v/v) ethanol (Wintermans and De Mots, 1965).
DNA- and RNA-Blot Analysis
Southern- and northern-blot analyses were performed as described by Van and Spalding (1999). Total RNA was purified with TRIzol reagent (Life Technologies, Gaithersburg, MD) from air-induced cells exposed to limiting CO2 (aeration with normal air) and Hybond N+ nylon transfer membrane (Amersham Pharmacia Biotech Inc., Piscataway, NJ) was used for blotting. After phoporimager analysis of each northern blot (Molecular Dynamics, Piscataway, NJ), total RNA amounts were normalized to hybridization with 25S and 5.8S rRNA (Marco and Rochaix, 1980) using ImageQuaNT (Molecular Dynamics).
Genetic Analyses
All matings were performed by crossing insertionally generated mutants with various strains (Table I) according to the protocol of Harris (1989). To isolate vegetative diploids, gametes from HCR mutants (sr-u-2-60) and CC1068 (nr-u-2-1) were induced under nitrogen stress, mated, and the mating mixture spread onto kanamycin-containing medium to select for expression of the plastid-encoded kanamycin resistance (nr-u-2-1) transmitted from the mating-type minus parent. Putative diploids (surviving colonies) were verified by selection for simultaneous expression of the plastid-encoded streptomycin resistance (sr-u-2-60) from the mating-type plus parent and by DNA quantity in flow cytometry (performed at the Iowa State University Cell Facility, Ames).
Complementation group analyses required construction of mating type minus strains of each HCR mutant (both new and previously described mutants). Mating type minus strains of cia-5, ca-1, pmp-1, and pgp-1 were generated by crossing with CC124 (Table I). CC1068 (Table I) was used for generating mating type minus strains from all new HCR mutants, except HCR95 and HCR105. After crossing each of the seven new HCR mutants and the four known mutants with each other, the progeny from each cross were tested for photoautotrophic growth in low CO2 (50–100 μL L−1). Because all HCR mutants required elevated CO2 for survival, wild-type colonies were observed in low CO2 only if the cross generated wild-type recombinant progeny.
Footnotes
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant nos. 97–35100–4210 and 99–35100–7569 to M.H.S.). This is journal paper no. J–19297 of project no. 3578 of the Iowa Agriculture and Home Economics Experiment Station (Ames) and was supported by the Hatch Act and State of Iowa funds.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010333.
LITERATURE CITED
- Badger MR, Kaplan A, Berry JA. Internal inorganic carbon pool of Chlamydomonas reinhardtii: evidence for a CO2 concentrating mechanism. Plant Physiol. 1980;66:407–413. doi: 10.1104/pp.66.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Spalding MS. CO2 acquisition, concentration and fixation in cyanobacteria and algae. In: Leegood RC, Sharkey TD, von Caemmerer S, editors. Photosynthesis: Physiology and Metabolism. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 369–397. [Google Scholar]
- Burow MD, Chen Z-Y, Mouton TM, Moroney JV. Isolation of cDNA clones induced upon transfer of Chlamydomonas reinhardtii cells to low CO2. Plant Mol Biol. 1996;31:443–448. doi: 10.1007/BF00021807. [DOI] [PubMed] [Google Scholar]
- Chen Z-Y, Lavigne MD, Mason CB, Moroney JV. Cloning and overexpressing of two cDNAs encoding the low-CO2-inducible chloroplast envelope protein LIP-36 from Chlamydomonas reinhardtii. Plant Physiol. 1997;114:265–273. doi: 10.1104/pp.114.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Grossman AR. Biosynthesis of carbonic anhydrase in Chlamydomonas reinhardtii during adaptation to low CO2. Proc Natl Acad Sci USA. 1984;81:6049–6053. doi: 10.1073/pnas.81.19.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz F, Grossman AR. Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell. 1994;6:53–63. doi: 10.1105/tpc.6.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix JD. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989;8:2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Garderstrom P, Samuelsson G. Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1996;93:12031–12034. doi: 10.1073/pnas.93.21.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Villand P, Garderstrom P, Samuelsson G. Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 1998;116:637–641. doi: 10.1104/pp.116.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Fukuzawa H, Tachiki A, Miyachi S. Structure and differential expression of two genes encoding carbonic anhydrase in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:9779–9783. doi: 10.1073/pnas.87.24.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S. cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii: regulation by environmental CO2 concentration. Proc Natl Acad Sci USA. 1990;87:4383–4387. doi: 10.1073/pnas.87.11.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Miura K, Ishizaki K, Kucho KI, Saito T, Kohinata T, Ohyama K. Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc Natl Acad Sci USA. 2001;98:5347–5352. doi: 10.1073/pnas.081593498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke RP, Kovar JL, Weeks DP. Intracellular carbonic anhydrase is essential to photosynthesis in Chlamydomonas reinhardtii at atmospheric levels of CO2. Plant Physiol. 1997;114:237–244. doi: 10.1104/pp.114.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty AM, Anderson JC, Spalding MH. A 36 kilodalton limiting-CO2 induced polypeptide of Chlamydomonas is distinct from the 37 kilodalton periplasmic carbonic anhydrase. Plant Physiol. 1990;93:116–121. doi: 10.1104/pp.93.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty AM, Spalding MH. Molecular and structural changes in Chlamydomonas under limiting CO2: a possible mitochondrial role in adaptation. Plant Physiol. 1996;111:1339–1347. doi: 10.1104/pp.111.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Ishida S, Muto S, Miyachi S. Structural analysis of periplasmic carbonic anhydrase 1 of Chlamydomonas reinhardtii. Eur J Biochem. 1993;214:9–16. doi: 10.1111/j.1432-1033.1993.tb17890.x. [DOI] [PubMed] [Google Scholar]
- Joy KW, Blackwell RD, Lea PJ. Assimilation of nitrogen in mutants lacking enzymes of the glutamate synthase cycle. J Exp Bot. 1992;43:139–145. [Google Scholar]
- Kaplan A, Reinhold L. CO2 concentrating mechanisms in photosynthetic microorganisms. Ann Rev Plant Physiol Mol Biol. 1999;50:539–570. doi: 10.1146/annurev.arplant.50.1.539. [DOI] [PubMed] [Google Scholar]
- Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G. A novel alpha-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J. 1998;17:1208–1216. doi: 10.1093/emboj/17.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC, Lea PJ, Hausler RE. Use of barley mutants to study the control of photorespiratory metabolism. Biochem Soc Trans. 1996;24:757–761. doi: 10.1042/bst0240757. [DOI] [PubMed] [Google Scholar]
- Marco E, Ohad N, Schwarz R, Lieman Hurwitz J, Gabay C, Kaplan A. High CO2 concentration alleviates the block in photosynthetic electron transport in an ndhB-inactivated mutant of Synechococcus sp. PCC7942. Plant Physiol. 1993;101:1047–1053. doi: 10.1104/pp.101.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco Y, Rochaix JD. Organization of the nuclear ribosomal DNA of Chlamydomonas reinhardtii. Mol Gen Genet. 1980;177:715–723. doi: 10.1007/BF00272684. [DOI] [PubMed] [Google Scholar]
- Marek LF, Spalding MH. Changes in photorespiratory enzyme activity in response to limiting CO2 in Chlamydomonas reinhardtii. Plant Physiol. 1991;97:420–425. doi: 10.1104/pp.97.1.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Husic HD, Tolbert NE, Kitayama M, Manuel LJ, Togasaki RK. Isolation and characterization of a mutant of Chlamydomonas reinhardtii deficient in the CO2 concentration mechanism. Plant Physiol. 1989;89:897–903. doi: 10.1104/pp.89.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Tolbert NE, Sears BB. Complementation analysis of the inorganic carbon concentration mechanism of Chlamydomonas reinhardtii. Mol Gen Genet. 1986;204:199–203. [Google Scholar]
- Ogawa T. Cloning and inactivation of a gene essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol. 1991;96:280–284. doi: 10.1104/pp.96.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol. 1992;99:1604–1608. doi: 10.1104/pp.99.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa H, Sonoda M, Katoh H, Ogawa T. The use of mutants in the analysis of the CCM in cyanobacteria. Can J Bot. 1998;76:1035–1042. [Google Scholar]
- Price GD, Badger MR. Isolation and characterization of the high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1989;91:514–525. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Sültemeyer D, Klughammer B, Ludwig M, Badger MR. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins and recent advances. Can J Bot. 1998;76:973–1002. [Google Scholar]
- Quarmby LM, Hartzell HC. Dissection of eukaryotic transmembrane signaling using Chlamydomonas. Trends Pharmacol Sci. 1994;15:343–349. doi: 10.1016/0165-6147(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Ramazanov Z, Mason CB, Geraghty AM, Spalding MH, Moroney JV. The low CO2-inducible 36 kD protein is localized to the chloroplast envelope of Chlamydomonas reinhardtii. Plant Physiol. 1993;101:1195–1199. doi: 10.1104/pp.101.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. Genetic modification of photorespiration. Trends Biochem Sci. 1982;7:171–174. [Google Scholar]
- Spalding MH. CO2 acquisition: adaptation to changing carbon availability. In: Rochaix J-D, Goldschmidt-Clermont M, Merchant S, editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 529–547. [Google Scholar]
- Spalding MH, Spreitzer RJ, Ogren WL. Carbonic anhydrase deficient mutant of Chlamydomonas requires elevated carbon dioxide concentration for photoautotrophic growth. Plant Physiol. 1983a;73:268–272. doi: 10.1104/pp.73.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH, Spreitzer RJ, Ogren WL. Reduced inorganic carbonic transport in a CO2-requiring mutant of Chlamydomonas reinhardtii. Plant Physiol. 1983b;73:273–276. doi: 10.1104/pp.73.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH, Winder TL, Anderson JC, Geraghty AM, Marek LF. Changes in protein and gene expression during induction of the CO2-concentrating mechanism in wild-type and mutant Chlamydomonas. Can J Bot. 1991;69:1008–1016. [Google Scholar]
- Suzuki K, Marek LF, Spalding MH. A photorespiratory mutant of Chlamydomonas reinhardtii. Plant Physiol. 1990;93:92–96. doi: 10.1104/pp.93.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Spalding MH. Adaptation of Chlamydomonas reinhardtii high-CO2-requiring mutants to limiting-CO2. Plant Physiol. 1989;90:1195–1200. doi: 10.1104/pp.90.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K, Spalding MH. Periplasmic carbonic anhydrase structural gene (Cah1) mutant in Chlamydomonas reinhardtii. Plant Physiol. 1999;120:757–764. doi: 10.1104/pp.120.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A, Garcia Reina G, Ramazanov Z. Regulation of the low-CO2-inducible polypeptides in Chlamydomonas reinhardtii. Planta. 1996;199:481–485. [Google Scholar]
- Villarejo A, Martinez F, Ramazanov Z. Effect of aminooxyacetate, an inhibitor blocking the glycolate pathway, on the induction of a CO2-concentrating mechanism and low-CO2-inducible polypeptides in Chlamydomonas reinhardtii (Chlorophyta) Eur J Phycol. 1997;32:141–145. [Google Scholar]
- Winder TL, Anderson JC, Spalding MH. Translational regulation of the large and small subunits of ribulose bisphosphate carboxylase/oxygenase during induction of the CO2-concentration mechanism in Chlamydomonas reinhardtii. Plant Physiol. 1992;98:1409–1414. doi: 10.1104/pp.98.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Quick WP, Bungard RA, Bailey KJ, Lea PJ, Leegood RC. The role of photorespiration during drought stress: an analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant Cell Environ. 1999;22:361–373. [Google Scholar]
- Wintermans JFGM, De Mots A. Spectrophotometric characteristics of chlorophyll and their pheophytins in ethanol. Biochem Biophys Acta. 1965;109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhang J, Weeks DP. The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2001;98:5341–5346. doi: 10.1073/pnas.101534498. [DOI] [PMC free article] [PubMed] [Google Scholar]