Abstract
The infectious component of hepatitis B (HB) virus (HBV), the Dane particle, has a diameter of ≈44 nm and consists of a double-layered capsid particle enclosing a circular, incomplete double-stranded DNA genome. The outer capsid layer is formed from the HB surface antigen (HBsAg) and lipid, whereas the inner layer is formed from the HB core Ag assembled into an icosahedral structure. During chronic infection HBsAg is expressed in large excess as noninfectious quasispherical particles and tubules with ≈22-nm diameter. Here, we report cryo-EM reconstructions of spherical HBsAg particles at ≈12-Å resolution. We show that the particles possess different diameters and have separated them into two predominant populations, both of which have octahedral symmetry. Despite their differing diameters, the two forms of the particle have the same mass and are built through conformational switching of the same building block, a dimer of HBsAg. We propose that this conformational switching, combined with interactions with the underlying core, leads to the formation of HBV Dane particles of different sizes, dictated by the symmetry of the icosahedral core.
Keywords: cryo-EM, subviral particle, virus structure
Hepatitis B (HB) virus (HBV) is a major cause of both acute and chronic liver disease. It is estimated that there are ≈350 million carriers of the virus, and a high proportion of these will develop serious liver diseases, including hepatocellular carcinoma. The double-layered mature infectious particle, known as the Dane particle, is an icosahedral structure comprising an inner core formed from 180 or 240 copies of the HB core antigen (HBcAg) arranged with triangulation (T) numbers 3 or 4 and an outer proteolipid HB surface Ag (HBsAg) layer. The HBsAg envelope glycoprotein of HBV occurs in three forms, denoted by L (large), M (medium) and S (small), which form a nested set of products sharing a common C-terminal domain, the SHBsAg polypeptide. The SHBsAg itself has a molecular mass of ≈24 kDa and is predicted to be intimately associated with lipid, having four or five membrane-spanning helices, and is found in two forms, glycosylated and unglycosylated, in approximately equal amounts. SHBsAg glycosylation is a single biantennary glycan at Asn-146 and raises its molecular mass to ≈27 kDa (1, 2). About 80% of HBsAg is in the S form, which assembles with M and L forms into both the outer surface of the Dane particle and smaller subviral particles. These HBsAg subviral particles, known as 22-nm particles because of their diameter, are stabilized by disulfide bonds (3).
HBsAg particles greatly outnumber mature virus particles and presumably act as decoys for the immune system. Although the 22-nm particles are predominately SHBsAg, tubular assemblies, also observed in the serum of chronic carriers, tend to be richer in LHBsAg. Because HBV subviral particles share important immunological determinants with mature virus, they can be used as effective immunogens. Indeed, expression of SHBsAg particles in yeast in the 1980s led to the development of the first recombinant human vaccine (4). Although these particles lack preS1 sequences, which are thought to contain the receptor-binding determinants of the virus, they are still effective in controlling HBV infection, and hence a common human cancer, and constitute the standard HB vaccine.
Previous studies of HBsAg particle structure have used visual comparison of electron micrographs to compare the morphology of SHBsAg particles isolated from recombinant yeast and the plasma of an HBV-infected human (5). It appears from this work that particles differ in size depending on their source and sample preparation, that they are empty, that their projection images are ovoid in some orientations, and that the surfaces of the particles contain small pores. We have used cryo-EM and 3D image reconstruction to investigate SHBsAg particle morphology in more detail. The SHBsAg used was expressed in a transgenic mouse (mSHBsAg), from which spherical particles similar to those purified from human serum or yeast were isolated. Like the previously studied forms of SHBsAg, the mSHBsAg particles showed two different size populations (1, 6). We used iterative cross-correlation against reference images to separate the whole dataset into small- and large-sized subsets and determined their symmetry by real-space averaging procedures and self-correlation analysis of the reconstructions (see Materials and Methods and Supporting Text and Figs. 5, 6, and 8, which are published as supporting information on the PNAS web site).
Materials and Methods
Sample Preparation. Mice of lineage TRE-HBs15 were crossed with those of lineage LT2. Lineage TRE-HBs15 contain the HBsAg coding region from the Galibert ayw hepatitis clone (7) under the control of a CMV promoter transcriptionally silent in the absence of a tetracycline regulatable transactivator (tTA). This transactivator is provided by the second lineage of mice. Lineage LT2 expresses tTA under the control of a liver-specific promoter (8). In the absence of doxycycline the double-transgenic mice produce HBsAg at a blood concentration of ≈10 mg/ml. After cross-breeding of the two lineages, the double-transgenic mice (25% of progeny) were selected and the highest expressors of HBsAg were used to obtain serum. One milliliter of such transgenic mouse serum was used to purify HBsAg particles by affinity chromatography. Anti-HBsAg IgG2 mAbs were purified from mouse ascites fluid on a protein A column and subsequently coupled to CNBr-activated Sepharose to produce a 5-ml affinity column. The transgenic mouse serum was loaded onto the PBS-washed column, followed by the addition of 1 ml of sodium phosphate buffer (pH 7.2). After 2-h incubation, the column was washed with 20 vol of sodium phosphate buffer (pH 7.2). mHBsAg particles were eluted in high salt by using 4 ml of phosphate buffer (pH 7.2) with 3.8 M MgCl2.
EM. Images of mSHBsAg and cAg particles were collected by using a 200-kV CM200 FEG electron microscope (FEI, Eindhoven, The Netherlands) under low-dose conditions at a magnification of 38,000×. Samples were cooled by plunging into liquid ethane and were visualized at a range of defocus values on holey carbon grids in vitreous ice. Micrographs showing no evidence of drift or astigmatism were digitized by using a PowerLook 3000 scanner (Umax UK, Milton Keynes, U.K.) on an 8.322-μm raster, corresponding to a 2.19-Å raster at the specimen. The defocus of each micrograph was determined from its radially averaged incoherent power spectrum average (9). Individual mSHBsAg particles were located interactively by using ximdisp software (mrc program suite) (10) and excised by using spider software (11).
There were two particular problems to be overcome in reconstructing mHBsAg: variable size and unknown symmetry. Because these problems cannot be considered separately, we undertook an iterative refinement of both. Size fractionation was initially by classification in imagic (12). This exercise yielded templates for cross-correlation in spider (11), allowing initial size fractionation. Initial size subsets were subjected to icosahedral reconstruction using the embl icosahedral programs (13) and, subsequently, to reconstruction using various symmetries in frealign (14) and gap (15). Direct comparison of maps, cross-correlation coefficient analysis of averaging procedures, and self-rotation functions were used to show that both large and small particles were octahedral (see Supporting Text and Fig. 6). Those reconstructions were then used again in size fractionation. Reconstructions were then computed by using frealign to 12.5-Å resolution. In a final iteration, maps were computed in spider (11) using Fourier terms to 12-Å resolution. The effective resolution of both maps is 15 Å when the 50% Fourier correlation criterion (16) is used and 12 Å when the less conservative criterion of Rosenthal and Henderson (17) is used. The map was scaled in reciprocal space to the Fourier transform of an atomic model, built for each of the small and large maps from two copies of bacteriorhodopsin placed within the asymmetric unit, that were then built into a full particle by the application of octahedral symmetry (see Fig. 9, which is published as supporting information on the PNAS web site). The hand of each of the reconstructions was determined absolutely by use of the method of Rosenthal and Henderson (17), with a -15° rotation of the sample holder between micrograph pairs (anticlockwise when viewed toward the microscope column). See Supporting Text for more details.
Performing Density Fits. Fitting of densities was done with gap (15), which also was used in the calculation of the volume of mSHBsAg particles to given thresholds (18).
Scanning Transmission EM (STEM). STEM was carried out in the Brookhaven National Laboratory STEM facility with tobacco mosaic virus (TMV) included as an internal control. Samples were prepared in different buffers (PBS/Hepes, pH 7.2-7.5) and for different dilutions of salt. The program pcmass (written by J. S. Wall, Brookhaven National Laboratory) was used to analyze the digital STEM images, always measuring masses for TMV and mHBsAg from the same image. The models used to measure masses were either a TMV mass per unit length model rod or a simple spherical model for the mHBsAg. For each buffer condition and sample dilution, multiple measurements were taken, and a mean and standard deviation were calculated for each of the TMV and mHBsAg datasets, a dataset being a collection of images taken with a particular dilution or buffer type. Details of the STEM operation and mass analysis were described in refs. 19 and 20.
Figures were drawn by using bobscript (21) and rendered in raster3d (22).
Results and Discussion
Structure Determination of mSHBsAg Particles. Although Crick and Watson predicted cubic symmetry in general for the capsid shells of viruses, allowing tetrahedral, octahedral, or icosahedral geometry of assembly (23), only icosahedral capsids have, in fact, been described. The natural assumption was, therefore, that SHBsAg particles are icosahedral. However, we were unable to see any evidence for fivefold symmetry in images of the particles, and attempts at icosahedral reconstruction were unsatisfactory (see Materials and Methods). Initial classification of the images suggested the existence of two populations of differently sized particles (see Fig. 5). The problem of reliably separating these from each other is bound up with the problem of determining their symmetry. We, therefore, took an iterative, coupled approach to size fractionation and symmetry determination (see Materials and Methods and Supporting Text and Figs. 6 and 7, which are published as supporting information on the PNAS web site). Our assumption that the particles belong to two discrete populations is consistent with the final statistical analysis (see Fig. 8), although we cannot rule out the presence of intermediate structures.
The icosahedron is one of the five Platonic solids, which allow a single kind of asymmetric unit to self-assemble into a quasi-spherical structure of defined size (24). The simplest Platonic symmetry is tetrahedral, which underlies the other cubic symmetries. We, therefore, simply used tetrahedral symmetry in determining initial reconstructions and then systematically tested for higher symmetry (see Materials and Methods). As a control, we used the same procedures with images of the HBV core particle. By using this approach, the core was unambiguously identified as icosahedral, whereas both small- and large-sized mSHBsAg particle subsets showed octahedral symmetry (see Supporting Text). This symmetry is confirmed by the high-resolution refinement of the structures, the presence of common density features between large and small particles, the decided handedness present in the maps, and the fact that a reconstruction of mSHBsAg using only the underlying tetrahedral symmetry is very similar to that obtained using octahedral symmetry but quite unlike that obtained with icosahedral symmetry. The total dataset consisted of 1,793 projection images, obtained at 26 different defocuses; these were separated into populations of 911 small particles and 882 large particles. The resolution of both the small- and large-particle reconstructions is ≈12 Å (see Materials and Methods). Electron-density maps were contoured with reference to density gradients in the maps. There was effectively no difference in the appearance of the particle surface among 1σ, 2σ, and 3σ contour levels (except that at 1σ, additional noise appeared in the background). One problem in EM image analysis is the determination of an absolute hand for reconstructions (17). In Supporting Text and Fig. 10, which are published as supporting information on the PNAS web site, we show hand-determination for mSHBsAg particles, which also demonstrates a significant signal-to-noise ratio in the data for both size particles.
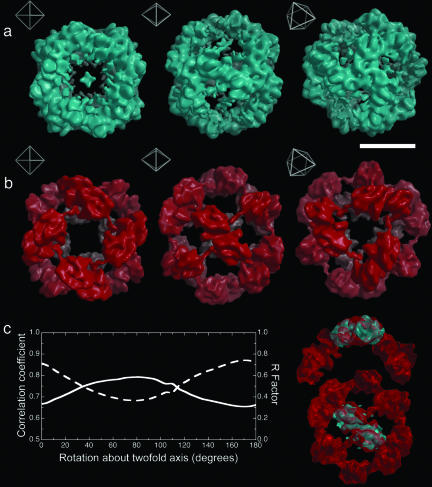
Overall Morphology of SHBsAg Reconstructions. Fig. 1. shows the reconstructions of the small and large mSHBsAg particles. Each particle is shown in three orientations to illustrate fully their structure and the relationship of the molecular structure to the octahedral symmetry. The electron density in the two particles appears to have distinctively different organizations, thus although both size particles feature strong twofold density at the twofold octahedral axes and a void around the fourfold axes, the smaller particle features strong three-legged density at the octahedral threefold axes, whereas the larger particles have a threefold hole. The arrangement of density in the larger particle also is noticeably looser than in the smaller particle, with well defined arms reaching between the twofold axes.
Fig. 1.
mHBsAg particle reconstructions. (a) Surface-rendered views (threshold 2.5 σ) of the small mHBsAg particle reconstruction at 12-Å resolution. Insets show the orientation of each image with respect to the octahedral symmetry of the particle. (Scale bar, 100 Å.) (b) As a for the large mHBsAg particle. (c) A plot of the real-space correlation coefficient (dashed line) and R factor (solid line) between the twofold blocks of the large and small reconstructions as they are rotated with respect to one another. The peak in correlation coefficient and trough in R factor indicate a rotation of ≈10°. On the right the twofold block from the small reconstruction (cyan) is shown fitted in this manner to that from the large reconstruction.
The clear similarity in the structure of the basic octahedral building block, a twofold symmetric piece of structure (which we term the twofold block), centered on the octahedral twofold axes, and therefore comprising two octahedral asymmetric units, suggests that the differently sized particles are assembled from the same number of protein subunits, arranged in a closely related fashion. Using this model as a starting point, we can define the relationship between the two structures in an objective, quantitative way by using density correlation or density consistency as a metric. The first degree of freedom to consider is the radial position of the twofold blocks along the octahedral twofold axes. On translation of the corresponding density for the smaller particles outward, there is an optimum match to the larger particle with a radial expansion of 10 Å. The second degree of freedom is the rotation of the twofold block around the octahedral twofold axis. The results for this are shown in Fig. 1c, suggesting that, in line with visual inspection, there is an ≈10° clockwise rotation of the building block in going from the small to large particles. The good correlation in density achieved by application of these two operations confirms the similarity apparent on visual inspection of Fig. 1 a and b.
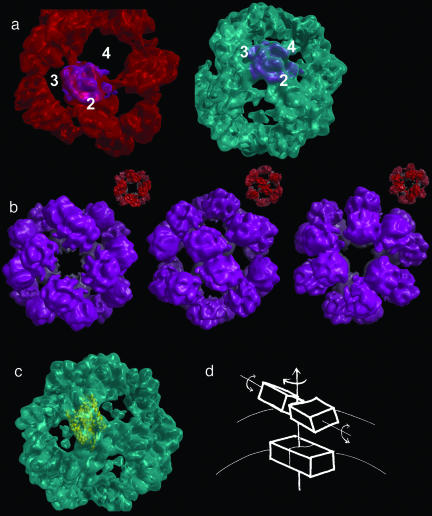
We were able further to improve the fits of the two asymmetric units over the fit found in this superposition of the building block by bisecting it and fitting each asymmetric unit alone (Fig. 2a). This exercise indicated that there is a rotation of up to 36° in the mutual arrangements of the asymmetric units within the building block of the small, compared with the large particle, equating to a shear at the twofold axis of the octahedron (in other words, a rotation about an axis perpendicular to the twofold axis itself). Because this rotation is about the major axis of the structure, the accuracy with which we can determine the magnitude of the shear is rather low; however, it does seem that there is some internal rearrangement within the twofold block. To underscore the improved quality of this fit of single asymmetric units to that of the twofold building blocks of the structures, we reassembled the large particle by applying octahedral symmetry to the small asymmetric unit fitted to the large-particle map (Fig. 2b). The overall relation between the position and orientation of the protein building block in the small and large particles is shown in Fig. 2d.
Fig. 2.
Comparison of asymmetric units. (a Left) The asymmetric unit of the small reconstruction (magenta) fitted to the large reconstruction (red). The numbers 2, 3, and 4 refer to the 2-, 3-, and 4-fold symmetry axes, respectively. (Right) The same feature in the same orientation and its native environment in the small reconstruction (cyan). (b) Theoretical reassembly of the large reconstruction from the small asymmetric unit fitted to the large one and shown in a. (Insets) The actual large reconstruction is shown in red for comparison. Three images are shown corresponding to the three orientations in Fig. 1 a and b. (c) The asymmetric unit of the small reconstruction fitted with two bacteriorhodopsin molecules (yellow ribbons). (d) The figure shows schematically how the twofold block for the small reconstruction can be superimposed on the large one by first translating by 10 Å, then rotating by 10°, and finally by applying a shear of ≈36° perpendicular to the twofold axis of the particle.
Composition of the Octahedral Asymmetric Unit. At ≈12-Å resolution, it is not possible to determine protein secondary structure or define precisely the boundary between closely associated protein subunits. However, in addition to the protein component, it has been established (25) that the particles contain some 40% by mass of lipid and also sugar (the latter arising from the glycosylation of approximately half of the subunits). We cannot unambiguously position either of these nonpolypeptide components of the structure. In particular, there is no clear density for a lipid bilayer, which supports the proposal from Satoh et al. (26) that, unlike in the mature virus particles, the small Ag particles contain the lipid in an unusual arrangement very closely intercalated with the protein. The glycosylation will be discussed further below. At the resolution that we have achieved, the most we can say is that the common core structural unit from which the mSHBsAg particle is assembled is a block that, due to its high density and globular form, is almost certainly made up mostly of protein, strongly associated into dimers around the octahedral twofold axis, with an apparently sigmoidal interface. One important question is, what is the molecular composition of the morphological unit that forms the protein building block of both the large and small particles? An established way to address this question is to superimpose a protein of similar molecular mass (24 kDa) and secondary structure composition (helical) (27-29). Using this approach, we have fitted bacteriorhodopsin, a 24-kDa helical protein, within the asymmetric portions of the reconstructions. The electron density in the octahedral asymmetric unit is sufficient to accommodate mass equivalent to two bacteriorhodopsin molecules. This fit suggests that the octahedral asymmetric unit common to the larger and smaller particles is a dimer of SHBsAg (Fig. 2c), i.e., the building block centered on the octahedral twofold axes in both particle sizes is composed of four SHBsAg subunits.
We have confirmed this composition by direct quantitative analysis of the map densities. Using a 2σ threshold, the electron-density volumes are [determined with gap (15, 18)] 1.82 × 106Å3 for the small particles and 1.80 × 106Å3 for the large particles, values that are indistinguishable within experimental error. Using the known partial specific volumes of protein and lipid (0.73 and 0.98 ml/g, respectively) and ratio of protein to lipid in the particle (60:40; ref. 25), this density is accounted for by 50 subunits in the large particles (estimated mass, 2.16 MDa) and 51 in the small particles (estimated mass, 2.18 MDa) i.e., two protein subunits in both octahedral asymmetric units. These data rule out a single subunit being the building block, and unless there are large regions of disorder, for which we have no evidence, three subunits per octahedral asymmetric unit also is unlikely.
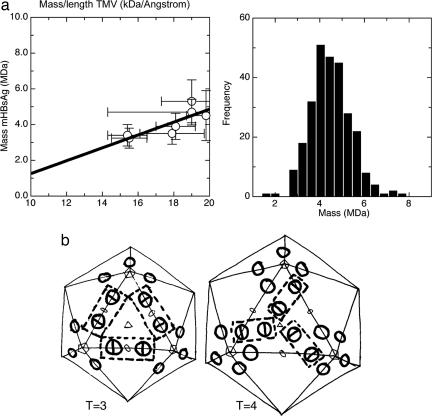
To achieve an independent confirmation of the protein stoichiometry, we used STEM to estimate the masses of the mHBsAg particles (see Materials and Methods). Inclusion of a standard sample (TMV) and multiple measurements with different buffers and salt concentrations allowed us to put these measurements on an absolute scale (Fig. 3a shows the expected linear regression). The mass population of the particles was unimodal, indicating a single type of mHBsAg particle with mass 2.38 MDa, in good agreement with the values estimated from the reconstruction (Fig. 3a). Assuming 50% glycosylation, the total theoretical protein mass would be 2.04 MDa, using the reported weight distribution between protein and lipid in the particles (25) and a dimer of two protein subunits in the octahedral asymmetric unit, which is in close agreement with the STEM measurements. Each one of our arguments leads to the conclusion that there is a dimer in the octahedral asymmetric unit; however, for neither the large nor the small particles is there a clear local twofold axis within the asymmetric unit of the particle. This absence may simply reflect the limited resolution of the analysis and the fact that the two subunits are very closely associated. However, because the environment of the two subunits within the octahedral asymmetric unit will be different, conformational differences between the subunits may contribute to the asymmetry of the electron density. It also is possible that this asymmetry is correlated with the observation that the particles tend to assemble with 50% of protein subunits glycosylated and that the two subunits may, therefore, be chemically as well as conformation-ally different. That is, the building block of the mHBsAg particle may be a heterodimer of one 24-kDa unglycosylated protein subunit and one 27-kDa glycosylated subunit.
Fig. 3.
Particle stoichiometry. (a) STEM analysis of the mHBsAg particles. The left-hand graph shows a plot of observed mass/length of TMV against observed mass of mHBsAg for a series of different sample dilutions and buffer conditions. The linear regression of observed masses with respect to one another allows us to control for variable backgrounds arising from finite salt and buffer contributions to scatter. For the known mass/length of TMV of 13.1 kDa/Å the mass of mHBsAg is 2.38 MDa. The right-hand graph shows the monomodal frequency of occurrence of different mHBsAg masses for one of the STEM datasets. (b) Schematic representation of possible T = 3 and T = 4 models for the Dane particle surface based on the assembly of sAg molecules observed for the small and large sAg particles. The T = 3 core is coated by a T = 3 surface where the twofold block (shown in broken line) binds across two HBcAg spikes (shown as circles) and is arranged in a manner similar to that found in the large sAg particle. The T = 4 core is coated by a T = 4 surface where the sAg molecules are arranged as observed in the small sAg particle.
Assembly of HBsAg Particles. We suggest that the final size of the assembly is controlled by a switch in the conformation of the portions of the octahedral subunit that are involved in intersubunit contacts. This switching must occur at a very early stage in the assembly, most likely at the initial interaction of two HBsAg molecules, to form the octahedral asymmetric unit. In fact, most spherical virus particles are made up from >60 identical copies of one or more distinct protein subunits, and such conformational switching is the most widely used assembly mechanism for such systems (15, 30, 31). It seems that the SHBsAg particles extend this model by forming pleomorphic octahedral, as well as (in the intact virus) pleomorphic icosahedral assemblies. One other subviral octahedral assembly was described in ref. 32, that formed by polyomavirus, where the pentameric capsomere of the virus can assemble into T = 1 or T = 7 icosahedral structures, or an octahedral structure consisting of 24 pentamers. Polyomavirus shows a level of pleomorphism similar to that of HBV: both form tubular structures as well as octahedral and icosahedral assemblies (33). Polyomavirus VP1 is able to assemble into these different structures as a result of changes in the way the pentamers link together, rather than changes in the basic building block of the assemblies (32, 33). The SHBsAg dimers seem to echo this principle.
Model for the Dane Particle Surface. Dane particles can occur in two sizes, according to the size of the underlying core assembly, which is predominantly T = 4 but also is found in T = 3 structures. From the known size of the core particles, the known thickness of a eukaryotic membrane, and the size of the sAg protein building block seen in our reconstructions (which we assume to interact directly with the tip of the dimeric spikes of the core, and in view of its antigenic properties, we assume to be oriented similarly in the Dane and HBsAg particles), we can calculate the likely diameter of the two classes of Dane particles to be ≈470 Å and 500 Å. Given the diameters of the small and large HBsAg particles based on our reconstructions, namely 200 and 230 Å, we can calculate, assuming a consistent packing density on the particle surface, the likely number of sAg protein subunits in the two classes of Dane particles. For the T = 4 particle, the numbers are 210-270 subunits for the packing densities seen in the two HBsAg particles. We assume, therefore, that there is a 1:1 stoichiometry between the core subunits and the HBsAg subunits. This ratio is consistent with the finding that one peptidomimetic of the large sAg will bind per cAg molecule (34). The size of the HBsAg building block, which contains four protein subunits, and the distance between the dimeric spikes of the cAg are consistent with the organization of the HBsAg shown in Fig. 3b. Interestingly, the nature of the T = 3 and T = 4 lattices is such that the block contacts in these models recapitulate the contacts seen in the large and small HBsAg particles, respectively. Thus, it appears that although the assembly of the Dane particle surface requires the inner core to act as a template to form icosahedral particles, the nature of the conformational switching required to form different-sized Dane particles also may be responsible for the size heterogeneity observed in the small octahedral assemblies of HBsAg.
Conclusion
Cryo-EM has revealed, unexpectedly, that HBV forms octahedral subviral particles rather than icosahedral ones, probably composed of 48 protein subunits. Remarkably, the subunits are capable of self-assembling to form two distinct octahedral structures and also are capable of assembling to form a larger icosahedral arrangement as found in the mature virus particles (classically quoted from EM studies as 44 nm in diameter, compared with 22 nm in diameter). The pleomorphic nature of the interactions between the dimers of dimers that modulate the size of the final mHBsAg particle presumably underlie also the switch to icosahedral assembly that is required to form the complete HBV particles. This mechanism appears to be an extension of the conformational switching mechanism recognized as the cornerstone to the assembly of many icosahedral virus particles (15, 30-32). Finally, the antigenic properties of these particles, many millions of doses of which are administered every year as a recombinant HB vaccine, confirm that the presentation of surface epitopes on the octahedral structures mimics that in the intact virus. This fact is in line with our model in which conformational switching, rather than major rearrangements in molecular orientation, controls the different assembly pathways that lead to either octahedral or icosahedral particles.
Supplementary Material
Acknowledgments
We thank Dr. Jenny Waters (Imperial College, London) for supplying the anti-HBsAg mAb; Dr. Joseph S. Wall for the pcmass program; and Drs. Peter Rosenthal and Richard Henderson for providing advice and help with the use of frealign. This work was supported by National Institutes of Health Grant P41-RR01777; the Department of Energy's Office of Biological and Environmental Research; and the Medical Research Council, United Kingdom. The Brookhaven National Laboratory STEM facility is a National Institutes of Health-supported resource center. D.I.S. is an Medical Research Council, United Kingdom, Research Professor, and R.J.C.G. is a Royal Society University Research Fellow.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HB, hepatitis B; HBV, HB virus; Ag, antigen; HBcAg, HB core Ag; HBsAg, HB surface Ag; LHBsAg, long HBsAg; MHBsAg, medium HBsAg; SHBsAg, small HBsAg; mSHBsAg, mouse SHBsAg; STEM, scanning transmission EM; TMV, tobacco mosaic virus.
Data deposition: The reconstructions of HBSsAg particles reported herein have been deposited in the Macromolecular Structure Database, www.ebi.ac.uk/msd/index.html (accession nos. EMD-1158 and EMD-1159).
References
- 1.Nassal. M. (1996) Curr. Top. Microbiol. Immunol. 214, 297-337. [DOI] [PubMed] [Google Scholar]
- 2.Mehta, A., Lu, X., Block, T. M., Blumberg, B. S. & Dwek, R. A. (1997) Proc. Natl. Acad. Sci. USA 94, 1822-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wounderlich, G. & Bruss, V. (1996) Arch. Virol. 141, 1191-1205. [DOI] [PubMed] [Google Scholar]
- 4.McAleer, W. J., Buynak, E. B., Maigetter, R. Z., Wampler, D. E., Miller, W. J. & Hilleman, M. R. (1984) Nature 307, 178-180. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi, M., Sugahara, K., Shiosaki, K., Mizokami, H. & Takeo, K. (1998) FEMS Microbiol. Lett. 165, 363-367. [DOI] [PubMed] [Google Scholar]
- 6.Blumberg, B. S., Alter, H. J. & Visnich, S. (1965) J. Am. Med. Assoc. 191, 541-546. [DOI] [PubMed] [Google Scholar]
- 7.Galibert, F., Mandart, E., Fitoussi, F., Tiollais, P. & Charnay, P. (1979) Nature 281, 646-650. [DOI] [PubMed] [Google Scholar]
- 8.Kistner, A., Gossen, M., Zimmermann, F., Jerecic, J., Ullmer, C., Lubbert, H. & Bujard, H. (1996) Proc. Natl. Acad. Sci. USA 93, 10933-10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferlenghi, I., Gowen, B., de Haas, F., Mancini, E. J., Garoff, H., Sjoberg, M. & Fuller, S. D. (1998) J. Mol. Biol. 283, 71-81. [DOI] [PubMed] [Google Scholar]
- 10.Crowther, R. A., Henderson, R. & Smith, J. M. (1996) J. Struct. Biol. 116, 9-16. [DOI] [PubMed] [Google Scholar]
- 11.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. & Leith, A. (1996) J. Struct. Biol. 116, 190-199. [DOI] [PubMed] [Google Scholar]
- 12.van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. (1996) J. Struct. Biol. 116, 17-24. [DOI] [PubMed] [Google Scholar]
- 13.Fuller, S. D., Butcher, S. J., Cheng, R. H. & Baker, T. S. (1996) J. Struct. Biol. 116, 48-55. [DOI] [PubMed] [Google Scholar]
- 14.Grigorieff, N. (1998) J. Mol. Biol. 277, 1033-1046. [DOI] [PubMed] [Google Scholar]
- 15.Grimes, J. M., Burroughs, J. N., Gouet, P., Diprose, J. M., Malby, R., Zientara, S., Mertens, P. P. & Stuart, D. I. (1998) Nature 395, 470-478. [DOI] [PubMed] [Google Scholar]
- 16.Gabashvili, I. S., Agrawal, R. K., Spahn, C. M. T., Grassucci, R. A., Svergun, D. I., Frank, J. & Penczek, P. (2000) Cell 100, 537-549. [DOI] [PubMed] [Google Scholar]
- 17.Rosenthal, P. B. & Henderson, R. (2003) J. Mol. Biol. 333, 721-745. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, R. J. C., Fucini, P., Connell, S., Nierhaus, K. H., Robinson, C. V., Dobson, C. M. & Stuart, D. I. (2004) Mol. Cell 14, 57-66. [DOI] [PubMed] [Google Scholar]
- 19.Wall, J. S., Hainfeld, J. F. & Simon, M. N. (1998) Methods Cell Biol. 53, 139-166. [DOI] [PubMed] [Google Scholar]
- 20.Wall, J. S. & Simon, M. N. (2001) Methods Mol. Biol. 148, 589-601. [DOI] [PubMed] [Google Scholar]
- 21.Esnouf, R. M. (1999) Acta Crystallogr. D 55, 938-940. [DOI] [PubMed] [Google Scholar]
- 22.Merritt E. A. & Murphy M. E. P. (1994) Acta Crystallogr. D 50, 869-873. [DOI] [PubMed] [Google Scholar]
- 23.Crick, F. R. & Watson, J. C. (1956) Nature 177, 474-475. [DOI] [PubMed] [Google Scholar]
- 24.Klug, A. (1969) Point Groups and the Design of Aggregates, Proceedings of the Nobel Symposium, eds. Engström, A. & Strandberg, B. (Nobel Foundation, Stockholm), Vol. 11, pp. 425-436. [Google Scholar]
- 25.Diminsky, D., Schirmbeck, R., Reimann, J. & Barenholz, Y. (1997) Vaccine 15, 637-647. [DOI] [PubMed] [Google Scholar]
- 26.Satoh, O., Imai, H., Yoneyama, T., Miyamura, T., Utsumi, H., Inoue, K. & Umeda, M. (2000) J. Biochem. 127, 543-550. [DOI] [PubMed] [Google Scholar]
- 27.Beckmann, R., Spahn, C. M., Eswar, N., Helmers, J., Penczek, P. A., Sali A., Frank, J. & Blobel, G. (2001) Cell 107, 361-372. [DOI] [PubMed] [Google Scholar]
- 28.Sonveaux, N., Conrath, K., Capiau, C., Brasseur, R., Goormaghtigh, E. & Ruysschaert, J. M. (1994) J. Biol. Chem. 269, 25637-25645. [PubMed] [Google Scholar]
- 29.Lambert, C. & Prange, R. (2001) J. Biol. Chem. 276, 22265-22272. [DOI] [PubMed] [Google Scholar]
- 30.Liddington, R. C., Yan, Y., Moulai, J., Sahli, R., Benjamin, T. L. & Harrison, S. C. (1991) Nature 354, 278-284. [DOI] [PubMed] [Google Scholar]
- 31.Conway, J. F., Wikoff, W. R., Cheng, N., Duda, R. L., Hendrix, R. W., Johnson, J. E. & Steven, A. C. (2001) Science 292, 744-748. [DOI] [PubMed] [Google Scholar]
- 32.Salunke, D. M., Caspar, D. L. D. & Garcea, R. L. (1989) Biophys. J. 56, 887-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker, T. S., Caspar, D. L. D. & Murakami, W. T. (1983) Nature 303, 446-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böttcher, B., Tsuji, N., Takahashi, H., Dyson, M. R., Zhao, S., Crowther, R. A. & Murray, K. (1998) EMBO J. 17, 6839-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.