Abstract
The eukaryotic minichromosome maintenance (MCM) family of proteins (MCM2–MCM7) is evolutionarily conserved from yeast to human. These proteins are essential for DNA replication. The signal transducer and activator of transcription proteins are critical for the signal transduction of a multitude of cytokines and growth factors leading to the regulation of gene expression. We previously identified a strong interaction between Stat1 and MCM5. However, the physiological significance of this interaction was not clear. We show here by chromatin immunoprecipitation (ChIP) analyses that the MCM5 protein, as well as other members of the MCM family, is inducibly recruited to Stat1 target gene promoters in response to cytokine stimulation. Furthermore, the MCM proteins are shown to move along with the RNA polymerase II during transcription elongation. We have also identified an independent domain in MCM5 that mediates the interaction between Stat1 and MCM5; overexpression of this domain can disrupt the interaction between Stat1 and MCM5 and inhibit Stat1 transcriptional activity. Finally, we used the RNA interference technique to show that MCM5 is essential for transcription activation of Stat1 target genes. Together, these results demonstrate that, in addition to their roles in DNA replication, the MCM proteins are also necessary for transcription activation.
Keywords: RNA polymerase II, DNA helicase, IFN-γ
The evolutionarily conserved eukaryotic minichromosome maintenance (MCM) family of proteins consists of six members: MCM2–MCM7 (reviewed in refs. 1 and 2). The molecular structure and in vitro analyses of these proteins suggest that they function as a DNA helicase (3); they form a heterohexamer complex that binds to DNA replication origins and moves along with the DNA polymerase during DNA replication elongation (4, 5). In addition to the hexamer complex, the MCM proteins also form subcomplexes containing some members of the family, such as MCM4/6/7 or MCM3/5 (3, 6–8). It has been suggested that these subcomplexes represent segments during the assembly of the hexameric MCM complex (9). The MCM proteins are also highly abundant, and their number far exceeds that of the replication origins in yeast (8, 10–12). These observations have led to the suggestion that the MCM proteins may play additional roles in other biological processes, such as DNA repair, chromatin remodeling, and transcription (2, 13).
The signal transducer and activator of transcription (STAT) family of transcription factors mediates a multitude of cytokine-regulated gene transcription (reviewed in refs. 14 and 15). In response to ligand binding to cell surface receptors, the STATs are activated through tyrosine phosphorylation, form dimers, enter the nucleus, and bind to specific DNA sequences for transcription activation. The transcriptional activity of STATs are mediated by the transcription activation domain (TAD) located in the C terminus of the molecule (16). The STAT TADs can function independently of the rest of the STAT molecule, and their activities rely on their interaction with other nuclear transcription coactivators, such as CBP/p300 [cAMP-responsive element-binding protein (17–23)]. We have previously identified that two members of the MCM family, MCM3 and MCM5, are among a group of nuclear proteins that specifically interacted with Stat1 through affinity purification (24). Gel filtration analyses showed that the MCM3/5 subcomplex coeluted with the active tyrosine-phosphorylated Stat1 when cells were stimulated with IFN-γ (25). Other interactions between members of the MCM family and proteins involved in transcription activation were also identified, including MCM2 with the RNA polymerase II (26, 27) and MCM7 with Rb (28). However, it is not clear whether these interactions result in the participation of MCM proteins in transcription or these transcription factors may modulate DNA replication.
In this report, we further investigated the functional importance of the interaction between Stat1 and MCM5. We used the ChIP analyses to show that the MCM5 protein, as well as other members of the MCM family, is inducibly recruited to Stat1 target gene promoters in response to cytokine stimulation. Furthermore, analyses of a Stat1 target gene locus showed that the MCM proteins move along with the RNA polymerase II during transcription elongation. We have also identified an independent domain in MCM5 that can disrupt the interaction between Stat1 and MCM5 and inhibit Stat1 transcriptional activity. Finally, we use the RNA interference (RNAi) technique to show that MCM5 is essential for transcription activation of Stat1 target genes. Together, these results demonstrate that, in addition to their roles in DNA replication, the MCM proteins are also necessary for transcription activation.
Materials and Methods
Cell Culture and Antibodies. U3A and 2fTGH cells [provided by G. Stark (Cleveland Clinic Foundation Research Institute, Cleveland, OH) and I. Kerr (Cancer Research UK, London)] and 293T cells (American Type Culture Collection) were maintained in DMEM supplemented with 10% bovine calf serum (Hy-Clone). Antibody against phosphoserine-Stat1 was from Upstate Biotechnology (Lake Placid, NY). Antibody against phosphotyrosine-Stat1 was from Cell Signaling Technology. Antibodies against Stat1 N terminus and Stat3 were from BD Biosciences (Franklin Lakes, NJ). Extracellular signal-regulated kinase 2 antibody was from Santa Cruz Biotechnology. For ChIP assays, antibodies against Stat3, Stat1 C terminus, RNA polymerase II (Pol II) were from Santa Cruz Biotechnology; antibodies against MCM2, 6, and 7 were kindly provided by Rolf Knippers (Universität Konstanz, Konstanz, Germany) and antibodies against MCM5 and MCM3 were from Bethyl Laboratories (Montgomery, TX). Recombinant human IFN-γ from Roche was used at 5 ng/ml.
ChIP Assay. ChIP experiments were performed as described in ref. 29 with the 2.5 μg of each of following antibodies: anti-Stat1C, anti-MCM2, anti-MCM3, anti-MCM5, anti-MCM6, anti-MCM7, anti-RNA Pol II, and anti-Stat3. For Fig. 1A with 2fTGH cells, cells from 10-cm dishes were used for each immunoprecipitation. For Fig. 1 A with U3A cells and Fig. 1B, lysate from one 15-cm dish was divided for three separate immunoprecipitations. For Figs. 1C and 2, the lysate from one 15-cm dish was used for one immunoprecipitation, and the precipitated DNA was used for three to five PCR experiments with primers specific to the target gene promoters and the IFN regulatory factor (IRF)-1 locus, respectively. The resulting PCR products were separated on 6% acylamide gel and quantitated by a Molecular Dynamics Storm PhosphorImager. Primer pair sequences include the following: ChIP PCR for human IRF-1 promoter, 5′-CTTCGCCGCTAGCTCTACAACAG-3′ and 5′-GCTCCGGGTGGCCTCGGTTCG-3′; for the middle of the IRF-1 gene, 5′-GAGGCACTCACGTTAACACAGA-3′ and 5′-CCTGAAGCCACACACTTTCTAA-3′; for the 5′ intergenic region of IRF-1, 5′-GCTGCGGAGCTTCATTTCT-3′ and 5′-AGGAGAGTGCTGATCCCATC-3′; for the 3′ UTR region of the IRF-1 gene, 5′-AGTGACCCCAGAAAAGCATAAC-3′ and 5′-CCCACTTTCCTTCACATTTCAC-3′; for the 3′ intergenic region of IRF-1, 5′-CAGCTCTCCATCCTGAAAGG-3′ and 5′-AGTCCTCAATTTCCCCATCC-3′; for the class II transactivator (CIITA) promoter, 5′-GCTATGATACTGGCCCCATC-3′ and 5′-CCTTAAGCCCTCCCTACACC-3′; for the transporter associated with antigen processing (TAP)1 promoter, 5′-ATCTGATTTCCACGCTTGCT-3′ and 5′-GGAAAGTCCCAGGAACAGG-3′; for guanylate-binding protein (GBP)1 promoter are, 5′-TGAGAAATCTTTAAACCCTCCC-3′ and 5′-TGGCTTCTAGCACTTCTGTGTC-3′.
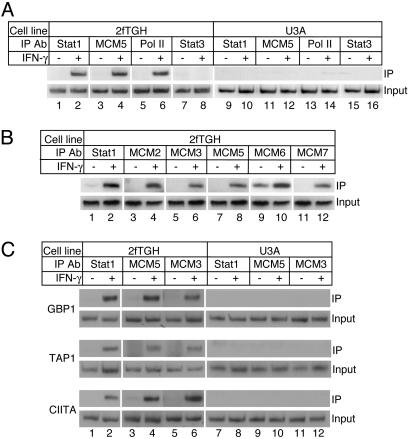
Fig. 1.
The MCM proteins are recruited to Stat1 target gene promoters in response to IFN-γ. (A) 2fTGH and U3A (a Stat1-null derivative of 2fTGH) cells were treated with IFN-γ for 30 min, and the IRF-1 promoter was analyzed by ChIP assays with the indicated antibodies. One representative result from two to 10 independent experiments is shown. IP, immunoprecipitation. (B) 2fTGH cells were treated with IFN-γ for 30 min, and the IRF-1 promoter was analyzed by ChIP assays with the indicated antibodies. (C) 2fTGH and U3A cells were treated with IFN-γ for 30 min (for TAP1) or 60 min (for GBP1 and CIITA) and analyzed by ChIP assays with the indicated antibodies.
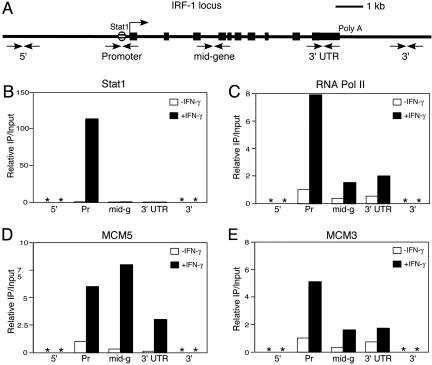
Fig. 2.
The MCM proteins move along with the RNA Pol II during transcription elongation. (A) The IRF-1 locus and the ChIP primer sets. The Stat1-binding site is indicated by a hatched oval. Exons are indicated by rectangles. Open rectangles indicate coding regions, and filled rectangles indicate UTRs. Paired arrows indicate the position of ChIP primer sets. Poly(A), polyadenylation site; 5′ and 3′, intergenic region 5′ or 3′ of the IRF-1 locus. (B–E) 2fTGH cells were treated with IFN-γ for 30 min and analyzed by ChIP assays with anti-Stat1 (B), anti-Pol II (C), anti-MCM5 (D), and anti-MCM3 (E) antibodies. 32P-labeled PCR products were separated on acrylamide gels and quantitated by a PhosphorImager. One representative result from two to six independent experiments is shown. Quantitation of one experiment was further confirmed by real-time PCR. Pr, promoter; mid-g, mid-gene; *, undetected.
GST Pull-Down Assays, Coimmunoprecipitation, and Western Blot Analyses. GST fusion proteins were purified from bacteria with glutathione–Sepharose beads (Amersham Pharmacia). In vitro translation reactions were done by using the TNT T7 system (Promega). Reaction mixtures containing 40 μl were incubated with 5 μl of GST fusion protein beads (5 μg of protein per μl of beads) in 1 ml of 20 mM Hepes, pH 7.9/10 mM KCl/140 mM NaCl/1 mM EDTA/0.1 mM Na3VO4/0.1% Nonidet P-40/13% glycerol (vol:vol) at 4°C overnight. For coimmunoprecipitation experiments, total cell lysates from IFN-γ-treated (30 min) cells or control cells were incubated overnight with 5 μg of anti-Stat1 antibodies (Santa Cruz Biotechnology) in PBS plus 0.1% Triton X-100. GST fusion proteins and immune complexes were separated by SDS/PAGE followed by autoradiography or Western blot analyses with chemiluminescence (DuPont/NEN).
Plasmid Constructions. GST-Stat1TAD was constructed as described in ref. 24. Stat1 expression plasmids were constructed as reported in ref. 30. cDNAs of MCM5 mutants were PCR amplified from a full-length MCM5 construct as described. Corresponding MCM5 mutant proteins are MCM5/550 (residues 550–734), MCM5/604 (residues 604–734), MCM5/666 (residues 666–734), and MCM5/696 (residues 696–734). Mutant MCM5 cDNAs were subcloned into the pRSET-C (Invitrogen) vector for in vitro translation. MCM5/550 cDNA was also subcloned into pEGFP-C2 (Clontech) vector for expression as a GFP fusion protein.
RNAi. RNAi for MCM5 was performed using the RNAi Human/Mouse Control kit from Qiagen (Valencia, CA). Two sets of human MCM5 small inhibitory RNA (siRNA) duplexes were synthesized by Qiagen: r(GGGUUACCAUCAUGGGCAU)dTdT/r(AUGCCCAUGAUGGUAACCC)dTdG and r(GCACGGGCUUCACCUUCAA)dTdT/r(UUGAAGGUGAAGCCCGUGC)dGdG. 293T cells were transfected with 0.25 μg of each MCM5 siRNA or 0.5 μg of the negative control scrambled siRNA according to the manufacturer's instructions. Cells were cultured for 72 h and then treated with IFN-γ for 2–4 h or left untreated. RNA and protein extraction were performed for Western blotting and RT-PCR analyses.
RNA Analyses. Total RNAs were prepared using TRIzol (Invitrogen). Real-time RT-PCR analyses were done on the prism 7900HT Sequence Detection system (Applied Biosystems) with the SYBR Green PCR kit from Applied Biosystems by following the manufacturer's instructions. The relative level of expression for a specific gene was calculated according to the equation REn = 2-(ΔCtn - ΔCt0), ΔCt = Cttest gene - CtGAPDH (where RE is the relative level of expression, Ct is the cycle threshold, n is the specific sample, 0 is the untreated wild-type, and test gene is IRF-1, CIITA, TAP1, GBP1, MCM5, or MCM3; see Fig. 4). Primer pair sequences for the real-time RT-PCR are hGAPDH, 5′-ATCAAGAAGGTGGTGAAGCA-3′ and 5′GTCGCTGTTGAAGTCAGAGGA-3′; hIRF-1, 5′CAAATCCCGGGGCTCATCTGG-3′ and 5′CTGGCTCCTTTTCCCCTGCTTTGT-3′; hCIITA, 5′CTCACGGGACTCTATGTCG-3′ and 5′TGTAGGGTACTTTGATGTCTGC-3′; hMCM3, 5′CGAGGAAGACCAGGGAATTT-3′ and 5′AGGCAACCAGCTCCTCAAAG-3′; hMCM5, 5′CCCATTGGGGTATACACGTC-3′ and 5′ACGGTCATCTTCTCGCATCT-3′; hGBP1, 5′CTAAGGAGAAAAAGAACAGACAAGG-3′ and 5′TAGGCTGTGTAATGGCAGAAA-3′; hTAP1, 5′TCTCCTCTCTTGGGGAGATG-3′ and 5′ATCCCGTCACCCACGAACT-3′.
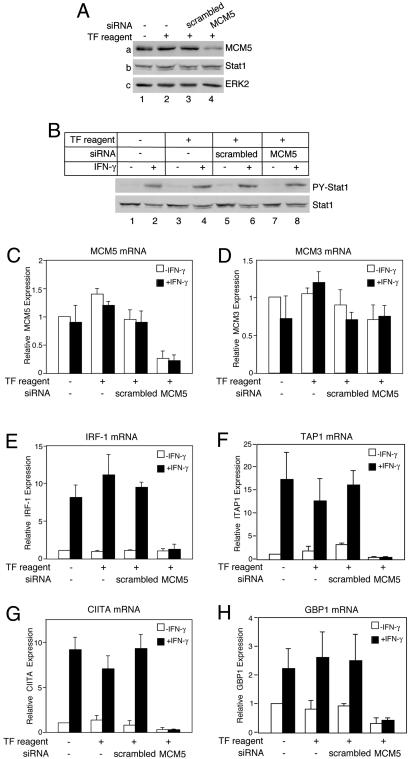
Fig. 4.
The MCM5 protein is essential for Stat1-mediated gene activation. 293T cells (1 × 105) were transiently transfected with a scrambled siRNA (0.5 μg) or two siRNAs against MCM5 (0.25 μg of each) for 72 h and analyzed as follows. (A) Whole-cell lysates were harvested for Western blotting analyses with the indicated antibodies. TF reagent, RNAiFect transfection reagent (Qiagen); scrambled, a negative control siRNA oligo with a scrambled nucleotide sequence. (B) Cells were either left untreated or treated with IFN-γ for 30 min followed by Western blotting. PY-Stat1, phospho-tyrosine Stat1. (C–H) Cells were either left untreated or treated with IFN-γ for 2 h (D–G) or 4 h (H) followed by real-time RT-PCR analyses of RNA expression of MCM5 (C), MCM3 (D), IRF-1 (E), TAP1 (F), CIITA (G), GBP1 (H), and an internal control GAPDH. Results shown are normalized against GAPDH and the mean ± SD of four to seven experiments.
Transfection Experiments. Transient transfection of U3A and 2fTGH cells was performed with Lipofectamine 2000 method according to the manufacturer's instructions (Invitrogen). Cells were treated 12 h after transfection with IFN-γ for 6 h or were left untreated. Luciferase assays were performed by using the dual-luciferase reporter system according to the manufacturer (Promega). All results shown were luciferase activities normalized against an internal control luciferase reporter of Renilla luciferase. For stable transfection with pEGFP-C2 and pEGFP-C2-M5/550 vectors, cells were selected in media containing 1 mg/ml G418. Clones with green fluorescent colors were picked and further screened by Western blotting using an anti-MCM5 antibody.
Results
The MCM Proteins Are Inducibly Bound to Stat1 Target Gene Promoters in Response to IFN-γ Stimulation. To directly demonstrate that MCM5 is involved in Stat1-mediated transcription activation, we first examined the promoter of a Stat1 target gene, IRF-1, for the presence of the MCM5 protein in response to IFN-γ stimulation. 2fTGH cells (31) were treated with IFN-γ for 30 min and analyzed by ChIP assays. Stat1 and RNA Pol II were detected on the IRF-1 promoter only in cells treated with IFN-γ (Fig. 1 A, lanes 2 and 6). Strikingly, MCM5 also interacted strongly with the IRF-1 promoter and was present only in cells that have been treated with IFN-γ (Fig. 1 A, lane 4). If the binding of MCM5 to the IRF-1 promoter was due to the presence of a DNA replication origin, then we should have also detected MCM5 on the IRF-1 promoter in untreated cells because treatment with IFN-γ for 30 min would not affect the asynchronously growing cell population. This result indicates that the binding of MCM5 to the IRF-1 promoter is induced by IFN-γ for transcription activation. Furthermore, in U3A cells (a 2fTGH derivative cell line that contains a Stat1-null mutation) (31), MCM5 was not detected on the IRF-1 promoter when neither Stat1 nor RNA Pol II was bound (Fig. 1 A, lanes 10, 12, and 14). As a negative control, another member of the STAT family, Stat3, was not detected on the IRF-1 promoter in either untreated or treated cells of both cell lines (Fig. 1 A, lanes 7, 8, 15, and 16), although a low level of tyrosine-phosphorylated Stat3 was detected by Western blotting in IFN-γ-treated cells (data not shown). These results clearly demonstrate that MCM5 is specifically recruited to a Stat1 target promoter for transcription activation in response to IFN-γ stimulation.
To further investigate the functional role of the MCM family in transcription activation, we performed ChIP assays with antibodies against MCM2, MCM3, MCM6, and MCM7. All of these MCM proteins were present on the IRF-1 promoter when cells were treated with IFN-γ (Fig. 1B, lanes 4, 6, 10, and 12) but were undetectable in untreated cells for MCM2, MCM3, and MCM7 or at low level for MCM6 (Fig. 1B, lanes 3, 5, 9, and 11). The binding of the MCM4 protein was not assessed because of the poor quality of the antibody. These results indicate that, although Stat1 can directly interact with only MCM5 (25), the whole MCM family of proteins is recruited to the IRF-1 promoter for transcription activation in response to IFN-γ stimulation.
To further show that the MCM proteins are involved in Stat1-mediated transcription activation in response to IFN-γ,we analyzed more Stat1 target gene promoters. As shown in Fig. 1C, Stat1 directly bound to the promoters of TAP1, GBP1, and CIITA when 2fTGH cells were treated with IFN-γ (Fig. 1C, lane 2). MCM3 and MCM5 were bound on these promoters only in IFN-γ-treated 2fTGH cells (Fig. 1C, lanes 4 and 6). However, MCM3 and MCM5 were not detected in the Stat1-deficient U3A cells (Fig. 1C, lanes 10 and 12).
All together, these results demonstrate that in response to IFN-γ signaling, Stat1 recruits the MCM proteins to the Stat1 target gene promoters for transcription activation.
The MCM Proteins Move Along with the RNA Pol II During Transcription Elongation. To further determine whether the MCM proteins are involved in transcription initiation or transcription elongation, we designed a series of ChIP primers covering the IRF-1 locus (Fig. 2 A) and analyzed the movement of Stat1, RNA Pol II, MCM3, and MCM5 along the IRF-1 gene. Stat1 was present at the IRF-1 promoter only when cells were treated with IFN-γ (Fig. 2B). RNA Pol II was present at the promoter region and in the middle and 3′ UTR regions of the IRF-1 gene when cells were treated with IFN-γ (Fig. 2C). However, RNA Pol II was not detected in the nontranscribing intergenic regions 5′ or 3′ of the IRF-1 locus (Fig. 2C). Strikingly, MCM5 had a profile similar to that of the RNA Pol II; i.e., not only was MCM5 on the IRF-1 promoter, it was also in the middle and 3′ UTR of the IRF-1 gene (Fig. 2D). MCM3 was also present in the middle and 3′ UTR of the IRF-1 gene (Fig. 2E), further suggesting that the whole MCM family travels along with RNA Pol II. MCM5 and MCM3 were not detected in the nontranscribing intergenic region either 5′ or 3′ to the IRF-1 locus (Fig. 2 D and E). Stat3 was not detected in any of these regions (data not shown).
These results strongly demonstrate that the MCM proteins are recruited to the IRF-1 promoter by Stat1 and, furthermore, move along with the transcribing RNA polymerase during transcription elongation.
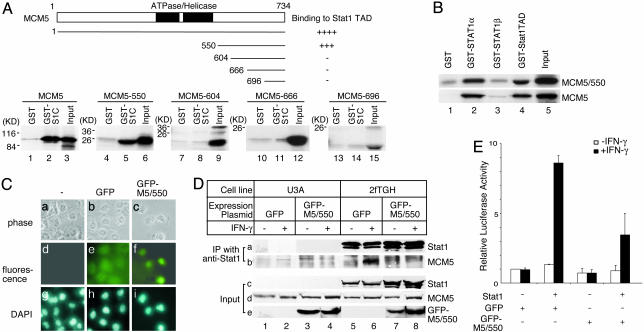
Functional Importance of the Interaction Between Stat1 and MCM5 for Transcriptional Activation. We have previously demonstrated that there is a direct and specific interaction between Stat1 and MCM5 (24, 25). To further test whether this interaction is essential for the Stat1 mediated transcription activation, we first looked for an independent interaction domain in MCM5 that is responsible for interacting with Stat1. Deletional mutants of MCM5 were translated in vitro and tested for their interactions with the Stat1 TAD in GST pull-down assays (Fig. 3A). The C-terminal region of MCM5 (MCM5/550, residue 550–734) could independently interact with Stat1 TAD (Fig. 3A, lane 5). Any further deletion of this domain resulted in a considerable loss of binding to Stat1 (Fig. 3A and data not shown). Interestingly, MCM5/550 does not include the ATPase/helicase domain, which is essential for DNA replication. Furthermore, this MCM5/550 domain could interact with Stat1 with the same specificity as the full-length MCM5; i.e., it interacted with the full-length Stat1α (Fig. 3B, lane 2) and Stat1 TAD directly (Fig. 3B, lane 4) but not with Stat1β (Fig. 3B, lane 3). Stat1β is an alternatively spliced form of Stat1 lacking the Stat1TAD (32). MCM5/550 could also interact with MCM3 directly (data not shown). These results indicate that the C-terminal region of the MCM5 protein can function as an independent domain for protein–protein interaction.
Fig. 3.
The specific interaction between Stat1 and MCM5 is necessary for Stat1 activity. (A) In vitro interaction between MCM5 mutants and Stat1 TAD. Full-length MCM5 and MCM5 deletional mutants, as indicated, were in-vitro-translated and labeled with 35S for GST pull-down assay with Sepharose-bead-bound GST or GST-Stat1TAD fusion proteins. Bound proteins were separated by SDS/PAGE and visualized by autoradiography. S1C, Stat1 TAD. (B) In vitro interaction between MCM5/550 and Stat1. Full-length MCM5 and MCM5/550 were in-vitro-translated and 35S-labeled for GST pull-down assays with Sepharose-bead-bound GST, GST-Stat1α, GST-Stat1β, or GST-Stat1 TAD. (C) Cellular localization of the MCM5/550 mutant. 2fTGH cells were stably transfected with pEGFP-C2 or pEGFP-C2-MCM5/550 expression vectors. Living green fluorescence was detected, and localization of nucleus was visualized by DAPI staining. (D) Disruption of interaction between endogenous Stat1α and MCM5 by overexpressed MCM5/550 protein. U3A or 2fTGH cells were transiently transfected with pEGFP-C2 or pEGFP-C2-MCM5/550 expression vectors. Cells were left untreated or were treated 12 h after transfection with IFN-γ for 30 min. Total cell lysates were immunoprecipitated with an anti-Stat1 antibody and subjected to SDS/PAGE followed by Western blotting. M5/550, MCM5/550. (E) Inhibition of Stat1 transcriptional activity by MCM5/550. U3A cells were cotransfected with pRcCMV-Stat1 and pEGFP-C2 or pEGFP-C2-MCM5/550 together with the 3xly6E-GAS luciferase reporter and an internal control Renilla luciferase reporter. Cells were left untreated or were treated 12 h after transfection with IFN-γ for 6 h and harvested for luciferase assay. Results are shown as the means ± SD of three independent experiments.
To further demonstrate that the interaction between Stat1 and MCM5 plays a role in Stat1-mediated gene activation, we tested whether the MCM5/550 domain could be used as a dominant-negative mutant to inhibit the interaction. We generated a GFP-MCM5/550 fusion protein to allow visualization of its subcellular localization. When 2fTGH cells were transfected with GFP alone, green fluorescence was diffusely detected throughout the whole cell (Fig. 3Ce). However, when GFP-MCM5/550 was expressed in the cells, green fluorescence was only detected in the nucleus (Fig. 3Cf), indicating that this GFP-MCM5/550 fusion is localized in the nucleus just as endogenous MCM proteins are (33). This nuclear localization of MCM5/550 was not affected by different methods of expression or specific cell types (data not shown). To test whether this MCM5/550 domain has any effect on the interaction between Stat1 and MCM5, we expressed GFP-MCM5/550 in U3A cells or 2fTGH cells (Fig. 3De, lanes 3, 4, 7, and 8). Cells were untreated or treated with IFN-γ for 30 min. Stat1 proteins in whole-cell lysates were immunoprecipitated only in 2fTGH cells (Fig. 3 Da and Dc, lanes 5–8) and not in the Stat1-null U3A cells (Fig. 3 Da and Dc, lanes 1–4). MCM5 was coimmunoprecipitated with Stat1 in 2fTGH cells with GFP expression and IFN-γ treatment (Fig. 3Db, lane 6). However, when MCM5/550 was overexpressed in 2fTGH cells, very little MCM5 was coprecipitated with Stat1 (Fig. 3Db, lane 8). This result indicates that the MCM5/550 domain can disrupt the in vivo interaction between Stat1 and MCM5 and potentially serve as a dominant-negative mutant. To examine whether the interaction between Stat1 and MCM5 is required for Stat1 activity, U3A cells were transiently transfected with Stat1, GFP, or GFP-MCM5/550 together with a Stat1-dependent luciferase reporter, 3xLy6E (30). IFN-γ treatment induced an ≈10-fold increase in luciferase activity in cells with GFP coexpression (Fig. 3E). However, in cells that contained GFP-MCM5/550, luciferase activity decreased ≈60% in response to IFN-γ (Fig. 3E). All together, these results demonstrate that the interaction between MCM5 and Stat1 is necessary for Stat1 transcription activity.
The MCM5 Protein Is Essential for Stat1 Target Gene Activation in Response to IFN-γ. To further prove that MCM5 is essential for Stat1-mediated transcriptional activation, we used the RNAi technique to “knock down” the level of endogenous MCM5. We generated two different siRNAs against MCM5. When used individually, each of these MCM5 siRNA could decrease the MCM5 protein level partially (data not shown). When both MCM5 siRNA oligos were used, they could significantly decrease the level of MCM5 protein (Fig. 4Aa, lane 4) and MCM5 RNA in 293T cells (Fig. 4C). The expression of Stat1 (Fig. 4Ab, lane 4) and the phosphorylation of Stat1 on Tyr-701 (Fig. 4B Upper, lane 8) were not affected by the MCM5 siRNA, indicating that the IFN-γ signaling pathway is not affected by the MCM5 knockdown. The lower band in the Stat1 blot is Stat1β (32). As a loading control, the expression of extracellular signal-regulated kinase 2 was not affected (Fig. 4Ac). In addition, the MCM5 siRNA did not affect the expression of MCM3 mRNA (Fig. 4D). The effect of MCM5 siRNA on the expression of several Stat1 target genes was analyzed by real-time RT-PCR. In nontransfected cells or cells transfected with the negative control scrambled siRNA, IFN-γ treatment could induce the expression of IRF-1, TAP1, CIITA, or GBP1 (Fig. 4 E–H). When the cells were transfected with the MCM5 siRNA, the IFN-γ-inducible expression of these genes were significantly inhibited (Fig. 4 E–H). These results strongly demonstrate that MCM5 is essential for Stat1-mediated gene activation.
Discussion
The MCM2–MCM7 family of proteins was initially identified as a group of genes essential for DNA replication (1). However, their abundance in cells has lead to the speculation of their function in other cellular processes that involve DNA (2). The identification of interactions between members of the MCM2–MCM7 family and proteins not known to be involved in DNA replication further supports the concept of a multifunction role for the MCM2–MCM7 proteins. The rapid induction of RNA synthesis mediated by Stat1 in response to IFN-γ stimulation provides us with a system to investigate the physiological importance of the interaction between Stat1 and MCM5 for transcription activation. Our results show that the binding of MCM proteins to the IRF-1 promoter is Stat1-dependent and IFN-γ-inducible (Fig. 1), indicating that these MCM proteins are part of the transcription process. In particular, although Stat1 interacts directly only with MCM5 (25), other members of the MCM family are also present on the IRF-1 promoter (Fig. 1B) as well as several other Stat1 target gene promoters (Fig. 1C), which would suggest that the whole hexameric MCM complex is recruited to a transcription start site through the specific interaction between a transcription activator and one member of the MCM family.
Furthermore, the presence of MCM5 and MCM3 in the middle of the IRF-1 gene (Fig. 2) suggests that the MCMs are also involved in transcription elongation. DNA helicases necessary for the initiation of transcription have been well defined. These helicases include the subunits of the transcription factor IIH complex, which specifically interact with RNA Pol II complex on the promoter (34, 35). The DNA helicase involved in transcription elongation has remained a mystery. Several DNA helicase-containing chromatin remodeling complexes, such as SWI/SNF (switch genes/sucrose nonfermentation), have been implicated in maintaining transcription by RNA Pol II (36). However, there is no demonstrated specific interaction between these complexes with the RNA Pol II. Our results would suggest that the MCM2–MCM7 proteins travel along with the RNA Pol II to unwind DNA during transcription elongation just as they do for DNA replication elongation (4, 5). The interaction between MCM2 and the Pol II C-terminal domain (26, 27) would provide an anchor and guide for the MCM2–MCM7 hexamer to move along the DNA template during transcription elongation.
The MCM2–MCM7 proteins are highly conserved from yeast to human (37). The conserved MCM box, containing the ATPase/helicase domain, is in the middle of all MCM proteins. The N- and C-terminal regions have no homology between members of the MCM2–MCM7 family within a single species. But for each individual member, there is sequence similarity between species in the terminal regions. It is conceivable that the N- and C-terminal regions are used for specific protein–protein interaction for the assembly of the MCM2–MCM7 hexamer. The diversity of the terminal regions could also provide interaction surfaces for other non-MCM proteins. In this report, we identified the C-terminal region of MCM5 (MCM5/550) as an independent domain that can directly interact with Stat1 (Fig. 3) and MCM3 (data not shown). This domain can interrupt the interaction between endogenous Stat1 and MCM5, resulting in inhibition of Stat1-mediated transcriptional activation. Although the in vitro interaction between MCM5 and Stat1 in GST pull down assays does not require tyrosine or serine phosphorylation (probably because of the high concentration of purified proteins), the in vivo interaction between endogenous Stat1 and MCM5 was only detectable in IFN-γ-treated cells (Fig. 3D), suggesting that the dimer form of Stat1 with serine phosphorylation may be necessary for Stat1 to interact with MCM5 at an optimal level to recruit MCM5 for transcription activation.
The functional necessity of MCM5 for Stat1 activity is further demonstrated by the RNAi knockdown of MCM5. By knocking down the level of MCM5 but not eliminating it, the RNAi technique allowed us to obtain viable cells that have a substantial reduction in endogenous MCM5 protein (Fig. 4). Our results clearly indicate that the MCM5 protein is essential for the induction of Stat1 target gene expression in response to IFN-γ stimulation. Because of the abundance of the MCM proteins and their excess compared with the number of replication origins, the MCM proteins may very well play a dual role for both DNA replication and transcription activation, two essential biological processes for all cells. In addition, the small amount of MCM5 proteins in the knock-down cells allowed the cells to proceed through the cell cycle (data not shown), suggesting that the “default” function of the MCMs are for DNA replication and they are only recruited for transcription through interaction with an activated transcription factor, which, in this study, was Stat1 in response to IFN-γ. Whether this recruitment and redistribution of MCMs can occur by other transcription factors remains to be further investigated.
Acknowledgments
We thank George Stark and Ian Kerr for the U3A cell lines, the Department of Microbiology and Immunology at the Weill Medical College of Cornell University for the use of the Applied Biosystems PRISM 7900HT Sequence Detection equipment, and all of the laboratory members for their help. This work was supported by National Institutes of Health Grant GM61652 and American Heart Association Grant 0455896T (to J.J.Z.).
Abbrevaitions: STAT, signal transducer and activator of transcription; MCM, minichromosome maintenance; siRNA, small inhibitory RNA; RNAi, RNA interference; TAD, transcription activation domain; Pol II, polymerase II; IRF, IFN regulatory factor; TAP, transporter associated with antigen processing; GBP, guanylate-binding protein; CIITA, class II transactivator.
References
- 1.Tye, B. K. (1999) Annu. Rev. Biochem. 68, 649-686. [DOI] [PubMed] [Google Scholar]
- 2.Forsburg, S. L. (2004) Microbiol. Mol. Biol. Rev. 68, 109-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishimi, Y. (1997) J. Biol. Chem. 272, 24508-24513. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio, O. M., Weinstein, D. M. & Bell, S. P. (1997) Cell 91, 59-69. [DOI] [PubMed] [Google Scholar]
- 5.Labib, K., Tercero, J. A. & Diffley, J. F. (2000) Science 288, 1643-1647. [DOI] [PubMed] [Google Scholar]
- 6.Holthoff, H. P., Baack, M., Richter, A., Ritzi, M. & Knippers, R. (1998) J. Biol. Chem. 273, 7320-7325. [DOI] [PubMed] [Google Scholar]
- 7.Thommes, P., Kubota, Y., Takisawa, H. & Blow, J. J. (1997) EMBO J. 16, 3312-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei, M., Kawasaki, Y. & Tye, B. K. (1996) Mol. Cell. Biol. 16, 5081-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tye, B. K. & Sawyer, S. (2000) J. Biol. Chem. 275, 34833-34836. [DOI] [PubMed] [Google Scholar]
- 10.Donovan, S., Harwood, J., Drury, L. S. & Diffley, J. F. (1997) Proc. Natl. Acad. Sci. USA 94, 5611-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J. & Fangman, W. L. (2001) Science 294, 115-121. [DOI] [PubMed] [Google Scholar]
- 12.Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., Bell, S. P. & Aparicio, O. M. (2001) Science 294, 2357-2360. [DOI] [PubMed] [Google Scholar]
- 13.Newlon, C. S. (1997) Cell 91, 717-720. [DOI] [PubMed] [Google Scholar]
- 14.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell. Biol. 3, 651-662. [DOI] [PubMed] [Google Scholar]
- 15.O'Shea, J. J., Gadina, M. & Schreiber, R. D. (2002) Cell 109, S121-S131. [DOI] [PubMed] [Google Scholar]
- 16.Darnell, J. E., Jr., Kerr, I. M. & Stark, G. M. (1994) Science 264, 1415-1421. [DOI] [PubMed] [Google Scholar]
- 17.Chrivia, J. C., Kwok, R. P. S., Lamb, N., Hagiwara, M., Montminy, M. R. & Goodman, R. H. (1993) Nature 365, 855-859. [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharya, S., Eckner, R., Grossman, S., Oldread, E., Arany, Z., D'Andrea, A. & Livingston, D. M. (1996) Nature 383, 344-347. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, J. J., Vinkemeier, U., Gu, W., Chakravarti, D., Horvath, C. M. & Darnell, J. E., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckner, R., Ewen, M. E., Newsome, D., Gerdes, M., DeCaprio, J. A., Lawrence, J. B. & Livingston, D. M. (1994) Genes Dev. 8, 869-884. [DOI] [PubMed] [Google Scholar]
- 21.Lu, B., Reichel, M., Fisher, D. A., Smith, J. F. & Rothman, P. (1997) J. Immunol. 159, 1255-1264. [PubMed] [Google Scholar]
- 22.Moriggl, R., Berchtold, S., Friedrich, K., Standke, G. J., Kammer, W., Heim, M., Wissler, M., Stocklin, E., Gouilleux, F. & Groner, B. (1997) Mol. Cell. Biol. 17, 3663-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulson, M., Pisharody, S., Pan, L., Guadagno, S., Mui, A. L. & Levy, D. E. (1999) J. Biol. Chem. 274, 25343-25349. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, J. J., Zhao, Y., Chait, B. T., Lathem, W. W., Ritzi, M., Knippers, R. & Darnell, J. E., Jr. (1998) EMBO J. 17, 6963-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DaFonseca, C. J., Shu, F. & Zhang, J. J. (2001) Proc. Natl. Acad. Sci. USA 98, 3034-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yankulov, K., Todorov, I., Romanowski, P., Licatalosi, D., Cilli, K., McCracken, S., Laskey, R. & Bentley, D. L. (1999) Mol. Cell. Biol. 19, 6154-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland, L., Gauthier, L., Bell-Rogers, P. & Yankulov, K. (2002) Eur. J. Biochem. 269, 5192-5202. [DOI] [PubMed] [Google Scholar]
- 28.Sterner, J. M., Dew-Knight, S., Musahl, C., Kornbluth, S. & Horowitz, J. M. (1998) Mol. Cell. Biol. 18, 2748-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, E., Henriksen, M. A., Schaefer, O., Zakharova, N. & Darnell, J. E., Jr. (2002) J. Biol. Chem. 277, 13455-13462. [DOI] [PubMed] [Google Scholar]
- 30.Wen, Z., Zhong, Z. & Darnell, J. E., Jr. (1995) Cell 82, 241-250. [DOI] [PubMed] [Google Scholar]
- 31.Muller, M., Laxton, C., Briscoe, J., Schindler, C., Improta, T., Darnell, J. E., Jr., Stark, G. R. & Kerr, I. M. (1993) EMBO J. 12, 4221-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindler, C., Fu, X.-Y., Improta, T., Aebersold, R. & Darnell, J. E., Jr. (1992) Proc. Natl. Acad. Sci. USA 89, 7836-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu, B., Burkhart, R., Schulte, D., Musahl, C. & Knippers, R. (1993) Nucleic Acids Res. 21, 5289-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzder, S. N., Sung, P., Bailly, V., Prakash, L. & Prakash, S. (1994) Nature 369, 578-581. [DOI] [PubMed] [Google Scholar]
- 35.Moreland, R. J., Tirode, F., Yan, Q., Conaway, J. W., Egly, J. M. & Conaway, R. C. (1999) J. Biol. Chem. 274, 22127-22130. [DOI] [PubMed] [Google Scholar]
- 36.Eisen, A. & Lucchesi, J. C. (1998) BioEssays 20, 634-641. [DOI] [PubMed] [Google Scholar]
- 37.Kearsey, S. E. & Labib, K. (1998) Biochim. Biophys. Acta 1398, 113-136. [DOI] [PubMed] [Google Scholar]