Abstract
Cystatins, an amyloid-forming structural superfamily, form highly stable, domain-swapped dimers at physiological protein concentrations. In chicken cystatin, the active monomer is a kinetic trap en route to dimerization, and any changes in solution conditions or mutations that destabilize the folded state shorten the lifetime of the monomeric form. In such circumstances, amyloidogenesis will start from conditions where a domain-swapped dimer is the most prevalent species. Domain swapping occurs by a rearrangement of loop I, generating the new intermonomer interface between strands 2 and 3. The transition state for dimerization has a high level of hydrophobic group exposure, indicating that gross conformational perturbation is required for domain swapping to occur. Dimerization also occurs when chicken cystatin is in its reduced, molten-globule state, implying that the organization of secondary structure in this state mirrors that in the folded state and that domain swapping is not limited to the folded states of proteins. Although the interface between cystatin-fold units is poorly defined for cystatin A, the dimers are the appropriate size to account for the electron-dense regions in amyloid protofilaments.
Keywords: amyloid/cystatin/domain swapping/molten globule
Introduction
The deposition of abnormal fibrillar protein aggregates or ‘amyloid’ has been observed in a wide range of tissues and is now associated not only with most neurodegenerative diseases but also with a large number of other pathologies. The more prominent include Alzheimer’s, the fourth highest cause of death in the developed world, and bovine spongiform encephalopathy (BSE), along with its associated new variant Creutzfeld–Jakob disease (CJD). Structurally, the protein molecules associated with amyloid vary greatly in their sequences and native folds. However, they assemble into remarkably similar fibrous structures (Sunde et al., 1997), implying a convergence of the mechanism of fibrillization. Although a detailed structural analysis of these fibres has not been feasible to date, existing data have shown that mature amyloid fibrils are straight, rigid and unbranched (Sunde and Blake, 1997) and have a diameter between 50 and 130 Å. X-ray diffraction data have led to a general model for amyloid fibrils as ‘cross-β’ structures where the backbone hydrogen bonds lie in the direction of the long axis of the fibril (Blake and Serpell, 1996). Three-dimensional reconstructions based on cryo-electron microscopy (EM) images of SH3 fibrils (Jimenez et al., 1999) reveal two pairs of protofilaments of ∼20 Å diameter, separated by ∼40 Å, that wind around a hollow core.
Understanding the dynamics of fibril formation is an even more challenging problem. So far, atomic force microscopy and EM have shown that fibrils appear in conjunction with the disappearance of spheres and protofibrillar intermediates (Harper et al., 1999; Jackson et al., 1999), which are 60–80 Å in diameter (Walsh et al., 1997). These late events are preceded by a number of determinant steps that decide the fate of the protein molecule. Relevant experimental data have been rare (Serio et al., 2000) and, as a result, models for the kinetics of fibril assembly are still developing. It is commonly believed that globular amyloidogenic proteins partially unfold and convert into alternatively folded conformations to self-assemble into fibrils (Carrell and Gooptu, 1998; Kelly, 1998). Three-dimensional domain swapping, the process in which a domain in a protein breaks its non-covalent bonds with the remainder of the molecule and its place is taken by the same domain of a second molecule, has been widely recognized as a candidate for one of the conversion steps (Ekiel et al., 1997; Schlunegger et al., 1997; Liu et al., 1998; Zegers et al., 1999). In common with amyloidogenesis, the nature of these association steps is of both high specificity and high affinity. Although the idea that these phenomena may be linked has been received with enthusiasm (Cohen and Prusiner, 1998; Purich and Allison, 2000), this is somewhat surprising considering that, so far, no three-dimensional domain swapping has been shown in an amyloidogenic protein.
Here, we present work on the cystatins, a family of structurally homologous cysteine proteinase inhibitors that are widely distributed in all higher organisms (Barrett, 1987). Their physiological role is vital and is believed to be tightly linked to the control of protein turnover and the defence of tissues against invasion by viruses and parasites. The L68Q variant of human cystatin C (hCC) is the causative agent of an amyloidotic disease, hereditary cystatin C amyloid angiopathy, where repeated haemorrhage, dementia, paralysis and eventual death is associated with cystatin deposits in cerebral blood vessels (Abrahamson, 1996). Cystatins have a number of properties that make them ideally suited to the study of amyloidogenesis at a molecular level. Compared with other amyloidogenic proteins, they are relatively small in size (11–13 kDa), both NMR and X-ray crystal structures are available and they have a well-defined folding pathway. Structurally, the two most studied families of the superfamily are highly homologous: each cystatin consists of a core, five-stranded antiparallel β-sheet wrapped around a central helix, as illustrated by the structure of chicken cystatin (cC) in Figure 1. Family II cystatins, e.g. hCC and its homologue cC, can be distinguished from family I cystatins, e.g. human cystatins A and B (hCA and hCB), by the presence of two disulfide bonds and an ∼20 residue insertion of irregular structure (Engh et al., 1993) between strands 3 and 4. Strong implications as to the mechanism of amyloidotic aggregation (Abrahamson and Grubb, 1994) came with the discovery that hCC (Ekiel and Abrahamson, 1996), hCA (Jerala and Zerovnik, 1999) and hCB (Zerovnik et al., 1997) have a propensity to form inactive dimers under pre-denaturing conditions and, moreover, that the L68Q variant of hCC partially dimerizes under physiological conditions. For each cystatin, NMR chemical shift changes indicate that dimerization involves no structural rearrangement of the main fold; changes are confined to the region containing the active site, the loop between strands 2 and 3 (Figure 1). The unexpectedly slow kinetics and high activation energy of dimerization led to the proposal that dimerization may be the result of a domain-swapping event rather than a more simple association reaction (Ekiel et al., 1997; Jerala and Zerovnik, 1999).
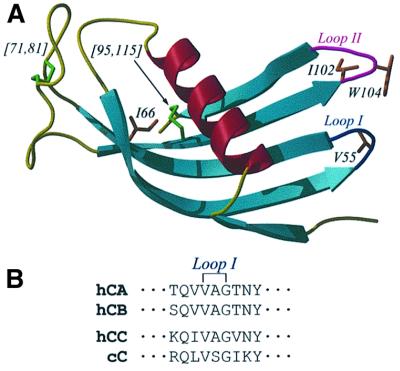
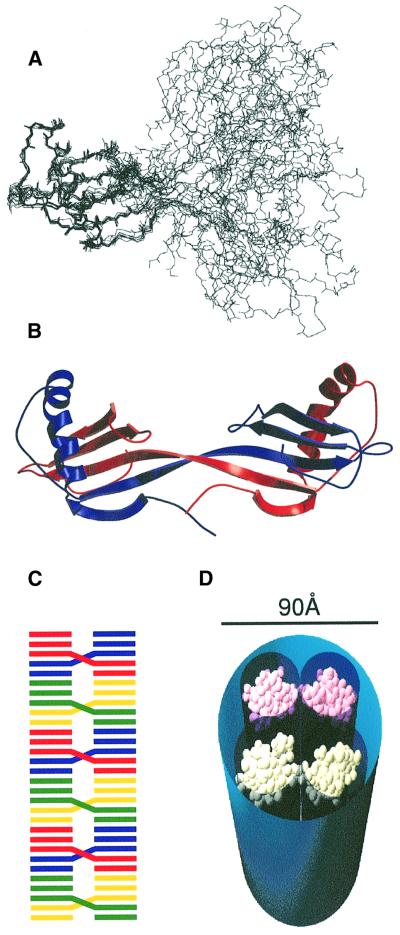
Fig. 1. (A) The three-dimensional structure of chicken cystatin (Engh et al., 1993), showing the classical cystatin monomer fold, created using the programs MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Murphy, 1994). In sequence order, strands 1–5 are from bottom to top. Coloured blue and magenta are the active site loops I and II, which connect strands 2 to 3, and 4 to 5, respectively. Also highlighted are the two disulfide bonds ([71,81] and [95,115]), the point mutations and the single tryptophan residue. (B) Sequence of the loop I regions of hCA, hCB, hCC and cC showing the conserved VXG motif present in all cystatins. The conserved valine corresponds to V55 in cC.
In order to determine whether domain swapping is a common feature of dimerization by members of the cystatin superfamily, we investigated the representative of each family that was most fully defined structurally in the monomeric state, namely hCA (family I) and cC (family II). Here we show that cystatin dimers from both of the main subfamilies are formed via a three-dimensional domain-swapping mechanism and, intriguingly, can achieve this whether or not their side chains are immobilized. We discuss the implications of domain swapping within a superfamily containing amyloidogenic proteins in the context of a molecular basis of the protofilaments observed in amyloid fibres.
Results and discussion
The dimerization reaction
Mechanistic data on the process of domain swapping (Hayes et al., 1999; Rousseau et al., 2001) are not as abundant as structures of domain-swapped proteins. Generally, dimerization by domain swapping differs from other dimerization reactions by having a very high kinetic barrier (due to the large conformational changes involved) and a small thermodynamic driving force (due to the similarity of the final structures) (Schlunegger et al., 1997). The transition between folded monomers and folded dimers of hCC and hCA is favoured by elevated but pre-denaturating temperatures or levels of denaturant (Ekiel and Abrahamson, 1996; Jerala and Zerovnik, 1999). Retention times on size-exclusion chromatography (Figure 2) reveal that a 1 mM cC solution rapidly undergoes such a transition at 80°C in the absence of denaturant or at 20°C in 4 M guanidinium chloride (GdmCl). Kinetic data (Figure 3B) show that the bimolecular rate constants for dimerization vary logarithmically with GdmCl concentration over the range measured (2.0–3.4 M), and extrapolate to a rate constant of ∼1 × 10–8 M–1 s–1 in the absence of denaturant, i.e. a half-time of ∼2000 years for a 1 mM solution. The GdmCl dependence of the rate constant of dimerization can also provide information on the structural integrity of the transition state since it directly reflects the solvent exposure of hydrophobic groups in this state relative to the folded monomer. The correlation between the slope, ln kobs/[denaturant], and solvent exposure has been widely documented in protein folding where this parameter is referred to as an m-value (Parker et al., 1995). In this case, the GdmCl ‘m-value’ for the dimerization reaction is 10 ± 2 M–1. If the transition state of the bimolecular reaction were as solvent exposed as the unfolded monomer, this value would be twice the observed dependence (Staniforth et al., 2000) of the unfolding equilibrium constant, i.e. 2mF/U, 17.2 ± 0.6 M–1. There fore, the structure of the transition state en route to the dimer is closer to the unfolded state than the folded state, indicating that considerable disruption of the structure is required for the monomer to dimer transition to occur. Furthermore, this transition state is less folded than the kinetic intermediate populated in the monomeric folded pathway, as also reported recently for the domain swapping of p13suc1 (Rousseau et al., 2001). This suggests that such folding intermediate species are not key elements for this type of oligomerization reaction. The linearity of the protein concentration dependence of the rates of these reactions was verified up to 450 µM, revealing that the reaction is limited by a bimolecular process and not, for example, the unfolding of the monomer. Rather, the rate is most likely to be limited by the association of predominantly unfolded proteins.
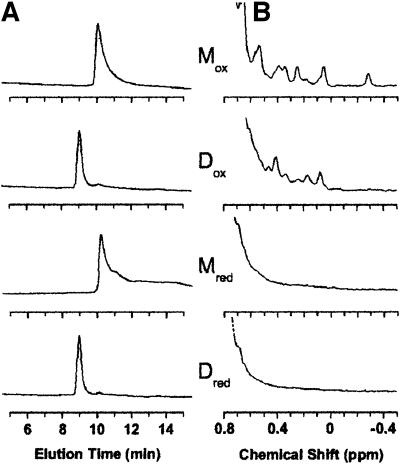
Fig. 2. (A) Size-exclusion chromatography of monomeric (M) and dimeric (D) cC samples (5 µM) under oxidizing (Mox and Dox) and reducing (Mred and Dred) conditions. Retention times of 9 and 10.2 min (corresponding to mol. wts of 30 000 and 15 000 Da, respectively) were observed for the dimeric and monomeric species, respectively. No significant increase in hydrodynamic radius is apparent on loss of side chain packing in the reduced proteins. (B) Upfield-shifted methyl region of 1H-NMR spectra under the conditions in (A), at protein concentrations of 150 µM, showing the retention of a folded conformation (Mox, Dox) and a molten-globule (Mred, Dred) conformation on dimerization for the oxidized and reduced proteins, respectively.
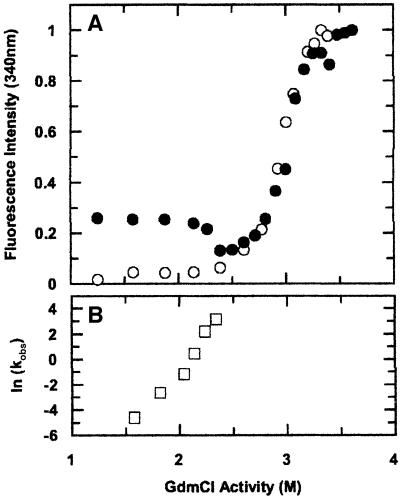
Fig. 3. (A) Unfolding profile of dimeric (open circles) and monomeric (filled circles) cC incubated at different GdmCl concentrations for 24 h and monitored using tryptophan fluorescence. Data for monomeric cC match those reported previously (Bjork and Pol, 1992) and are superimposable on those of dimeric cC at GdmCl activity >2.5 M. Below this activity, conversion of monomer to dimer does not reach equilibrium within the incubation period. (B) The GdmCl dependence of the observed bimolecular rate constant for dimerization (kobs, M–1 s–1), determined from the time dependencies of the unique NMR signals. The logarithmic dependence of kobs has a slope of 10 ± 2 M–1 The equivalent dependence of a bimolecular reaction via the unfolded state would be 17.2 ± 0.6 M–1 i.e. twice that of the unfolding equilibrium constant (Staniforth et al., 2000).
In order to determine the stability of the dimer with respect to the monomeric state of cC, measurements of the fraction of monomer to dimer needed to be made as a function of protein concentration. Two observations led us to conclude that, at least in the case of cC, the reaction is irreversible in favour of the dimer. The first was that no residual monomer was ever detectable at the end points of the kinetic reactions. Also, unfolding profiles of monomeric and dimeric forms of cC show the transition from monomer to dimer after a prolonged incubation period (Figure 3A), but not from dimer to monomer, even at concentrations of cC as low as 600 nM. Thermodynami cally, therefore, the dimer is the more stable species under the accessible experimental conditions, and the folded monomer is a kinetic trap, albeit a highly stable species (Staniforth et al., 2000) out of which the transition is immeasurably slow in the absence of solvent perturbation.
Structure of the dimers
The dimers of cC, like those of hCC (Ekiel et al., 1997), hCA (Jerala and Zerovnik, 1999), and hCB (Zerovnik et al., 1997), are symmetric on the NMR time scale, since the monomer and dimer 1H-15N HSQC spectra contain the same number of peaks with no evidence of peak duplication. Also, similarly to other cystatins, chemical shift perturbation on dimerization of cC is limited to the region containing the active site (see Figure 1), indicating that the cystatin fold is preserved in the dimer. In order to determine whether domain swapping has occurred, isotope-filtered NOESY spectra (Otting et al., 1986) were recorded separately for samples of hCA and cC dimers. These experiments were recorded for samples containing dimers made from a 1:1 mixture of uniformly 15N-labelled and unlabelled monomers, which therefore contained 25% labelled–labelled, 50% labelled–unlabelled and 25% unlabelled–unlabelled dimers. The appropriate processing of such spectra yields a sub-spectrum where predominantly only NOESY cross-peaks resulting from intermolecular interactions are observed. For hCA (Figure 4A), cross-peaks were observed between hydrogens on strand 2 and those on strand 3 in the dimer; these interactions reproduce original contacts present in the native monomers. In conjunction with evidence from chemical shift data showing that the cystatin-fold unit remains unperturbed, this proves that the dimer is domain swapped with a hinge region corresponding to the loop between strands 2 and 3. Each cystatin-fold unit in the domain-swapped dimer is thus constituted by strand 1, the α-helix and strand 2 from one monomer, and strands 3–5 from the other monomer. The equivalent experiment on cC was inconclusive owing to the insensitivity of this type of experiment and to the considerably increased tumbling time for this dimer (τc ∼25 ns for cC versus 14 ns for hCA) which results in lower signal:noise ratios for this protein. cC monomers also show unexpectedly long tumbling times. An undecoupled standard two-dimensional homonuclear NOESY experiment, which does not suffer from the substantial relaxation problems of isotope filtering, was recorded on a sample containing cC dimers prepared from a 1:1 mixture of uniformly 13C/15N-labelled and unlabelled monomers. The splitting pattern predicted for intermolecular interactions between labelled and unlabelled molecules is very different from that for intramolecular interactions due to the fact that only the former can contain correlations connecting labelled and unlabelled atoms. The pattern and intensities of components within an individual cross-peak can therefore be used to distinguish inter- from intramolecular nuclear Overhauser effects (NOEs), but the identification is limited to cross-peaks with sufficient chemical shift resolution. Two of the intermolecular NOEs that confirm that cC is domain swapped in the same way as hCA are shown in Figure 4B. These NOEs are from F27 in the helix to I107 in strand 5, reproducing a contact observed in the native monomer, similarly to the hCA experiment above.
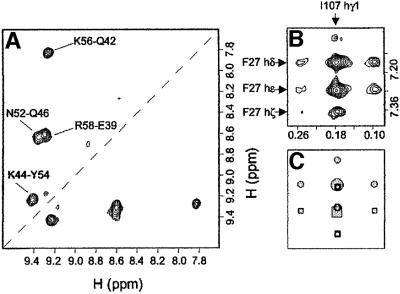
Fig. 4. (A) Amide–amide region from a 100 ms isotope-filtered NOESY experiment (Otting et al., 1986) on an hCA dimer made from a 1:1 mixture of uniformly 15N-labelled and unlabelled monomers, showing the sum of the signal from components where the filters are on–off and where they are off–on. Thus, only intermolecular dNN NOEs are observed in this region of the spectrum. The labelled cross-peaks are cross-strand NOEs between strand 2 and strand 3. (B) Section from a 100 ms NOESY experiment on a cC dimer made from a 1:1 mixture of uniformly 13C,15N-labelled and unlabelled monomers. Illustrated are two intermolecular NOE cross-peaks from the side chain of F27 to that of I107. Components of the cross-peaks where magnetization has been transferred from H(13C) to H(12C) and vice versa flank the central H(12C) to H(12C) component. (C) A schematic representation of (B), showing the expected peak pattern at the S/N ratio recorded in (B). The circle and square symbols represent the cross-peaks of F27hδ and F27hε, respectively. The unfilled symbols represent components that are unresolvable from more intense signals.
The structure of the more tractable hCA dimer therefore contains the essential features of domain swapping in the cystatin superfamily. Resonance assignment showed that the vast majority of the residues in the dimer retain the same chemical shift and, hence, the same structural environment, as illustrated by the 1H-15N HSQC spectrum. Therefore, in order to restrain the structure of each cystatin-fold unit within the dimeric assembly, it was possible to use structural restraints (NOEs and hydrogen bonds) from data collected on a monomeric sample (Martin et al., 1995). Where chemical shift differences between monomer and dimer had been observed, the monomer restraints were omitted from the calculation. Torsional angle constraints for the dimer were calculated using TALOS (Cornilescu et al., 1999) and incorporated. Backbone torsional constraints in the cystatin-fold unit showed the expected close similarity with data calculated for the monomer, whereas those in the hinge region changed from typical turn values to extended strand values. In addition, the intermolecular NOEs observed in the isotope-filtered NOESY experiment were used along with two additional hydrogen bonds restraints (Jerala and Zerovnik, 1999). An ensemble of structures was then calculated using a total of 1803 NOE, 52 hydrogen bonding and 170 torsional constraints. The overlay of structures and a representative example are shown in Figure 5A and B, respectively. Units corresponding to the cystatin fold overlay very closely (backbone r.m.s.d. of 0.50 Å) and are indistinguishable from the ensemble of cystatin A monomers reported previously (Martin et al., 1995). In contrast, the angular relationship between cystatin-fold units in the dimer is not well defined. However, there is no evidence that this angle should be well defined as neither the N-terminal strands nor the loop II regions pack together. Although there are some chemical shift changes in these regions (Jerala and Zerovnik, 1999), these are likely to result from subtle changes in the relationships of these regions with strands 2 and 3 rather than cystatin-fold unit contact. Therefore, the only determinant of the angular relationship between the cystatin-fold units is the β-sheet twist as strand 2 becomes strand 3.
Fig. 5. (A) Ensemble of the eight lowest energy structures of the domain-swapped hCA dimer. The well-defined regions (T13–Q46, N52–P74 and L80–F98) of a cystatin-fold unit overlay with an r.m.s.d. to the mean structure of 0.50 and 1.02 Å for the backbone and heavy atoms, respectively. (B) Ribbon representation of the minimized averaged dimer structure of hCA, created using the programs MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Murphy, 1994). The component monomers are coloured blue and red. The closed interface is formed between strands 2 and 3. Loop I in the monomer merges these elements into a continuous strand in the dimer. The secondary structure elements comprise β1, G5–P11; α, E15–T31; β2/3, Q42–A59; β4, K63–S72; and β5, V81–K89. (C) Schematic representation of a possible longitudinal assembly of cystatin dimers into a continuous β-sheet structure where dimers are connected via an interface between strands 1 and 5. Such an assembly has the potential to be favoured energetically and kinetically in comparison with the equivalent association of monomers due to the increased effective concentration of monomers in the dimer. (D) A space-filling model illustrating how four cystatin-fold units, made up of two domain-swapped dimers of the type shown in (B), can be fitted into the cross-section of a generic fibril. In this model, the fibril would be formed by the assembly of further domain-swapped dimers onto each of the visible dimers in the manner illustrated in (C).
Identifying whether the two cystatin-fold units always have the same relative positions or can move between numerous positions is difficult to determine experimentally using X-ray or NMR methods, especially in such symmetric systems. The problems for both techniques are well illustrated in chemokine oligomerization (Lubkowski et al., 1997; Czaplewski et al., 1999), where crystallization selects single conformers from an ensemble, and averaging in NMR parameters prohibits unambiguous interpretation. Following the submission of this paper, a crystal structure of the dimer of hCC was published (Janowski et al., 2001). This cystatin forms a domain-swapped dimer in an analogous way to cC and hCA, confirming the generality of this phenomenon within the superfamily. This crystal form has selected a conformer that lies within the ensemble of calculated hCA dimer structures which has a 180° twist between strands 2 and 3 and an angle of 115° between cystatin-fold units. As in hCA, the core cystatin fold is retained (overall r.m.s.d. of 0.58 Å for 86 common Cα atoms with cC monomer).
The driving force for dimerization
The high stability of the cC domain-swapped dimer, the formation of which is irreversible under the accessible experimental conditions, contrasts the general case where the difference in free energy between monomers and domain-swapped dimers is small (Schlunegger et al., 1997). Since the interface between the cystatin-fold units in the dimer is so small and the overall fold is retained, the hinge region must account fully for any favourable energy changes. Furthermore, the feature of the hinge region must be common to cC, hCC, hCA and hCB since they all domain swap similarly despite the very low level of overall sequence conservation. The most likely source of stability is the alleviation of distortion in the turn connecting strands 2 and 3 in cC, hCA and hCB monomers (Stubbs et al., 1990; Engh et al., 1993; Martin et al., 1995), manifested in the ψ angle of the highly conserved valine residue in the VXG motif (Figure 1), or its neighbours, being in an unfavoured region in the Ramachandran plot. The VXG signature region of cystatins is situated close to the active site residues in complexes with target proteinases (Stubbs et al., 1990), where backbone distortion may be advantageous.
Domain swapping and amyloidogenesis
In the context of working models of the molecular organization of amyloid fibres, domain swapping in the cystatin superfamily dimers is an attractive candidate mechanism for cementing the building blocks that make up highly stable fibrils. Dimerization through this domain-swapping mechanism causes a considerable extension of β-strands 2 and 3, thereby closely tying the β-sheets of the two cystatin-fold units in a strongly hydrogen-bonded intermolecular relationship. Represented in the hCA ensemble are dimers that would present extended β-strands at the interface with adjacent dimers in a fibril (strands 1 against strands 5 in the next dimer) and would provide a means of propagation of a large, stable β-sheet structure (Figure 5C). The end-to-end distance in the calculated dimer structures ranges from ∼60 to 80 Å, which matches closely the widths of fibres observed for many amyloidogenic proteins (Sunde and Blake, 1997), including those of cystatins (Jensson et al., 1987). This relationship is illustrated schematically in Figure 5D where we have inserted two domain-swapped hCA dimers into the cross-section of a fibril comprising four protofilaments, based on the electron density distribution observed in SH3 fibrils (Jimenez et al., 1999). Fibres with cross-sections exhibiting two or six protofilaments arranged in pairs have also been observed recently (Saibil, 2001). The properties of the EM-derived cross-sections highlighted here are the arrangement of electron-dense protofilaments of tubular appearance into a fibril where they are separated by regions of surprisingly low electron density. The dimers fit into the electron density with a single cystatin unit for each protofilament and fittingly account for the lack of density observed between protofilaments.
It is plausible that the association of pairs of protofilaments may also involve domain swapping in which interfaces other than that identified above are used (e.g. the loop between strands 3 and 4 on the opposite end of the cystatin-fold unit). Models based on dimer structures show that such tetramers could be formed without steric clashes and without breaking the [C71,C81] disulfide bond in the long unstructured loop between strands 3 and 4 in cC (our unpublished results). However, a mechanism that could give rise to a tetramer equally could generate a much more extended domain-swapped multimer, which leads to fibril formation via a superhelix coiling in the direction of fibril propagation. Currently, though, there is no information available for tetramers of cystatins or for the morphology of cystatin fibrils at this level of resolution.
Mutations can induce or inhibit domain swapping
In contrast to the wild-type protein, where the rate of dimerization is immeasurably slow, the cC variant containing the mutation I66Q (the cC version of the hCC disease-causing mutation) dimerizes spontaneously and rapidly at room temperature in the absence of denaturant (Table I). The destabilization of the cystatin fold caused by the replacement of this core hydrophobic residue reduces the free energy barrier between the ground and transition state for dimerization and therefore removes the ability of the monomer to act as a prolonged kinetic trap. In order to demonstrate that dimerization could also be inhibited, two mutations replacing hydrophobic residues with charged residues were inserted into cC near the interface between cystatin-fold units in wild-type dimers (Table I). Proteins containing the substitutions V55D (on loop I in the monomer and at the cystatin-fold unit interface between strands 2 and 3 in the dimer) or I102K (on loop II, adjacent to loop I) remained monomeric under all conditions used to dimerize the wild-type protein. Although loop II is not directly involved in the cystatin-fold unit interface (Figure 5B), it is perturbed on dimerization in the wild-type protein. The 1H-NMR chemical shift changes indicate that the spatial proximity of the I102 and W104 side chains is removed on dimerization, and the fluorescence of W104, which is already reduced in the folded monomer relative to the unfolded state, is quenched further (Figure 3A).
Table I. Oligomeric states of cC variants.
| Refolded | Heat treated | |
|---|---|---|
| Wild-typeoxidized | monomer | dimer |
| Wild-typereduced | dimer | dimer |
| I66Q | dimer | dimer |
| V55D | monomer | monomer |
| I102K | monomer | monomer |
The predominant oligomeric state for wild-type and mutant cCs, following refolding from 7 M GdmCl then storage for 24 h at 20°C and after heat treatment at 90°C for 30 min, at protein concentrations in the range 5–500 µM. The oligomeric states were determined using characteristic 1H-NMR resonances at low and high temperatures and size-exclusion chromatography.
Oligomerization in a denatured state
A number of amyloidotic proteins are unstable under physiological conditions and remain effectively unfolded as monomers (e.g. α-synuclein that causes Parkinson’s disease and the yeast prion Sup35). It has been suggested that these proteins may require the formation of an initial nucleus made up of a number of unfolded molecules before they can rearrange into the folded β-structures that make up the amyloid fibre (Serio et al., 2000). The following results on cC suggest that folded, globular proteins can populate intermediate species that are similar to the extent that structural plasticity is retained within stable assemblies. On disulfide reduction, cC forms an unusually well-defined, monomeric, molten-globule state (Staniforth et al., 2000). The position of the disulfide bond that controls the folded state/molten-globule state switch in cC (C95–C115) is adjacent to I66 (Figure 1). This molten-globule state is on the folded state side of the major folding transition state and retains the secondary structure content of the folded state of the oxidized protein, but has not formed tight packing of its side chains. The molten-globule state is less stable than the folded oxidized protein by ∼8 kcal/mol but remains unusually highly stable against unfolding (ΔGF/U = –7.8 kcal/mol). Surprisingly, the reduced protein dimerizes in the absence of denaturant similarly to the folded forms of cystatins despite its only partially folded state (Figure 2). On refolding from its GdmCl-denatured state, the reduced protein rapidly forms a molten-globule monomer (kobs = 100 s–1), following which it slowly but spontaneously dimerizes (kobs = 3 × 10–1 M–1 s–1). The rate constant for dimerization of the reduced protein is broadly in line with that predicted above for the oxidized protein in the absence of denaturant, corrected for the difference in fold stability. Importantly, the dimer formed at the end point of the reaction is also a molten globule. Hence, despite its side chains being mobile, the molten-globule state of cC behaves as if it were simply a destabilized but folded cystatin, where the monomer is a kinetic trap with an appropriately shortened lifetime. The implications of this are that the molten-globule state must retain the essential features of the folded state, and that domain swapping is possible in states that are not folded in the traditional sense. This observation in turn raises the question of whether side chains are mobilized or immobilized in amyloid fibres.
Conclusion
The commonality of the domain-swapping behaviour shown above for hCA and cC, for hCC (Janowski et al., 2001) and, by implication, for hCB and the molten-globule state of cC, reveals this event as a general feature of the cystatin superfamily fold. hCC and cC are known to be amyloidogenic, and solution conditions that stimulate amyloidogenesis correlate with those that stimulate domain-swapping dimerization more rapidly. The domain-swapping reaction is slow relative to folding/unfolding rates and requires a high level of exposure of hydrophobic side chains in the transition state, implying that the reaction is rate-limited by the association of predominantly unfolded proteins. The free energy of this state can be estimated to be ∼11 kcal/mol compared with a ΔGunfolding of 15.6 kcal/mol (Staniforth et al., 2000). Whether domain swapping is retained in the cystatin fibrils and whether it occurs generally in amyloid fibres remains open to question. For example, monellin, a two-chain plant protein that shares the same fold but low sequence homology with cystatins, forms dimers and aggregates into amyloid-like fibres upon heating (Konno et al., 1999), despite being cleaved in the hinge loop (Murzin, 1993) involved in the cystatin domain-swapping process. However, it cannot be assumed that cystatin and monellin fibres form amyloid in the same way given the variations in morphology that are observed even within the same species. Domain swapping is a means of generating very strong intermolecular interactions and is a good candidate for the connections between protofilaments in a fibril. The cystatin-fold units within domain-swapped dimers have the appropriate dimensions to constitute the electron-dense regions of protofilaments, and would present β-rich surfaces for propagation into a continuous β-structure proposed in fibres. This would be achievable with or without side chain immobilization.
Materials and methods
Expression, purification and dimerization of hCA and CC
Recombinant hCA (mol. wt = 11 000 Da) was expressed in Escherichia coli (BL21-DE3) using the pET3A system and purified from the soluble cell extract using a combination of ion-exchange (Q-Sepharose in 20 mM Tris–HCl pH 8.0, elution with a gradient of 0–0.3 M NaCl) and size-exclusion (Superdex 75) chromatographies. Recombinant cC (mol. wt = 13 100 Da) was expressed in E.coli (TG1) using the pIN-III-OmpA system and purified from a periplasmic extract using papain affinity chromatography (Anastasi et al., 1983) followed by size-exclusion chromatography (Superdex 75) under denaturing conditions (50 mM K3PO4, 0.5 M KCl pH 11.5). Single (15N), double (15N–13C) and triple (15N–13C–2H) uniformly labelled proteins were prepared using minimal media (Venters et al., 1995). Dimerization was achieved by heating hCA and cC to temperatures of 85°C for 20 min and 80°C for 1 h, respectively, at protein concentrations >200 µM followed by rapid cooling to 4°C. Dimeric proteins were purified from residual monomer by size-exclusion chromatography (Superdex 75). Site-directed mutants (I66Q, V55D and I102K) of cC were constructed using the QuikChange™ mutagenesis kit (Stratagene, CA).
Kinetic measurements
The denaturation profile of cC was recorded using the intrinsic tryptophan fluorescence (Schimadzu RF5301PC, λex = 290 nm, λem = 340 nm) of both monomeric and dimeric solutions (5 and 100 µM, respectively, in 50 mM NaPO4 pH 7.0, 20°C) pre-incubated at different GdmCl concentrations for 24 h. Fluorescence is plotted as a function of GdmCl activity (Parker et al., 1995), where activity is derived from GdmCl molarity by the relationship: activity = (7.5*[GdmCl])/([GdmCl] + 7.5). The GdmCl dependence of the rate constants for dimerization (kobs) for both oxidized and reduced cC was determined from the time dependence of resonance intensity in 1H-NMR spectra. Assuming dimerization occurs via the diffusion-limited collision of two partially unfolded molecules, the free energy of the partially unfolded state relative to the folded state can be estimated as ΔG = RTln[(kobs/kbim)1/2], where kobs is the dimerization rate, and kbim is the collision rate (∼108 M–1 s–1). Dimerization of oxidized cC was initiated by addition of folded monomer to solutions of GdmCl (2–3.4 M) in 50 mM Na3PO4 pH 7.0, 25°C to a protein concentration of 150 µM. The linearity of kobs versus protein concentration was verified between 50 and 450 µM at [GdmCl] = 2.8 M. Dimerization of reduced cC was initiated by the refolding of reduced and GdmCl-denatured cC into 20 mM dithiothreitol, 50 mM NaPO4 pH 7.0, 25°C (1.1 M GdmCl) to a protein concentration of 150 µM. This concentration of reducing agent is sufficient to maintain both disulfide bonds reduced (Bjork and Ylinenjarvi, 1992). Size-exclusion chromatography retention times of monomeric and dimeric cC were determined using a BIOSEP-SEC 3000 HPLC column (Phenomenex, CA) under the above buffer conditions at protein concentrations of 5 µM.
NMR spectroscopy
Resonance assignment experiments for fully 15N/13C/2H-labelled hCA and fully 15N/13C-labelled cC dimers were collected on a Bruker DRX 600 MHz spectrometer using a triple resonance z-gradient probe, along with the NOESY experiments for identifying intermonomer NOEs. In addition, a trosy-HN(CA)CB (Salzmann et al., 1999) and 100 ms NOESY were run at 800 MHz for hCA dimer and cC dimer, respectively. The triple resonance NMR and NOE assignment experiments were run at 308 K for hCA and 328 K for cC using a combination of HNCA (H 164 ms, N 24 ms, C 24 ms), HNCO (H 164 ms, N 24 ms, C 24 ms), HN(CA)CB (H 82 ms, N 24 ms, C 6.4 ms), 27 ms TOCSY-HSQC (H 164 ms, H 17 ms, N 24 ms) and 100 ms NOESY-HSQC (H 164 ms, H 17 ms, N 24 ms). Intermonomer NOEs were identified for hCA dimer in 15N-isotope-filtered (H 328 ms, H 21 ms) and for cC dimer in unfiltered 100 ms NOESY experiments (H 328 ms, H 33 ms), on samples prepared from a 1:1 mixture of labelled and unlabelled monomers. The acquired data were processed and analysed using the program Felix2000 (MSI, San Diego) on Silicon Graphics workstations.
Structure calculation
Distance restraints (NOEs and hydrogen bonds) were taken from the data used for the hCA monomer structure (Martin et al., 1995), except those involving residues V48–G50 and N77–L80 where chemical shift perturbation was observed following resonance assignment. The NOEs were specified as intermolecular and intramolecular according to domain-swapped topology inferred from isotope filtering experiments. An additional hydrogen bond restraint was included between 48 and 50 based on evidence from protection experiments on the dimer (Jerala and Zerovnik, 1999). Dihedral restraints (φ and ψ) were determined using the 1Hα, 15N, 13Cα, 13Cβ and 13C′ chemical shifts and the program TALOS (Cornilescu et al., 1999). Where TALOS gave a ‘poor’ match, the experimental φ dihedral angle was taken from the data used for the hCA monomer structure calculation. In addition, monomer hCA χ1 constraints were used. TALOS dihedrals were entered with bounds of φ ± 2 σ, where σ is standard deviation.
Fifty structures were calculated as described by Nilges (1997) using the simulated annealing protocol of XPLOR (Brünger, 1992). Eight of the lower energy structures with no NOE constraint violations exceeding 0.5 Å or torsional constraint violations exceeding 5° were selected for the representative ensemble. The well-defined regions (T13–Q46, N52–P74 and L80–F98) of cystatin-fold units within the ensemble overlay with an r.m.s.d. to the mean structure of 0.50 and 1.02 Å for the backbone and heavy atoms, respectively.
Acknowledgments
Acknowledgements
We would like to thank Manal El-Baghdady for assistance with cystatin constructs, the Wellcome Trust, BBRSC, HEFCE and the Slovenian Ministry of Science and Technology for financial support, the Royal Society (R.A.S.) and Lister Institute (J.P.W.) for personal fellowships, MSI for providing FELIX, and the staff at the BBSRC 800 MHz NMR service at Cambridge. The Krebs Institute for Biomolecular Research is a designated BBSRC Centre and a member of NESBIC.
References
- Abrahamson, M. (1996) Molecular basis for amyloidosis related to hereditary brain hemorrhage. Scand. J. Clin. Lab. Invest., 56, 47–56. [PubMed] [Google Scholar]
- Abrahamson M. and Grubb,A. (1994) Increased body temperature accelerates aggregation of the Leu-68→Gln mutant cystatin C, the amyloid-forming protein in hereditary cystatin C amyloid angiopathy. Proc. Natl Acad. Sci. USA, 91, 1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasi A., Brown,M.A., Kembhavi,A.A., Nicklin,M.J.H., Sayers,C.A., Sunter,D.C. and Barrett,A.J. (1983) Cystatin, a protein inhibitor of cyteine proteinases—improved purification from egg-white. Biochem. J., 211, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A.J. (1987) The cystatins—a new class of peptidase inhibitors. Trends Biochem. Sci., 12, 193–196. [Google Scholar]
- Bjork I. and Pol,E. (1992) Biphasic transition curve on denaturation of chicken cystatin by guanidinium chloride—evidence for an independently unfolding structural region. FEBS Lett., 299, 66–68. [DOI] [PubMed] [Google Scholar]
- Bjork I. and Ylinenjarvi,K. (1992) Different roles of the 2 disulfide bonds of the cysteine proteinase inhibitor, chicken cystatin, for the conformation of the active protein. Biochemistry, 31, 8597–8602. [DOI] [PubMed] [Google Scholar]
- Blake C. and Serpell,L. (1996) Synchrotron X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous β-sheet helix. Structure, 4, 989–998. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR Version 3.1: A system for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Carrell R.W. and Gooptu,B. (1998) Conformational changes and disease—serpins, prions and Alzheimer’s. Curr. Opin. Struct. Biol., 8, 799–809. [DOI] [PubMed] [Google Scholar]
- Cohen F.E. and Prusiner,S.B. (1998) Pathologic conformations of the prion proteins. Annu. Rev. Biochem., 67, 793–819. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Czaplewski L.G. et al. (1999) Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1 α, MIP-1 β and RANTES—characterization of active disaggregated chemokine variants. J. Biol. Chem., 274, 16077–16084. [DOI] [PubMed] [Google Scholar]
- Ekiel I. and Abrahamson,M. (1996) Folding-related dimerization of human cystatin C. J. Biol. Chem., 271, 1314–1321. [DOI] [PubMed] [Google Scholar]
- Ekiel I. et al. (1997) NMR structural studies of human cystatin C dimers and monomers. J. Mol. Biol., 271, 266–277. [DOI] [PubMed] [Google Scholar]
- Engh R.A., Dieckmann,T., Bode,W., Aueswald,E.A., Turk,V., Huber,R. and Oschkinat,H. (1993) Conformational variability of chicken cystatin—comparison of structures determined by X-ray diffraction and NMR spectroscopy. J. Mol. Biol., 234, 1060–1069. [DOI] [PubMed] [Google Scholar]
- Harper J.D., Wong,S.S., Lieber,C.M. and Lansbury,P.T. (1999) Assembly of Aβ amyloid protofibrils: an in vitro model for a possible early event in Alzheimer’s disease. Biochemistry, 38, 8972–8980. [DOI] [PubMed] [Google Scholar]
- Hayes M.V., Sessions,R.B., Brady,R.L. and Clarke,A.R. (1999) Engineered assembly of intertwined oligomers of an immunoglobulin chain. J. Mol. Biol., 285, 1857–1867. [DOI] [PubMed] [Google Scholar]
- Jackson G.S., Hill,S.F., Joseph,C., Hosszu,L., Power,A., Waltho,J.P., Clarke,A.R. and Collinge,J. (1999) Multiple folding pathways for heterologously expressed human prion protein. Biochim. Biophys. Acta, 1431, 1–13. [DOI] [PubMed] [Google Scholar]
- Janowski R., Kozak,M., Jankowska,E., Grzonka,Z., Grubb,A., Abrahamson,M. and Jaskolski,M. (2001) Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain-swapping. Nature Struct. Biol., 8, 316–320. [DOI] [PubMed] [Google Scholar]
- Jensson O. et al. (1987) Hereditary cystatin C (γ-trace) amyloid angiopathy of the CNS causing cerebral hemorrhage. Acta Neurol. Scand., 76, 102–114. [DOI] [PubMed] [Google Scholar]
- Jerala R. and Zerovnik,E. (1999) Accessing the global minimum conformation of stefin A dimer by annealing under partially denaturing conditions. J. Mol. Biol., 291, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Jimenez J.L., Guijarro,J.L., Orlova,E., Zurdo,J., Dobson,C.M., Sunde,M. and Saibil,H.R. (1999) Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J., 18, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.W. (1998) The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol., 8, 101–106. [DOI] [PubMed] [Google Scholar]
- Konno T., Murata,K. and Nagayama,K. (1999) Amyloid-like aggregates of a plant protein: a case of a sweet-tasting protein, monellin. FEBS Lett., 454, 122–126. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Liu Y.S., Hart,P.J., Schlunegger,M.P. and Eisenberg,D. (1998) The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1 Å resolution. Proc. Natl Acad. Sci. USA, 95, 3437–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkowski J., Bujacz,G., Boque,L., Domaille,P.J., Handel,T.M. and Wlodawer,A. (1997) The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions. Nature Struct. Biol., 4, 64–69. [DOI] [PubMed] [Google Scholar]
- Martin J.R., Craven,C.J., Jerala,R., Kroonzitko,L., Zerovnik,E., Turk,V. and Waltho,J.P. (1995) The 3-dimensional solution structure of human stefin A. J. Mol. Biol., 246, 331–343. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) RASTER3D version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. (1993) Sweet-tasting protein monellin is related to the cystatin family of thiol proteinase inhibitors. J. Mol. Biol., 230, 689–694. [DOI] [PubMed] [Google Scholar]
- Nilges M. (1997) Ambiguous distance data in the calculation of NMR structures. Fold. Des., 2, S53–S57. [DOI] [PubMed] [Google Scholar]
- Otting G., Senn,H., Wagner,G. and Wuthrich,K. (1986) Editing of 2D 1H-NMR spectra using X half-filters—combined use with residue-selective 15N labeling of proteins. J. Magn. Reson., 70, 500–505. [Google Scholar]
- Parker M.J., Spencer,J. and Clarke,A.R. (1995) An integrated kinetic-analysis of intermediates and transition states in protein folding reactions. J. Mol. Biol., 253, 771–786. [DOI] [PubMed] [Google Scholar]
- Purich D.L. and Allison,R.D. (2000) Handbook of Biochemical Kinetics. Academic Press, San Diego, CA, pp. 213–214.
- Rousseau F., Schymkowitz,J.W.H., Wilkinson,H.R. and Itzhaki,L.S. (2001) Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc. Natl Acad. Sci. USA, 98, 5596–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H.R. (2001) Cryo electron microscopy of amyloid fibrils. FASEB J., in press. [DOI] [PubMed] [Google Scholar]
- Salzmann M., Wider,G., Pervushin,K., Senn,H. and Wütrich,K. (1999) TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J. Am. Chem. Soc., 121, 844–848. [Google Scholar]
- Schlunegger M.P., Bennett,M.J. and Eisenberg,D. (1997) Conformational changes and disease—serpins, prions and Alzheimer’s. Adv. Protein Chem., 50, 61–122. [DOI] [PubMed] [Google Scholar]
- Serio T.R., Cashikar,A.G., Kowal,A.S., Sawicki,G.J., Moslehi,J.J., Serpell,L., Arnsdorf,M.F. and Lindquist,S.L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science, 289, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Staniforth R.A., Dean,J.L.E., Zhong,Q., Zerovnik,E., Clarke,A.R. and Waltho,J.P. (2000) The major transition state in folding need not involve the immobilization of side chains. Proc. Natl Acad. Sci. USA, 97, 5790–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs M.T., Laber,B., Bode,W., Huber,R., Jerala,R., Lenarcic,B. and Turk,V. (1990) The refined 2.4 Å X-ray crystal structure of recombinant human stefin B in complex with the cyteine proteinase papain—a novel type of proteinase–inhibitor interaction. EMBO J., 9, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M. and Blake,C. (1997) The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv. Protein Chem., 50, 123–159. [DOI] [PubMed] [Google Scholar]
- Sunde M., Serpell,L.C., Bartlam,M., Fraser,P.E., Pepys,M.B. and Blake,C.C.F. (1997) Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol., 273, 729–739. [DOI] [PubMed] [Google Scholar]
- Venters R.A., Huang,C.C., Farmer,B.T., Trolard,R., Spicer,L.D. and Fierke,C.A. (1995) High level 2H/13C/15N labeling of proteins for NMR studies. J. Biomol. NMR, 5, 339–344. [DOI] [PubMed] [Google Scholar]
- Walsh D.M., Lomakin,A., Benedek,G.B., Condron,M.M. and Teplow,D.B. (1997) Amyloid β-protein fibrillogenesis—detection of a protofibrillar intermediate. J. Biol. Chem., 272, 22364–22372. [DOI] [PubMed] [Google Scholar]
- Zegers I., Deswarte,J. and Wyns,L. (1999) Trimeric domain-swapped barnase. Proc. Natl Acad. Sci. USA, 96, 818–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerovnik E., Jerala,R., Kroonzitko,L., Turk,V. and Lohner,K. (1997) Characterization of the equilibrium intermediates in acid denaturation of human stefin B. Eur. J. Biochem., 245, 364–372. [DOI] [PubMed] [Google Scholar]