Abstract
DNA glycosylases initiate base excision repair (BER) through the generation of potentially harmful abasic sites (AP sites) in DNA. Human thymine-DNA glycosylase (TDG) is a mismatch-specific uracil/thymine-DNA glycosylase with an implicated function in the restoration of G·C base pairs at sites of cytosine or 5-methylcytosine deamination. The rate-limiting step in the action of TDG in vitro is its dissociation from the product AP site, suggesting the existence of a specific enzyme release mechanism in vivo. We show here that TDG interacts with and is covalently modified by the ubiquitin-like proteins SUMO-1 and SUMO-2/3. SUMO conjugation dramatically reduces the DNA substrate and AP site binding affinity of TDG, and this is associated with a significant increase in enzymatic turnover in reactions with a G·U substrate and the loss of G·T processing activity. Sumoylation also potentiates the stimulatory effect of APE1 on TDG. These observations implicate a function of sumoylation in the controlled dissociation of TDG from the AP site and open up novel perspectives for the understanding of the molecular mechanisms coordinating the early steps of BER.
Keywords: DNA repair/SUMO conjugation/thymine-DNA glycosylase
Introduction
DNA bases are inherently unstable chemical structures that are susceptible to damage by reactive agents of endogenous or environmental origin. To minimize the mutagenic and cytotoxic consequences of such damage, nature has evolved a seemingly simple DNA repair system for the selective replacement of irregular bases. Base excision repair (BER) is initiated by damage-specific DNA glycosylases that recognize and remove the aberrant bases. DNA glycosylases can be grouped into mono- and bifunctional enzymes according to their reaction mechanisms (reviewed in Schärer and Jiricny, 2001). Mono functional glycosylases hydrolyse the N-glycosidic bond connecting the base with the deoxyribose moiety of the nucleoside through a nucleophilic attack mediated by an activated water molecule. This generates an abasic site (AP site) intermediate for further processing by a sequence of damage-general reactions. These involve DNA strand incision 5′ to the AP site by an (AP)-endonuclease, trimming of the resulting 5′ end by an (AP)-lyase or an endonuclease, DNA resynthesis by a DNA polymerase and strand sealing by a DNA ligase (reviewed in Nilsen and Krokan, 2001).
BER is associated with the generation of potentially harmful AP sites. These unstable intermediates can give rise to DNA strand breaks or interfere with DNA and RNA metabolism, all reflected in their cytotoxicity and mutagenicity (Lindahl, 1990). Thus, for BER to be beneficial to cells, relay mechanisms must operate to coordinate the dissociation of DNA glycosylases from the product AP sites with the recruitment of the downstream acting (AP)-endonuclease. Such a ‘passing the baton’ concept for BER was proposed on the basis of structural and biochemical considerations involving the uracil-DNA glycosylase, human (AP)-endonuclease (APE1/HAP1) and DNA polymerase β (Lindahl and Wood, 1999; Mol et al., 2000; Wilson and Kunkel, 2000). The model invokes that an upstream acting enzyme imposes a specific distortion on a DNA substrate that is then recognized and bound by a downstream factor, thereby actively displacing the former enzyme. This structure-based concept is particularly attractive for the damage-general steps of BER where it finds additional support by evidence for physical interactions between the key enzymes involved (Caldecott et al., 1994; Kubota et al., 1996; Prasad et al., 1996; Bennett et al., 1997). However, regarding the transition from the damage-specific to the damage-general steps of BER, the model seems inadequate; it fails to accommodate the biochemical and structural heterogeneity of the many different DNA glycosylases. Thus, alternative mechanisms for a coordinated handover of the AP site from the DNA glycosylase to the (AP)-endonuclease will have to be evaluated.
Human thymine-DNA glycosylase (TDG) is a monofunctional DNA glycosylase that excises thymine and uracil from G·T and G·U mismatched oligonucleotide substrates as well as 3,N4-ethenocytosine from double-stranded DNA. The implicated biological role of this glycosylase is thus the restoration of G·C base pairs at sites of cytosine or 5-methylcytosine deamination (G·U; G·T) or alkylation (G·εC) (reviewed in Hardeland et al., 2001). An interesting property of TDG is its inability to turn over in base release assays in vitro. When incubated with a G·T or a G·U substrate, it binds the mismatch and hydrolyses the thymine/uracil but then fails to dissociate from the resulting AP site (Waters et al., 1999; Hardeland et al., 2000). Although such product inhibition is not unusual among DNA glycosylases (Miao et al., 1998; Petronzelli et al., 2000; Hill et al., 2001; Nilsen et al., 2001), it seems particularly strong in the case of TDG. Crystal structure analyses of substrate-bound Mug, a conserved Escherichia coli homologue of TDG (Gallinari and Jiricny, 1996), showed that the enzyme establishes rigid hydrogen bonding interactions with the Watson–Crick face of the guanine opposite the AP site (Barrett et al., 1998, 1999). This mechanistic principle appears to apply also to the human enzyme (Hardeland et al., 2000) and most probably prevents its release from the product AP site. AP site binding can be viewed as a biologically important activity of DNA glycosylases, shielding the potentially harmful repair intermediate until the enzyme acting downstream in the pathway is in place to proceed with the repair process. Consistent with this are reports on the ability of human (AP)-endonuclease, APE1, to stimulate the enzymatic turnover of TDG (Waters et al., 1999) and other DNA glycosylases (Parikh et al., 1998; Hill et al., 2001; Nilsen et al., 2001; Vidal et al., 2001; Yang et al., 2001). However, at least in the cases of human TDG and OGG1, this stimulation is best explained by simple competition of the DNA glycosylase and the (AP)-endonuclease for the AP site (Vidal et al., 2001).
We set out to identify proteins that physically interact with human TDG and actively modulate its turnover in base release assays. Yeast two-hybrid screening revealed the human ubiquitin-like modifiers SUMO-1 and SUMO-3 as specific interaction partners of TDG. Three SUMO proteins have been identified in mammalian cells that, according to their amino acid sequence divergence, can be grouped into two subclasses, SUMO-1 and SUMO-2/3. All SUMOs are known to modify target proteins covalently by an enzymatic pathway analogous to ubiquitin conjugation. Unlike ubiquitylation, however, SUMO conjugation does not lead to protein degradation but provokes other, apparently target protein-specific effects, all modulating biological activities in different ways (reviewed in Melchior, 2000; Muller et al., 2001). We established that the SUMO proteins not only interact with but also covalently modify TDG in a reversible and mutually exclusive manner. SUMO conjugation induces a dramatic reduction in the DNA-binding affinity of the enzyme. This causes an enhanced turnover of TDG in base release assays with a G·U substrate and, at the same time, a complete loss of its G·T processing ability. Sumoylation also significantly potentiates the stimulatory effect of APE1 on the G·U processing efficiency of TDG. These consequences are consistent with a function for SUMO conjugation in the displacement of TDG from the product AP site.
Results
Physical interaction of human TDG with ubiquitin-like proteins
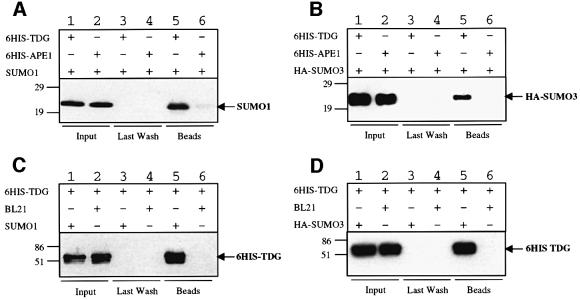
In a yeast two-hybrid screen for proteins interacting with human TDG, we repeatedly isolated clones from a HeLa two-hybrid library that encoded the small ubiquitin-like proteins SUMO-1 or SUMO-3. These interactions were specific by two-hybrid standards and were verified further by means of affinity pull-down experiments. Recombinant His6-tagged TDG protein immobilized on nickel affinity beads could specifically retrieve recombinant SUMO-1 (Figure 1A) and SUMO-3 (Figure 1B) proteins from extracts of E.coli cells. No binding of either of the SUMO proteins was observed in controls with APE1-coated beads. Vice versa, affinity beads coated with protein extracts from E.coli cells expressing either SUMO-1 or SUMO-3 precipitated recombinant TDG, while control beads covered with extract of vector control cells failed to do so (Figure 1C and D). Since these results were reproduced in experiments with purified SUMO proteins (data not shown), it is established that human TDG engages in a direct physical interaction with the ubiquitin-like proteins SUMO-1 and SUMO-3.
Fig. 1. Physical interaction of human TDG with SUMO-1 and SUMO-3. Co-precipitation of purified recombinant human SUMO-1 (A) and SUMO-3 (B) proteins with human TDG. Ni2+-NTA beads coated with 1 µg of His6-tagged human TDG protein (lanes 1, 3 and 5) or an equimolar amount of human APE1 (lanes 2, 4 and 6) were incubated with extracts of E.coli cells expressing either human SUMO-1 or HA-tagged human SUMO-3 protein. Input (2%), last wash (100%) and bead fractions (100%) were separated by 15% SDS–PAGE and the SUMO proteins detected by western blotting with antibodies against SUMO-1 (A) and the HA-tag (B). SUMO-1 and SUMO-3 precipitated with the TDG (lanes 5) but not with the APE1 beads (lanes 6). Co-precipitation of human TDG with SUMO-1 (C) and SUMO-3 (D) proteins. Affi-Gel®10 beads coated with E.coli BL21 extract containing either SUMO-1 or SUMO-3 (lanes 1, 3 and 5) or no recombinant protein as a control (BL21, lanes 2, 4 and 6) were incubated with recombinant human TDG. Western blotting with a TDG antibody revealed specific binding of TDG to SUMO-1 and SUMO-3 (lanes 5). No binding of TDG to control beads was detected (lanes 6). The input shown (lanes 1 and 2) corresponds to 10% of the total amount of TDG used in the experiment.
TDG is the target for modification by ubiquitin-like proteins
Since small ubiquitin-like proteins are known to modify other cellular proteins covalently, we wondered whether TDG is a target for SUMO attachment. Sumoylation is a highly dynamic, ATP-dependent and reversible process involving activating E1 (AOS1/UBA2) and conjugating E2 enzymes (UBC9) as well as deconjugating ULP isopeptidases (reviewed in Melchior, 2000; Muller et al., 2001). The modification is usually detectable as a mobility shift of the target protein in SDS–PAGE.
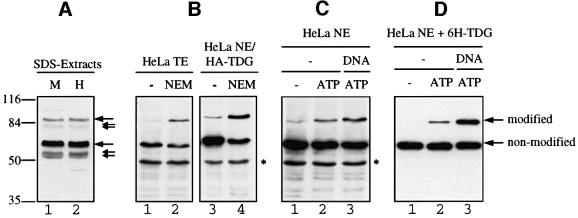
Western blotting of human MRC5 and HeLa cell lysates prepared by direct boiling in SDS buffer revealed the existence of at least two proteins cross-reacting with a TDG-specific polyclonal antibody, one migrating at ∼60 kDa and another at ∼86 kDa (Figure 2A). The predominant 60 kDa signal is known to represent endogenous TDG protein (Neddermann et al., 1996), but the identity of the 86 kDa protein was obscure. Western blotting of extracts from HeLa cells expressing a haemagglutinin (HA)-tagged version of human TDG produced the same pattern of cross-reacting proteins with both HA-specific and TDG-specific monoclonal antibodies, confirming that the 86 kDa protein represented a variant of endogenous TDG (Figure 2B). Additional signals at around 55 kDa were detected with the polyclonal antibody only. These may reflect TDG degradation in the extract (Neddermann and Jiricny, 1993; Neddermann et al., 1996) and/or products of shifted translation starts due to alternative splicing (Um et al., 1998). Correspondingly smaller species (81 kDa) also appeared to be associated with the 86 kDa form of TDG (Figure 2A). The same pattern of TDG variants was detectable in extracts from several different human cell lines (data not shown) and was also observed in extracts of mouse cells (Um et al., 1998).
Fig. 2. TDG modification in cell extracts. Extracts of MRC5 and HeLa cells were subjected to SDS–PAGE and western blot analysis with specific polyclonal (A) and monoclonal (B, C and D) antibodies against human TDG. (A) Analysis of 10 µl of MRC5 (M) and HeLa (H) extracts prepared by direct lysis of 107 cells in SDS buffer. Immunostaining revealed two major TDG-specific signals at ∼60 and ∼86 kDa (large arrows) and a few minor signals representing faster migrating forms of the protein (small arrows). (B) Total cell extract (TE) from HeLa cells or nuclear extract (NE) from HeLa cells overexpressing HA-tagged TDG were prepared in the absence or presence of 5 mM NEM. A 100 µg aliquot of TE and 50 µg of NE proteins were analysed by western blotting with antibodies against TDG (lanes 1 and 2) and against the HA tag (lanes 3 and 4). In both cases, the 86 kDa but not the 60 kDa form of TDG appeared stronger when the cell extracts were prepared in the presence of NEM. The asterisk indicates an unspecific protein detected by the secondary anti-rat IgG antibody used. (C) HeLa nuclear extract (NE) prepared in the absence of NEM showed only a faint band of the 86 kDa form of TDG (lane 1). De novo modification of endogenous TDG was detectable following a short incubation of the extract with 10 mM ATP (lane 2). This reaction was stimulated further by the presence of a 60mer G·U heteroduplex DNA substrate (lane 3). (D) Recombinant His6-tagged TDG (6H-TDG) was incubated with HeLa nuclear extract in the absence or presence of 10 mM ATP and G·U mismatched DNA. Western blotting revealed that the recombinant protein was modified in vitro in an ATP-dependent manner (lanes 1 and 2), and that this reaction was stimulated significantly when TDG was pre-bound to a 60mer G·U heteroduplex substrate (lane 3).
In total extracts of HeLa cells prepared by standard NP-40 lysis procedures, the low mobility form of TDG was detectable reproducibly only when the cysteine protease inhibitor N-ethylmaleimide (NEM) was added to the lysis buffer (Figure 2B). NEM is a potent inhibitor of ULP isopeptidases (Li and Hochstrasser, 1999). Since SUMO conjugation is a reversible and ATP-dependent process, we tested if we could restore modified TDG in HeLa nuclear extract that was prepared in the absence of NEM. Indeed, modified endogenous TDG reappeared after a short incubation of the extract at 30°C but only when the extract was supplemented with ATP (Figure 2C). Interestingly, the addition of heteroduplex (G·U) or homoduplex DNA (data not shown) further stimulated TDG modification. Thus, endogenous TDG exists in at least two different states, one representing the original translation product of an apparent mass of 55–60 kDa and another one carrying a cysteine protease-sensitive modification that gives rise to a mobility shift of 20–25 kDa in SDS–PAGE. Both observations are compatible with TDG being modified reversibly by ubiquitin-like proteins.
To facilitate further biochemical experimentation, we made an attempt to modify recombinant TDG protein in vitro, taking advantage of the modifying capacity of HeLa nuclear extract. Incubation of recombinant TDG with HeLa nuclear extract indeed yielded substantial amounts of the modified enzyme. This reaction was most efficient at 30°C, was ATP dependent and was stimulated significantly when the recombinant glycosylase was pre-bound to heteroduplex or homoduplex DNA (Figure 2D). Under these conditions, the relative amount of modified TDG remained constant (40–50%) over a two-log range of TDG concentrations, suggesting a concerted and dynamic action of conjugating and deconjugating activities in the HeLa nuclear extract.
SUMO-1 and SUMO-2/3 are modifiers of human TDG
Three distinct ubiquitin-like proteins, SUMO-1, SUMO-2 and SUMO-3, have been identified in mammalian cells. SUMO-1 shares ∼46% identity with SUMO-2 and SUMO-3, while SUMO-2 and SUMO-3 are 95% identical and, thus, form a distinct subfamily of SUMO proteins (Saitoh and Hinchey, 2000). We made use of specific antibodies recognizing either SUMO-1 or SUMO-2/3 polypeptides to reveal the identity of the TDG modifier.
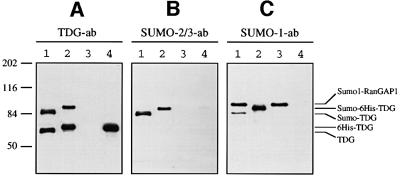
We enriched endogenous TDG by immunoprecipitation (IP) using magnetic beads coated with a TDG-specific monoclonal antibody. Subsequent western blotting with a TDG-specific polyclonal antibody revealed that both non-modified and modified TDG were present in such IP fractions (Figure 3A, lane 1) but not in control IPs with bovine serum albumin (BSA)-coated beads (Figure 3A, lane 3). We also modified His6-tagged recombinant TDG protein in HeLa nuclear extract and co-purified the resulting products by Ni-NTA–agarose and heparin– Sepharose chromatography. This yielded a mixture of >95% pure TDG protein, with modified and non-modified forms represented in an approximate ratio of 2:3 (Figure 3A, lane 2).
Fig. 3. Human TDG is modified by SUMO-1 and SUMO-2/3. Modification of TDG by SUMOs was examined by western blotting of protein fractions with a polyclonal anti-TDG antibody (A), a polyclonal anti-SUMO-2/3 antibody (B) and a monoclonal anti-SUMO-1 antibody (C). Lane 1, 5 µl of TDG immunoprecipitated from HeLa nuclear extracts; lane 2, 0.1 µl of the in vitro modified and purified recombinant His6-tagged TDG protein (0.4 M NaCl fraction); lane 3, 5 µl of the immunoprecipitate obtained with BSA-coated beads as a negative control; lane 4, 50 ng of pure recombinant His6-tagged TDG. The low mobility band in lanes 1 and 3 of (C) is probably the abundant SUMO-1-modified RanGAP1 protein that shows some affinity for the magnetic beads used for immunoprecipitation (Saitoh and Hinchey, 2000).
Incubation of an equivalent blot with a specific antibody against the SUMO-2/3 peptides resulted in the appearance of two discrete bands at positions corresponding to the modified native and recombinant TDG proteins (Figure 3B, lanes 1 and 2). No cross-reacting proteins were precipitated in the BSA control (Figure 3B, lane 3). Strikingly, probing of the western blot with a monoclonal antibody directed against SUMO-1 produced essentially the same result (Figure 3C, lanes 1 and 2), revealing that human TDG is subject to modification by both SUMO-1 and SUMO-2/3.
SUMO attachment involves a single C-terminal lysine of TDG
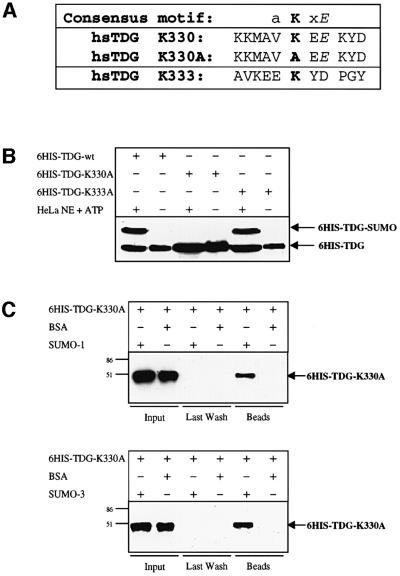
SUMO conjugation involves the formation of an isopeptide bond between the C-terminal glycine of the SUMO peptide and an ε-amino group of a lysine residue of the acceptor protein. Sumoylation sites identified in various target proteins defined the weak consensus acceptor motif aKxE, where a is an aliphatic amino acid. Human TDG is a protein of 410 amino acids with a highly conserved catalytic core domain and less conserved N- and C-terminal domains of unknown function (Hardeland et al., 2000). Of its 37 lysine residues, only one, the C-terminal Lys330, is located within a sequence matching the consensus motif (Figure 4A). Thus, we mutagenized this putative acceptor Lys330 and also the neighbouring Lys333 (Figure 4A). Mutation of Lys330 to alanine (K330A) produced a TDG that was unable to undergo SUMO conjugation in vitro, whereas mutation of Lys333 to alanine (K333A) had no effect on the protein’s sumoylation competence (Figure 4B). Transient expression of these TDG variants in HeLa cells confirmed the complete absence of SUMO conjugation to the K330A mutant under physiological conditions (data not shown). Both lysine mutants were unaffected in DNA glycosylase or mismatch binding activities (data not shown) and also retained their abilities to interact specifically with the SUMO-1 and SUMO-3 proteins (Figure 4C). These results establish that Lys330 is the major site for SUMO-1 and SUMO-2/3 attachment, but is not required for the physical interaction of TDG with the modifier proteins.
Fig. 4. Mapping of the sumoylation acceptor site within human TDG. (A) A four amino acid consensus sumoylation motif has been proposed (a = aliphatic amino acid). The amino acid sequence surrounding the candidate acceptor lysine K330 of human TDG is shown. K330A is the sequence resulting from mutagenesis of K330 to alanine; K333 is a conserved residue adjacent to K330 that was mutated to alanine as a control. (B) A 100 ng aliquot of purified His6-tagged wild-type or mutant (K330A, K333A) TDG proteins was subjected to modification by incubation with 25 µg of HeLa nuclear extract (NE) in the presence of 10 mM ATP. Western blotting revealed efficient sumoylation of the wild-type and the K333A TDG variants but no detectable modification of the K330 mutant. (C) The modification-deficient K330A mutant can still interact with SUMO-1 and SUMO-3. His6-tagged K330A mutant protein (600 ng) was incubated together with 20 µl of SUMO-1, SUMO-3 or BSA-coated affinity beads for co-precipitation. Subsequent immunoblotting of input, last wash and bead fractions revealed a specific retention of the mutant TDG protein (arrows) on the SUMO-1 and SUMO-3 but not on the BSA beads. The input represents 16% of the total amount of mutant TDG protein used. All western blots shown were performed with the polyclonal anti-TDG antibody.
Sumoylation changes the enzymatic properties of TDG
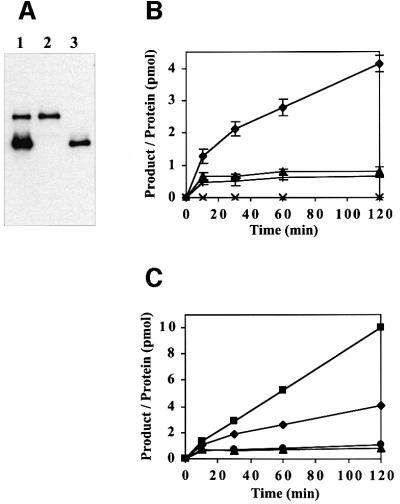
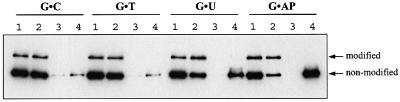
To assess the effect of sumoylation on the DNA-binding properties of TDG, we modified preparative amounts of recombinant His6-tagged TDG by incubation with nuclear extract of HeLa cells and subsequently repurified the glycosylase by Ni2+ affinity and heparin–Sepharose chromatography. This yielded highly pure TDG protein with a ratio of modified versus non-modified forms of ∼1:3 (Figure 6A). To test DNA binding, the mix of TDG proteins was incubated with an excess of biotinylated DNA substrate immobilized on magnetic streptavidin beads. The substrates used were homoduplex DNA and heteroduplexes with a G·T or G·U mismatch or a G·AP site. After washing, the fractions of bound and unbound proteins were analysed by SDS–PAGE and western blotting. Consistent with previous observations (Hardeland et al., 2000), the non-modified TDG showed weak interactions with homoduplex DNA and the G·T heteroduplex, and strong binding to the G·U and G·AP substrates (Figure 5). Sumoylated TDG, however, was unable to bind detectably to any of the DNA substrates analysed under a variety of different conditions. Thus sumoylation significantly reduces the DNA-binding affinity of TDG.
Fig. 6. Substrate nicking activities of modified and non-modified TDG. (A) Modified and non-modified TDG proteins (lane 1) were isolated by an SDS–gel extraction and renaturation procedure. Immunoblot analysis of equal amounts of the two eluates confirmed a perfect separation of the two TDG variants (lanes 2 and 3). (B) The substrate processing activity of both TDG proteins was analysed with a standardized nicking assay (Hardeland et al., 2000). The reactions were carried out in a 100 µl volume with 10 pmol of G·T or G·U substrates and 0.5 pmol of TDG protein. Product formation was assessed in at least three independent experiments and the results are plotted as ratios of nicked DNA molecules produced per enzyme (pmol product/pmol TDG) as a function of time. While non-modified TDG processed G·T (+) and G·U (triangles) mismatches in a typical single turnover reaction, the modified protein was inactive on the G·T substrate (×) but showed enhanced processing of the G·U substrate (diamonds). (C) When G·U substrate nicking assays were performed in the presence of 0.5 pmol of APE1, the processing efficiency of modified TDG was enhanced further by at least a factor of 3 (squares). No such stimulatory effect of APE1 was measurable for non-modified TDG (circles).
Fig. 5. DNA-binding properties of modified and non-modified TDG. In vitro sumoylated and purified TDG protein consisting of modified versus non-modified forms in a ratio of 1:3 was subjected to a DNA-binding assay. The protein mix was incubated with an excess of G·C-, G·T-, G·U- or G·AP site-containing 60mer oligonucleotides immobilized on magnetic beads. Input (1), unbound (2), last wash (3) and bound protein (4) fractions were analysed by western blotting with the polyclonal anti-TDG antibody. Unmodified wild-type TDG bound with low affinity to G·C and G·T substrates and showed increased affinity for G·U and G·AP site DNA. The modified form did not bind detectably to any of these DNA substrates.
Next, we investigated the substrate processing properties of modified and non-modified TDG. To separate the two forms from each other, we subjected the purified mix of TDG proteins to preparative SDS–PAGE and gel-extracted the two variants according to a previously described protocol (Neddermann and Jiricny, 1993) (Figure 6A). In subsequent nicking assays with G·T and G·U mismatched substrates (Hardeland et al., 2000), the gel-recovered non-modified TDG performed equally well as the untreated recombinant protein (not shown). Thus, the modification and purification procedure did not harm the activity of the enzyme. The gel-recovered modified TDG was still an active DNA glycosylase but, strikingly, with an altered substrate spectrum. It failed to act on the G·T mismatch but its G·U processing activity was significantly enhanced. After 2 h of incubation, 1 pmol of modified TDG excised ∼4 pmol of uracil from the G·U substrate (Figure 6B), while non-modified TDG processed only about half a molar equivalent of the substrate. Thus, unlike the activity of non-modified TDG that levelled off in a plateau reflecting the characteristic product inhibition kinetics of the enzyme (Hardeland et al., 2000), the modified protein continued to process substrate with a slow but steady turnover rate of 0.027/min.
Human APE1 was found to exert a stimulatory effect on the G·T processing activity of TDG. However, a significant turnover (>2-fold) was observed only when APE1 was added at high molar excess over TDG and after long incubation (Waters et al., 1999). The highest stimulation gained by a 100-fold excess of APE1 protein yielded an ∼3-fold turnover of TDG in a 1 h reaction. Since this is remarkably close to what we observed with our sumoylated TDG protein in the absence of APE1 (Figure 6B), we wondered whether these results reflect the peak rate of the enzyme or whether both modification and addition of APE1 protein could add up to a further increase in turnover. We tested the G·U processing activity of TDG in standard nicking assays and found that the modified but not the non-modified enzyme was stimulated by the presence of APE1 protein. As little as an equimolar amount of APE1 increased the turnover rate of the sumoylated glycosylase by 3-fold (0.079/min), while the activity of non-modified TDG was unaffected (Figure 6C).
These observations demonstrate that sumoylation of TDG induces a significant alteration of its enzymatic properties and modulates the way in which it cooperates with the (AP)-endonuclease acting immediately downstream in the BER pathway.
Discussion
The results of this study establish a functional interaction of the human TDG with the ubiquitin-like proteins SUMO-1 and SUMO-3. This interaction was detectable both as non-covalent physical association of the proteins and as covalent post-translational modification of TDG by the SUMO polypeptides. We identified a single lysine residue in the C-terminus of TDG that mediates the conjugation of both SUMO-1 and SUMO-2/3. Mutation of this lysine to alanine abolished the protein’s modification competence, while neither its non-covalent interaction with the SUMO proteins nor its DNA binding and glycosylase activities were affected. In vitro sumoylation of recombinant TDG altered its enzymatic properties: DNA substrate binding affinity was reduced to undetectable levels; the enzyme became incapable of acting as a thymine-DNA glycosylase on a G·T substrate but showed enhanced uracil processing on a G·U substrate; and the modification significantly potentiated the (AP)-endonuclease-stimulated turnover of TDG. In the light of a previous study that implicated a requirement for a high affinity TDG–substrate interaction for G·T processing but less so for G·U processing (Hardeland et al., 2000), we conclude that the selective loss of G·T processing and the gain in enzymatic turnover on a G·U substrate are both consequences of the reduction in the DNA-binding affinity imposed on TDG by sumoylation.
An appreciable number of mammalian, viral and yeast proteins have been discovered to undergo modification by SUMO-1. These can be either nuclear or cytoplasmic proteins falling into different and seemingly unrelated functional categories, and current evidence suggests that the consequences of SUMO-1 conjugation are target protein specific; some proteins were found to change their subcellular localization whereas others alter their enzymatic properties or acquire resistance to proteasomal degradation (reviewed in Melchior, 2000; Muller et al., 2001). Frequently, such effects are mediated through or accompanied by changes in protein–protein or protein– DNA interactions upon SUMO-1 conjugation or deconjugation. Owing to the fact that only a few SUMO-2/3 targets have been discovered, the specific role of this modification is not clear. At least two targets, TDG and the transcriptional transactivator IE2-p86 (Hofmann et al., 2000), are now known to be modified by either SUMO-1 or SUMO-2/3 proteins in a mutually exclusive manner. The physiology of the respective conjugation pathways could be different; SUMO-1 was found to be constitutively conjugated to target proteins and hardly detectable as a free polypeptide in cells, while most of SUMO-2/3 protein existed in a free form and only became attached to substrates in response to environmental stress (Saitoh and Hinchey, 2000). These observations provoke the hypothesis that SUMO-1 and SUMO-2/3 attachment might serve the same purpose but reflect modifications under different physiological conditions.
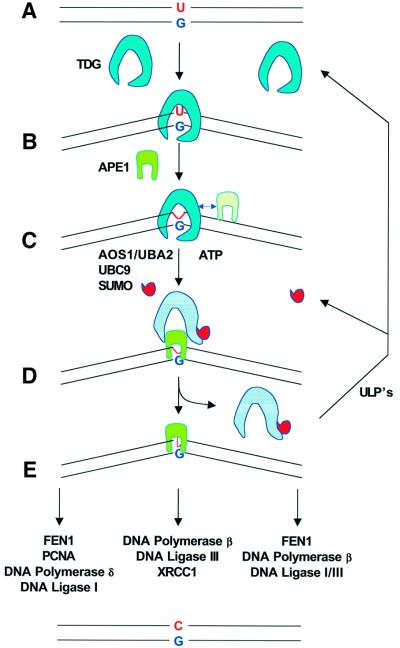
What possible role can we assign to sumoylation of TDG on the basis of the obvious changes induced in its enzymatic properties? As most DNA glycosylases do, TDG binds with high affinity to the product of its reaction, the AP site (Schärer et al., 1998; Waters et al., 1999; Hardeland et al., 2000). This apparent AP site shielding could be an integral part of DNA glycosylase function protecting the cells from the devastating effects of an uncontrolled processing of the unstable repair intermediates. It is therefore likely that mechanisms are in place to coordinate the dissociation of the DNA glycosylase with the association of the (AP)-endonuclease catalysing the subsequent step in the repair process. Our discoveries open up a novel perspective for the understanding of this coordination issue. We propose a model for TDG-mediated BER that assigns an active role to sumoylation in the release of the DNA glycosylase from the AP site (Figure 7). We postulate that unmodified TDG represents the glycosylase in a high affinity state, ready to bind and process a DNA substrate. Our data show that this is the predominant form of TDG present in nuclei of different human cell lines. Upon encountering its DNA substrate, the non-modified glycosylase hydrolyses the mismatched base and remains bound to the AP site. In this state, TDG is an optimal target for modification as supported by the finding that SUMO conjugation is stimulated under conditions where the protein is bound to DNA. The immediate consequence of sumoylation is a reduction in DNA-binding affinity such that the glycosylase dissociates from the AP site, a process that may be accelerated by the presence of the APE1 protein. The observation that a stoichiometric amount of APE1 protein was sufficient to enhance the turnover of sumoylated TDG significantly would argue in favour of an active displacement process mediated by physical interactions between modified TDG, the DNA substrate and APE1 protein. However, despite some considerable efforts, we failed to produce convincing evidence for a physical association of TDG with APE1, neither in the presence nor in the absence of DNA. Thus, such an interaction is either non-existent, too weak or too short-lived to be detectable under the conditions of our assays. Once sumoylated TDG has dissociated from the AP site, APE1 steps in and coordinates the subsequent steps of the BER reaction (Wilson and Kunkel, 2000). Since sumoylation is a reversible and dynamic process, rapid deconjugation of the liberated sumoylated TDG by an appropriate ULP protease follows to restore the active high affinity state of the glycosylase (Yeh et al., 2000).
Fig. 7. Model for coordination of the initial steps of BER via sumoylation-dependent dissociation of TDG from the AP site. Endogenous TDG is mainly non-modified. In this high affinity state (A), the glycosylase detects and binds a G·U, G·T or any other relevant substrate in DNA (B) and immediately hydrolyses the base to be replaced (C). After hydrolysis, the enzyme remains tightly bound to the AP site and is, in this DNA-bound state, a preferential substrate for SUMO conjugation. This modification, catalysed by the AOS1/UBA2 and UBC9 enzymes (D), is likely to induce a conformational change in TDG that results in a reduction of its DNA-binding affinity and ultimately in its dissociation from the AP site. This process may be coordinated by the presence of the APE1 protein, the AP-endonuclease acting downstream of TDG in the repair process. The resulting APE1–AP site–DNA complex (E) is then channelled into an appropriate BER pathway. Finally, the liberated modified TDG protein is recycled by a ULP protease-mediated de-conjugation reaction.
In other cases, SUMO conjugation was shown to induce changes in protein localization and stability or to modulate protein–protein interactions. Our present data argue against a role for sumoylation in the localization or stabilization of TDG. Cell fractionation experiments failed to produce evidence for a differential subcellular distribution of non-modified and modified TDG protein, and localization studies by immunofluorescence microscopy did not reveal an accumulation of TDG in PML bodies or in any other specific structure within the cells (data not shown). PML bodies are subnuclear protein complexes of particular relevance because they are preferential localization sites of sumoylated nuclear proteins (Muller et al., 2001). Also, the K330A mutation did not affect the subcellular localization pattern of the protein (data not shown). As regards stabilization, we found that non-modified TDG is the predominant endogenous form of the protein, which is inconsistent with sumoylation being important for protecting TDG from proteasome-mediated degradation.
Another recurring theme of sumoylation appears to be the direct or indirect regulation of transcription factors (Kim et al., 1999; Chakrabarti et al., 2000; Muller et al., 2000; Poukka et al., 2000). It is therefore noteworthy that TDG was found to interact physically and functionally with the nuclear receptors RAR and RXR (Um et al., 1998) and the thyroid transcription factor TTF1 (Missero et al., 2001). The specific role of TDG in this context is not clear and is particularly difficult to understand because in one case (RAR/RXR) interaction with TDG is associated with stimulation of transcriptional transactivation whereas in the other case (TTF1) it mediates transcriptional repression. Whether or not sumoylation plays a role in the modulation of this transcription-associated activity of TDG is unclear at this point and warrants further investigation.
In conclusion, most DNA glycosylases are inhibited by products of the AP site in vitro. This suggests a general requirement for mechanisms that regulate the displacement of DNA glycosylases from their reaction products under physiological conditions. We show here for TDG that sumoylation could provide an appropriate and attractive solution to this problem. It fulfils the key requirements for a sufficiently robust regulatory mechanism; it is a specific, controlled, reversible and highly dynamic process. Since we could identify putative acceptor site motifs in other human DNA glycosylases, it is possible that SUMO conjugation, rather than being a TDG-specific phenomenon, is an integral part of the reaction mechanism of many DNA glycosylases. This, however, remains to be established.
Materials and methods
Antibodies, expression constructs and recombinant proteins
See supplementary data available at The EMBO Journal Online.
Cell culturing and protein extractions
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium and MRC5 cells in Nutrition Mix F-10 (HAM) medium with Glutamax-I, both containing 10% fetal bovine serum and antibiotics, at 37°C in a 5% CO2 incubator. A stable TDG-overexpressing HeLa cell line was established by transfection of pPRS210 and subsequent isolation of clones resistant to 0.8 µg/ml puromycin.
For nuclear extract preparations, cells were harvested by gentle trypsination and centrifugation. Cytoplasmic proteins were separated from the nuclei by standard methods and the nuclei pelleted by centrifugation (2000 g, 4°C, 10 min). The nuclei were resuspended in 0.5 vol. of low salt buffer [20 mM HEPES pH 7.6, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.02 M KCl, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1× complete™ protease inhibitors (Roche Diagnostics)], and an equal volume of high salt buffer (low salt buffer supplemented with 0.8 M KCl) was added slowly with stirring. After incubation for 30 min at 4°C, the suspension was centrifuged (20 000 g, 4°C, 30 min) and the supernatant dialysed (2× 1 h, 4°C in 20 mM HEPES pH 7.6, 10% glycerol, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 1× EDTA-free complete™ protease inhibitors). After centrifugation (20 000 g, 4°C, 20 min), the soluble proteins (nuclear extract) were frozen in liquid nitrogen.
Total cell extracts were prepared by incubation of 108 cells in 2 ml of single detergent lysis buffer (50 mM Na-phosphate pH 8.0, 125 mM NaCl, 1% NP-40, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, 1× complete™ protease inhibitors) for 30 min on ice. Insoluble matter was removed by centrifugation (20 000 g, 4°C, 15 min) and the soluble proteins dialysed, recentrifuged and frozen in liquid nitrogen. For some experiments, nuclear and total cell extracts were prepared in the presence of 5 mM NEM. Protein concentrations of all native extracts were measured by the Bradford method (Bio-Rad). Total protein extraction under denaturing conditions was carried out with HeLa or MRC5 cells. A total of 107 cells grown in and attached to 9 cm tissue culture dishes were washed once with 1× phosphate-buffered saline (PBS), lysed directly in 200 µl of 2× SDS lysis buffer (120 mM Tris–HCl pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, 0.01% Bromphenol Blue, 8 M urea) and scraped off the dishes. The suspension was then incubated for 10 min at 96°C.
In vitro modification and purification of modified recombinant TDG
For SUMO modification of endogenous TDG, 45 µg of HeLa nuclear extract in 20 µl of 1× sumoylation buffer (50 mM Tris–HCl pH 8.0, 5% glycerol, 5 mM MgCl2, 1 mM DTT, 10 mM ATP) were incubated for 30 min at 30°C. Where indicated, 15 pmol of a 60mer DNA duplex (Hardeland et al., 2000) was added to the reaction.
For in vitro modification of recombinant TDG, 50 ng (0.9 pmol) of pure TDG protein were incubated with 15 µg of HeLa nuclear extract in a total of 10 µl of 1× sumoylation buffer for 30 min at 30°C. Modification of DNA-bound TDG was examined by pre-incubation of 0.9 pmol of TDG with 5 pmol of a 60mer oligonucleotide duplex (Hardeland et al., 2000) for 15 min at 30°C. The sumoylation reactions were stopped by addition of 2× SDS sample buffer and boiling of the samples. For purification of in vitro modified TDG, 60 µg of recombinant TDG were incubated with 7.5 mg of HeLa nuclear extract in a total volume of 800 µl of 1× sumoylation buffer for 1 h at 4°C. Then, 50 µl of equilibrated Ni-NTA–agarose (Qiagen) were mixed with 800 µl of 2× binding buffer (1× binding buffer: 20 mM HEPES pH 8.0, 1 M NaCl, 10% glycerol, 1 mM imidazole, 0.1% Tween-20, 5 mM β-mercaptoethanol, 1 mM PMSF, 1× protease inhibitor cocktail EDTA-free) and added to the modification reaction. After 1 h incubation at 4°C, the agarose beads were collected in a 1.5 ml reaction tube by gentle centrifugation (110 g, 4°C, 1 min) and washed twice with 1 ml of binding buffer. Subsequent washes were carried out with binding buffer containing increasing concentrations of imidazole (2× 1 ml of 5 mM imidazole, 2× 500 µl of 10 mM imidazole and 2× 250 µl of 20 mM imidazole). Bound proteins were eluted with 3× 100 µl of binding buffer containing 1 M imidazole. A 250 µl aliquot of the Ni-NTA eluate was then diluted with 1.65 ml of dilution buffer (20 mM HEPES pH 8.0, 10% glycerol, 0.1% Tween-20, 1 mM EDTA, 5 mM β-mercaptoethanol, 1 mM PMSF, 1× complete™ protease inhibitors), mixed with 50 µl of buffer-equilibrated (dilution buffer + 125 mM NaCl) heparin–Sepharose (Pharmacia™) and incubated for 1 h at 4°C. After pelleting the heparin–Sepharose by centrifugation (110 g, 4°C, 1 min), proteins were eluted sequentially with dilution buffer containing stepwise increasing concentrations of NaCl (2× 1 ml of 0.1 M NaCl, 2× 500 µl of 0.2 M NaCl, 2× 200 µl of 0.4 M NaCl, 2× 200 µl of 0.5 M NaCl, 2× 200 µl of 1 M NaCl). Modified TDG eluted at slightly lower NaCl concentrations (0.4 and 0.5 M) than the unmodified protein (0.4, 0.5 and 1 M). The 0.4 and 0.5 M NaCl fractions were pooled and stored at –80°C. This procedure yielded 800 µl of 0.2 µg/µl total TDG protein consisting of 65% non-modified and 35% modified forms. To separate the modified and non-modified forms of TDG, 20 µl of the TDG mixture were separated by 7.5% SDS–PAGE and the respective protein bands eluted from the gel and renatured as described in Neddermann and Jiricny (1993).
Immunoprecipitation and western blotting
Monoclonal anti-TDG antibody or BSA was covalently coupled to M-280 tosylactivated Dynabeads® (DYNAL®) according to the manufacturer’s protocol. All steps were performed in the absence of DTT or β-mercaptoethanol. Washed and pre-equilibrated beads (∼24 × 106 beads) were incubated with 200 µl of 6 µg/µl nuclear extract (prepared in the presence of 5 mM NEM) in a total of 400 µl of incubation buffer (20 mM HEPES pH 7.6, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.2 M KCl, 1 mM DTT, 1 mM PMSF and 1× complete™ protease inhibitors) for 1 h at 4°C. After three washes with 500 µl of incubation buffer containing increasing KCl concentrations of 0.4, 0.6 and 0.8 M, bound proteins were eluted by boiling the beads in 20 µl of 2× SDS sample buffer for 5 min at 99°C. For western blotting, proteins were separated in 7.5 or 10% SDS–polyacrylamide gels, transferred to a nitrocellulose membrane (Protran® Nitrocellulose Transfer Membrane, Schleicher & Schuell, Germany) and incubated with the respective antibodies. All antibodies were diluted in TBS-T containing 5% dry milk as blocking reagent. The dilutions were: 1:10 000 for the rabbit polyclonal anti-TDG antiserum, and 1:1000 for the rat monoclonal anti-TDG antibody, the rat monoclonal anti-HA antibody, the mouse monoclonal anti-SUMO-1 antibody and the rabbit polyclonal anti-SUMO-2/3 antiserum. Signals were detected with the enhanced chemiluminescent (ECL™) substrate system (Amersham Pharmacia Biotech).
TDG–SUMO co-precipitations
For co-precipitation of SUMO-1 with human TDG, 1 µg of purified recombinant human TDG or an equimolar amount of recombinant human APE1 was added to 20 µl of interaction buffer-equilibrated magnetic Ni2+-NTA–agarose beads (Qiagen). Following 30 min of incubation at 4°C under gentle shaking, the tubes were placed on a magnetic separator and the beads washed in 2× 200 µl of interaction buffer (50 mM sodium phosphate pH 8.0, 10 mM imidazole, 300 mM NaCl, 0.08% NP-40, 5 mM β-mercaptoethanol). A 9 µg aliquot of E.coli BL21(DE3) extract derived from cells expressing either SUMO-1, SUMO-3 or no human protein was then added to the beads in 200 µl of interaction buffer. After incubation for 45 min at 4°C, the beads were washed in 6× 200 µl and 1× 20 µl (last wash) of interaction buffer and then transferred to a new reaction tube. Bound proteins were eluted by boiling the beads in 20 µl of 2× SDS sample buffer at 95°C for 2 min and analysed further by 15% SDS–PAGE and western blotting.
For co-precipitation of human TDG with SUMO-1 and SUMO-3, crude protein extract from E.coli BL21(DE3) cells expressing either recombinant SUMO-1, SUMO-3 or no human protein was covalently coupled to Affi-Gel®10 beads (Bio-Rad) following the manufacturer’s protocol. A 20 µl aliquot of protein-coated Affi-Gel beads was centrifuged in a 1.5 ml reaction tube (110 g, 1 min, 4°C) and washed in 3× 100 µl interaction buffer (50 mM sodium phosphate, 300 mM NaCl, 5 mM β-mercaptoethanol, 0.4% NP-40). The beads were then incubated with 600 ng of TDG protein (wild-type, mutant or in vitro modified variants) in 50 µl of interaction buffer for 1 h at 4°C under gentle rotation. Subsequent washing/centrifugation (110 g, 1 min, 4°C) cycles with 4× 200 µl and 1× 20 µl of interaction buffer were performed to remove unbound TDG. Bound proteins were eluted by boiling the beads in 20 µl of 2× SDS sample buffer at 95°C for 2 min and analysed further by 10% SDS–PAGE and western blotting.
DNA binding and base release assays
The DNA-binding ability of TDG was tested with double-stranded 60mer oligonucleotide substrates as described (Hardeland et al., 2000) except that the G strand was 5′ modified with biotin. A 20 pmol aliquot of this substrate was bound to 20 µl of streptavidin Dynabeads® M-280 (Dynal®) according to the manufacturer’s protocol. The substrate-coated Dynabeads were equilibrated in 1× binding buffer (50 mM Tris–HCl pH 8.0, 10% glycerol, 120 mM NaCl, 0.5 mg/ml BSA, 0.2% NP-40, 1 mM EDTA, 1 mM DTT, 1× complete™ protease inhibitors) and incubated with 2 µl of the fraction containing purified non-modified and modified TDG in a total volume of 40 µl for 30 min at 4°C. After three washes at 4°C, the reactions were stopped by addition of 20 µl of 2× SDS sample buffer and heating for 5 min at 95°C. Aliquots (10 µl) of relevant fractions were subjected to 10% SDS–PAGE and western blotting.
The catalytic activity of TDG was monitored by means of a standardized nicking assay (Hardeland et al., 2000). The reactions were performed in 100 µl of 1× nicking buffer (50 mM Tris–HCl pH 8.0, 1 mM DTT, 0.1 mg/ml BSA, 1 mM EDTA, 0.05 U of Ugi), containing 10 pmol of substrate DNA and 0.5 pmol of gel-extracted non-modified or modified TDG. In an alternative setup, we added 0.5 pmol of APE1 to the reactions. After the indicated times of incubation at 37°C, 20 µl aliquots were withdrawn and the reactions were stopped by addition of 1 M NaOH to a concentration of 90 mM and heating for 10 min at 99°C. The reaction products were analysed by denaturing gel electrophoresis (Hardeland et al., 2000).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Tomas Lindahl for critical reading of the manuscript and to Marc Bentele and Teresa Lettieri and Orlando Schärer for stimulating discussions. We also thank Margaret Fäsi for technical assistance and Josef Hinchey for the gift of the SUMO-2/3 antiserum. This study was supported by grants from the Sussella Stiftung, Zürich (U.H. and P.S.) and the Swiss National Science Foundation (P.S. and R.S.).
References
- Barrett T.B., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G·T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- Barrett T.E., Schärer,O.D., Savva,R., Brown,T., Jiricny,J., Verdine,G.L. and Pearl,L.H. (1999) Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J., 18, 6599–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R., Wilson,D.R., Wong,D. and Demple,B. (1997) Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc. Natl Acad. Sci. USA, 94, 7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott K.W., McKeown,C.K., Tucker,J.D., Ljungquist,S. and Thompson,L.H. (1994) An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol., 14, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S.R., Sood,R., Nandi,S. and Nucifora,G. (2000) Post translational modification of TEL and TEL/AML1 by SUMO-1 and cell-cycle-dependent assembly into nuclear bodies. Proc. Natl Acad. Sci. USA, 97, 13281–13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P. and Jiricny,J. (1996) A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature, 383, 735–738. [DOI] [PubMed] [Google Scholar]
- Hardeland U., Bentele,M., Jiricny,J. and Schär,P. (2000) Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem., 275, 33449–33456. [DOI] [PubMed] [Google Scholar]
- Hardeland U., Bentele,M., Lettieri,T., Steinacher,R., Jiricny,J. and Schär,P. (2001) Thymine DNA glycosylase. Prog. Nucleic Acid Res. Mol. Biol., 68, 235–252. [DOI] [PubMed] [Google Scholar]
- Hill J.W., Hazra,T.K., Izumi,T. and Mitra,S. (2001) Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res., 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Floss,S. and Stamminger,T. (2000) Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol., 74, 2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.H., Choi,C.Y. and Kim,Y. (1999) Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc. Natl Acad. Sci. USA, 96, 12350–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Nash,R.A., Klungland,A., Schär,P., Barnes,D.E. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- Li S.J. and Hochstrasser,M. (1999) A new protease required for cell-cycle progression in yeast. Nature, 398, 246–251. [DOI] [PubMed] [Google Scholar]
- Lindahl T. (1990) Repair of intrinsic DNA lesions. Mutat. Res., 238, 305–311. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- Melchior F. (2000) SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol., 16, 591–626. [DOI] [PubMed] [Google Scholar]
- Miao F., Bouziane,M. and O’Connor,T.R. (1998) Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res., 26, 4034–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missero C., Pirro,M.T., Simeone,S., Pischetola,M. and Di Lauro,R. (2001) The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem., 276, 33569–33575. [DOI] [PubMed] [Google Scholar]
- Mol C.D., Izumi,T., Mitra,S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- Muller S., Berger,M., Lehembre,F., Seeler,J.S., Haupt,Y. and Dejean,A. (2000) c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem., 275, 13321–13329. [DOI] [PubMed] [Google Scholar]
- Muller S., Hoege,C., Pyrowolakis,G. and Jentsch,S. (2001) SUMO, ubiquitin’s mysterious cousin. Nature Rev. Mol. Cell. Biol., 2, 202–210. [DOI] [PubMed] [Google Scholar]
- Neddermann P. and Jiricny,J. (1993) The purification of a mismatch-specific thymine DNA glycosylase from HeLa cells. J. Biol. Chem., 268, 21218–21224. [PubMed] [Google Scholar]
- Neddermann P., Gallinary,P., Lettieri,T., Schmid,D., Truong,O., Hsuan,J.J., Wiebauer,K. and Jiricny,J. (1996) Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem., 271, 12767–12774. [DOI] [PubMed] [Google Scholar]
- Nilsen H. and Krokan,H.E. (2001) Base excision repair in a network of defence and tolerance. Carcinogenesis, 22, 987–998. [DOI] [PubMed] [Google Scholar]
- Nilsen H., Haushalter,K.A., Robins,P., Barnes,D.E., Verdine,G.L. and Lindahl,T. (2001) Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J., 20, 4278–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronzelli F., Riccio,A., Markham,G.D., Seeholzer,S.H., Stoerker,J., Genuardi,M., Yeung,A.T., Matsumoto,Y. and Bellacosa,A. (2000) Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J. Biol. Chem., 275, 32422–32429. [DOI] [PubMed] [Google Scholar]
- Poukka H., Karvonen,U., Janne,O.A. and Palvimo,J.J. (2000) Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl Acad. Sci. USA, 97, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Singhal,R.K., Srivastava,D.K., Molina,J.T., Tomkinson,A.E. and Wilson,S.H. (1996) Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem., 271, 16000–16007. [DOI] [PubMed] [Google Scholar]
- Saitoh H. and Hinchey,J. (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem., 275, 6252–6258. [DOI] [PubMed] [Google Scholar]
- Schärer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- Schärer O.D., Nash,H.M., Jiricny,J., Laval,J. and Verdine,G.L. (1998) Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases. J. Biol. Chem., 273, 8592–8597. [DOI] [PubMed] [Google Scholar]
- Um S., Harbers,M., Benecke,A., Pierrat,B., Losson,R. and Chambon,P. (1998) Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem., 273, 20728–20736. [DOI] [PubMed] [Google Scholar]
- Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters T.R., Gallinari,P., Jiricny,J. and Swann,P.F. (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem., 274, 67–74. [DOI] [PubMed] [Google Scholar]
- Wilson S.H. and Kunkel,T.A. (2000) Passing the baton in base excision repair. Nature Struct. Biol., 7, 176–178. [DOI] [PubMed] [Google Scholar]
- Yang H., Clendenin,W.M., Wong,D., Demple,B., Slupska,M.M., Chiang,J.H. and Miller,J.H. (2001) Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res., 29, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E.T., Gong,L. and Kamitani,T. (2000) Ubiquitin-like proteins: new wines in new bottles. Gene, 248, 1–14. [DOI] [PubMed] [Google Scholar]