Abstract
Viruses represent an attractive system with which to study the molecular basis of mRNA capping and its relation to the RNA transcription machinery. The RNA-dependent RNA polymerase NS5 of flaviviruses presents a characteristic motif of S-adenosyl-l-methionine-dependent methyltransferases at its N-terminus, and polymerase motifs at its C-terminus. The crystal structure of an N-terminal fragment of Dengue virus type 2 NS5 is reported at 2.4 Å resolution. We show that this NS5 domain includes a typical methyltransferase core and exhibits a (nucleoside-2′-O-)-methyltransferase activity on capped RNA. The structure of a ternary complex comprising S-adenosyl-l-homocysteine and a guanosine triphosphate (GTP) analogue shows that 54 amino acids N-terminal to the core provide a novel GTP-binding site that selects guanine using a previously unreported mechanism. Binding studies using GTP- and RNA cap-analogues, as well as the spatial arrangement of the methyltransferase active site relative to the GTP-binding site, suggest that the latter is a specific cap-binding site. As RNA capping is an essential viral function, these results provide a structural basis for the rational design of drugs against the emerging flaviviruses.
Keywords: crystal structure/flavivirus/GTP/RNA capping/RNA polymerase
Introduction
The cap is a unique structure found at the 5′-end of viral and cellular eukaryotic mRNA (Bisaillon and Lemay, 1997; Furuichi and Shatkin, 2000). It is critical for both mRNA stability and binding to the ribosome during translation. mRNA capping is a co-transcriptional modification resulting from a series of three chemical reactions (Shuman, 2001). The 5′-triphosphate of the mRNA is first converted to a diphosphate by an RNA triphosphatase. The second reaction is a transfer of a guanosine monophosphate (GMP) moiety from GTP to the 5′-diphosphate RNA by a guanylyltransferase to yield G5′-ppp5′-N. In a third reaction utilizing S-adenosyl-l-methionine (AdoMet) as the methyl donor, the transferred guanosine moiety is methylated by a (guanine-N7)-methyltransferase (N7MTase) to yield 7MeG5′-ppp5′-N (cap 0 structure). A second methylation reaction catalysed by a (nucleoside-2′-O-)-methyltransferase (2′OMTase) occurs on the first nucleotide 3′ to the triphosphate bridge to yield 7MeG5′-ppp5′-NMe (cap 1 structure). Adjacent nucleotides 3′ to the first one can also be 2′-O-methylated to various extents. The order of the methyltransfer reactions is variable, and in some viruses GTP is methylated at its N7 position before being transferred to the RNA 5′-end (Ahola and Kaariainen, 1995; Furuichi and Shatkin, 2000).
Few enzymes involved in the RNA capping pathway have been structurally characterized. The crystal structures of two cellular RNA triphosphatases from yeast and mouse have been determined at 2.0 and 1.6 Å resolution, respectively. These structures provide a mechanistic insight into the first reaction in the RNA capping pathway catalysed by prototypic enzymes of the metal-dependent and -independent RNA triphosphatase family, respectively (Lima et al., 1999; Changela et al., 2001).
Structural insights into the DNA virus PBCV-1 (Chlorella virus) guanylyltransferase in complex with GTP have revealed mechanistic determinants of the second reaction of the RNA capping pathway (Hakansson et al., 1997). The crystal structure of the double-stranded RNA Reovirus core at 3.6 Å resolution comprises a subunit of the λ2 protein as the sole example of a structurally defined RNA virus guanylyltransferase (Reinisch et al., 2000). It revealed that the guanylyltransferase domain adopts a different fold from that of the Chlorella virus enzyme, but provided no structural information about GTP binding.
The crystal structure of three RNA cap MTases involved in the third reaction of the RNA capping pathway has been determined. They are the 2′OMTase VP39 of the double-stranded DNA vaccinia virus (Hodel et al., 1996), and the N7MTase and 2′OMTase domains of core protein λ2 of Reovirus, a double-stranded RNA virus (Reinisch et al., 2000). RNA cap MTases share a common fold with a vast family of AdoMet-dependent MTases comprising small-molecule MTases, protein MTases, DNA (adenine-N6-), (cytosine-5-) and (cytosine-N4-) MTases, rRNA (adenine-N6-) MTases and rRNA 2′OMTases (Fauman et al., 1999; Bugl et al., 2000; Wang et al., 2000). Although they methylate different substrates, they all share a common catalytic core domain consisting of a seven-stranded β-sheet surrounded by six α-helices. Depending on the size of the substrate, target recognition domains might be appended to the catalytic core (Fauman et al., 1999). Alternatively, the MTase core domain can be integrated in a multidomain protein as is the case in protein λ2 of Reovirus (Reinisch et al., 2000), or in a protein complex. The only common feature of AdoMet-dependent MTases at the sequence level is a short motif involved in AdoMet binding (Ingrosso et al., 1989).
Vaccinia virus VP39 represents the sole structurally characterized viral MTase for which 2′-O-methyltransfer specificity has been ascertained (Barbosa and Moss, 1978). The crystal structure of the Reovirus λ2 protein showed the physical organization of two structurally related AdoMet-dependent mRNA MTase domains (Reinisch et al., 2000). Based on the spatial arrangement of the subdomains and the known methylation pathway, Reinisch et al. (2000) proposed an assignment of the N7MTase and 2′OMTase activities to respective domains of λ2; however, a sequence- and structure-based analysis of MTase substrate specificities suggested that this assignment should be swapped around (Bujnicki and Rychlewski, 2001). This proposition cannot be tested due to the difficulty of isolating enzymatically active λ2 protein.
Many viruses replicate in the cytoplasm of their eukaryotic host. Since cellular RNA capping is localized in the nucleus, these viruses often encode their own capping enzymes while relying on the host translation machinery for gene expression. Although RNA cap structures originating from viral or cellular enzymes are often identical, the physical organization of genes, subunit composition, structure, and catalytic mechanisms of enzymes from the capping apparatus differ significantly in fungi, metazoans, protozoa and viruses (Shuman, 2001). This diversity makes RNA capping an attractive target for drug design.
The group Flaviviridae comprises important human pathogens such as West Nile, Dengue and Yellow Fever viruses. Their single-stranded RNA genome is of positive polarity, and is capped with a cap 1 structure (Chambers et al., 1990). Little is known about the components of the flavivirus RNA capping machinery. RNA triphosphatase activity has been described for West Nile virus only, and has been mapped to the C-terminus of non-structural protein 3 (NS3) (Wengler, 1993). The guanylyltransferase has not yet been identified, and sequence analysis has revealed the presence of the characteristic motif of AdoMet-dependent MTases within the N-terminal domain of NS5, the viral RNA-dependent RNA polymerase (Koonin, 1993).
Flaviviruses are emerging mosquito-born agents that are expanding their distribution across the world. Dengue virus, an agent responsible for haemorrhagic fever, infects more than 50 million people every year, with an increasing incidence in the world’s tropical regions (Rigau-Perez et al., 1998; Isturiz et al., 2000). Likewise, the recent introduction of West Nile virus in North America may be an important milestone in the evolving history of this virus, as exemplified by outbreaks in the New York area (Anderson et al., 1999) and the subsequent geographic extension of its endemic zone (Enserink, 2001). Several regions of Europe re-witnessed West Nile virus infection in the late 1990s (Hubalek and Halouzka, 1999; Lvov et al., 2000). As inhibitors of viral replicases currently meet with clinical success, the capping and polymerization apparatus of flaviviruses remains a poorly defined but attractive therapeutical target.
In this report we present the crystal structure at 2.4 Å resolution of the N-terminal MTase domain of Dengue virus NS5 in complex with the product of the methylation reaction, S-adenosyl-l-homocysteine (AdoHcy). Enzym atic analysis and the structural conservation of characteristic amino acid residues in the active site demonstrate that this domain functions as a 2′OMTase. In addition, we found a GTP-binding site located on an N-terminal appendage of the MTase domain. This site demonstrates novel folding and achieves specific binding of guanine in an original manner.
Results
Structure determination and model quality
Based on sequence alignments, hydrophobicity profiles and secondary structure predictions, we delineated a putative globular domain at the N-terminus of NS5. This domain comprised the MTase signature within a consensus MTase domain (Fauman et al., 1999) and preceeded the known nuclear localization sequence (NLS; Forwood et al., 1999) (Figure 1A). Cloning of the corresponding DNA fragment of Dengue virus type 2 (New Guinea strain) in several expression vectors was performed, and tested for protein expression in Escherichia coli. The isolated domain was overexpressed as a soluble protein (His6-tagged) and subsequently purified (see Materials and methods). The 33-kDa recombinant protein comprised residues 1–296 and was named NS5MTaseDV (protein NS5, MTase domain, Dengue virus). It crystallized with lithium sulfate as precipitant (see Materials and methods) in space group P3121, with cell parameters a = b = 111.6 Å, c = 56.2 Å. The crystal structure of NS5MTaseDV was determined using multi-wavelength anomalous dispersion (MAD). A bound Hg2+ ion was the anomalous scatterer. Like most structurally characterized MTases (Fauman et al., 1999), NS5MTaseDV is monomeric both in solution and in the crystal. An atomic model consisting of residues 7–267 was built in the 2.8 Å experimental electron density map and refined to a resolution of 2.4 Å. The remaining amino acids (268–296) were not observed in the electron density map, although examination of the crystal packing showed that there would be enough space to accommodate these missing residues. Mass spectrometry analysis of dissolved NS5MTaseDV crystals showed that the C-terminus missing in the structure is present in the crystallized protein. Thus, the C-terminal part appears to be flexible, probably due to the absence of the polymerase domain of NS5. The crystallographic R-factor is 23.6% for the data between 30 and 2.4 Å. Most of the residues (88%) lie in the most favoured region of the Ramachandran plot. Data collection and refinement statistics are reported in Table I.
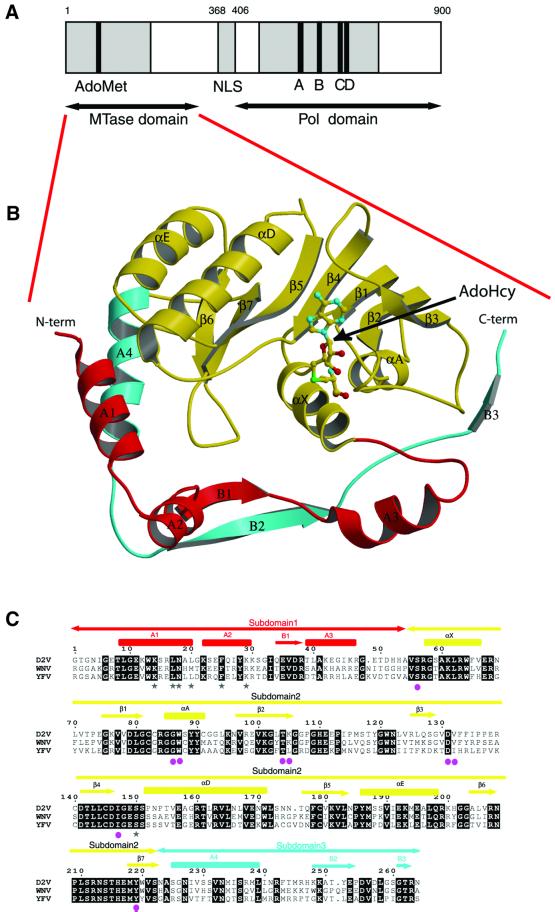
Fig. 1. General features of NS5MTaseDV. (A) Predicted domain structure of Dengue protein NS5. The putative N-terminal methyltransferase domain is shown in grey. The position of the AdoMet-binding motif I (residues 77–86) described by Koonin (1993) is highlighted in black. The region of the C-terminal polymerase domain containing motifs I to VIII of positive-strand RNA virus RNA polymerases (Koonin, 1991) is marked in grey. Motifs A to D, shared by RNA-dependent polymerases (Poch et al., 1989), are shown in black. AdoMet, S-adenosyl-l-methionine; NLS, nuclear localization sequence; Pol, polymerase. (B) Crystal structure of NS5MTaseDV in complex with AdoHcy. A ball-and-stick representation is used for AdoHcy, whereas NS5MTaseDV is drawn as a ribbon. The N-terminal subdomain of NS5MTaseDV (residues 7–54) is coloured red. The core subdomain (residues 55–222) has a typical AdoMet-dependent MTase topology and is coloured yellow. The C-terminal part of NS5MTaseDV (residues 223–267) is coloured cyan. The figure was generated using MOLSCRIPT (Kraulis, 1991) and rendered using RASTER3D (Merrit and Murphy, 1994). (C) Sequence alignment of flavivirus NS5MTases coloured according to the ribbon representation of NS5MTaseDV in (B). NS5MTase domains from Dengue virus type 2 New Guinea isolate (D2V), West Nile virus New York isolate (WNV) and Yellow Fever 17D (YFV) were aligned using Clustal_W (Thompson et al., 1994) and rendered using ESPript (Gouet et al., 1999). Secondary structures (α-helices and β-strands) of subdomains 1, 2 and 3 are indicated above the alignment and coloured in red, yellow and cyan, respectively. Helices and strands are named using Greek letters inside the core domain (subdomain 2), and roman letters outside (subdomains 1 and 3). Amino acids involved in GTP binding (see text) are indicated by a grey star below aligned sequences, and amino acids interacting with AdoHcy are indicated by a pink sphere.
Table I. Crystallization, data collection, structure solution and refinement statistics.
| Data set | Hg(CN)2 remote | Hg(CN)2 peak | Hg(CN)2 inflection | Native | GDPMP soak | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Resolution range (Å)a | 30–2.8 (2.95–2.8) | 30–2.8 (2.95–2.8) | 30–2.8 (2.95–2.8) | 30–2.4 (2.53–2.4) | 30–2.9 (3.06–2.9) | |
| Wavelength (Å) | 0.83211 | 1.00474 | 1.00850 | 0.933 | 0.933 | |
| Cell parameters (Å) | a = b = 111.6, c = 56.2 | a = b = 112.2, c = 56.5 | ||||
| Unique reflectionsb | 10 159 | 10 220 | 10 198 | 16 248 | 9310 | |
| I/(σ(I)) | 14.7 | 15.7 | 15.6 | 17.4 | 16.5 | |
| Rsym (%)a,c | 6.1 (30.4) | 5.5 (24.5) | 8.1 (30.5) | 5.1 (35.8) | 4.1 (27.7) | |
| Completeness (%)a,b | 99.7 (99.7) | 99.9 (99.9) | 99.8 (99.8) | 99.6 (99.6) | 99.9 (99.9) | |
| Multiplicitya,b | 4.9 (3.9) | 7.0 (6.2) | 7.0 (6.1) | 5.5 (5.2) | 4.1(4.1) | |
| MAD analysis | ||||||
| No. of sites | 1 | |||||
| Phasing power (acentrics/centrics) | 0.91/0.77 | 0.61/0.47 | ||||
| Rcullis (acentrics/centrics) | 0.87/0.86 | 0.96/0.94 | ||||
| Rcullis anomalous | 0.89 | 0.95 | ||||
| FOMmlphare (30–2.8 Å) | 0.36 | |||||
| FOMdm (30–2.4 Å) | 0.87 | |||||
| Refinement statistics | ||||||
| Resolution range (Å) | 30–2.4 | 30–2.9 | ||||
| No. of reflections (F>0) | 16 049 | 10 251 | ||||
| Rcrystd | 23.6 | 21.9 | ||||
| Rfreee | 25.0 | 25.0 | ||||
| R.m.s. deviations | ||||||
| Bonds (Å) | 0.009 | 0.008 | ||||
| Angles (°) | 1.485 | 1.418 |
aValues indicated in parentheses are for the highest resolution shell.
bUnique reflections, completeness and multiplicity assuming that F+ and F– are not equivalent.
cRsym = Σ|I – <I>|/Σ I, where <I> is the average intensity over symmetry equivalent reflections.
dRcryst = Σ||Fobs| – |Fcalc||/Σ|Fobs|. All data were used with no sigma cut-off. Summation is over the data used for refinement.
eRfree = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where Fobs are test set amplitudes (5% of the data) not used in the refinement.
Description of the structure and MTase fold identification
NS5MTaseDV has an overall globular fold with approximate dimensions of 55 × 45 × 40 Å, and can be subdivided into three subdomains (Figure 1B and C). The core subdomain 2 (in yellow, residues 55–222) folds into a seven-stranded β-sheet surrounded by four α-helices. This core structure closely resembles the catalytic domain of other AdoMet-dependent MTases (Fauman et al., 1999) and will be discussed later. Appended to the core are an N-terminal extension (subdomain 1 in red, residues 7–54) and a C-terminal extension (subdomain 3 in cyan, residues 223–267). The N-terminal subdomain comprises a helix– turn–helix motif followed by a β-strand and an α-helix. The C-terminal subdomain consists of an α-helix and two β-strands, and is located between subdomains 1 and 2.
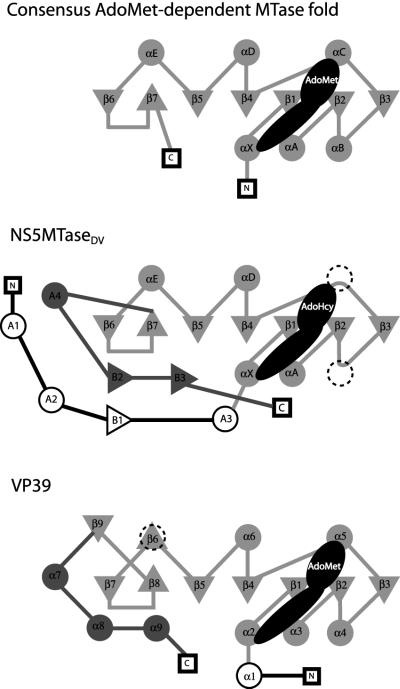
The consensus topology of the catalytic domain of Ado Met-dependent MTases, compared with NS5MTaseDV and the closely related mRNA cap MTase VP39, is shown in Figure 2. The consensus topology consists of a twisted mixed β-sheet comprising seven strands flanked by three α-helices on each side of the sheet (Fauman et al., 1999). The NS5MTaseDV core subdomain 2 adopts the depicted consensus fold with two major differences (Figures 1B and 2). First, helix B between β-strands 2 and 3 is virtually absent. It consists of only one turn immediately following β-strand 2, as in the DNA MTase M.TaqI (Schluckebier et al., 1997; Fauman et al., 1999). Secondly, helix C between β-strands 3 and 4 is completely missing in NS5MTaseDV, as it is again in M.TaqI, in another DNA MTase M.HhaI (O’Gara et al., 1996), in the protein MTase CheR (Djordjevic and Stock, 1997) and in a small-molecule MTase glycine-N-MTase (Fu et al., 1996). After β-strand 4, NS5MTaseDV matches the MTase consensus fold exactly. Comparing the NS5MTaseDV core domain with existing MTase structures, the nearest structural neighbours are the prokaryotic E.coli MTase FtsJ (Bugl et al., 2000) (r.m.s.d. of 2.4 Å for the 155 Cα atoms defined as structurally equivalent using the DALI server; Holm and Sander, 1993), M.TaqI (Schluckebier et al., 1997), the C-terminal domain of the fibrillarin homologue Mj0687 from the archaebacteria Methanococcus jannaschii (Wang et al., 2000) and the vaccinia virus MTase VP39 (Hodel et al., 1996). A comparison of the topologies of VP39 and NS5MTaseDV is discussed in detail below.
Fig. 2. Topological diagram of the consensus fold of AdoMet-dependent MTases (Fauman et al., 1999), NS5MTaseDV and VP39 (Hodel et al., 1996). Triangles represent strands and circles represent helices. The positions of the N- and C-termini are represented by squares. The elements of the consensus AdoMet-dependent MTase fold are coloured in light grey. Those that are replaced by different elements (loops or β-strand) in NS5MTaseDV or VP39 are dashed. AdoMet or AdoHcy bound at the C-terminus of the β-sheet is shown in black. The shape and dimension indicate which secondary structural elements are included in its contact zone. N- or C-terminal appendages of the MTase core in both NS5MTaseDV and VP39 are shown in black and dark grey, respectively.
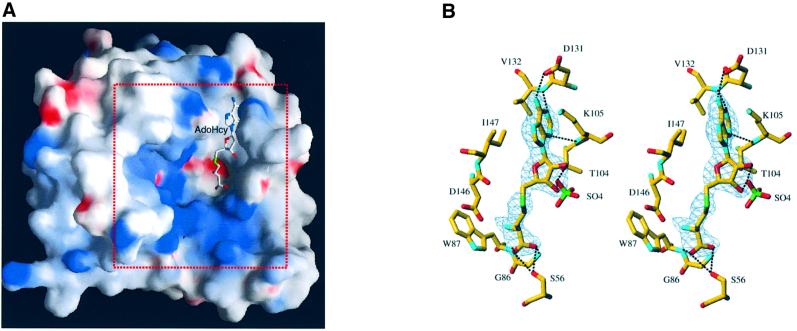
All the proteins mentioned above display a central cleft between β-strands 1 and 4. This cleft has been described as the active site where AdoMet and the substrate bind, and where the methyltransfer occurs (Fauman et al., 1999). Such a cavity is also present in the NS5MTaseDV structure (Figure 3A). During the refinement procedure, the Fo – Fc map revealed a residual strong density in this putative active site pocket. An AdoHcy molecule, a co-product of the methyltransfer reaction, was identified, modelled in this density and refined (Figure 3B). As neither AdoMet nor AdoHcy was added during crystallization, AdoHcy originated from E.coli and co-purified with NS5MTaseDV. The binding of AdoHcy relies on a network of hydrogen bonds (Figure 3B) and van der Waals interactions, similar to the described consensus interactions of MTase proteins (Fauman et al., 1999). The adenine is accommodated within a hydrophobic pocket defined by the side chains of Thr104, Lys105, Val132 and Ile147, and stabilized by hydrogen bonds with a side-chain oxygen of Asp131, and main-chain atoms of Lys105 and Val132 (all residues from β-strands 2, 3 and 4; see also Figure 1C). The ribose moiety is bridged via a water molecule to main-chain atoms of Gly106 and Glu111 (not shown), and to the side-chain oxygen of Thr104. The ribose is also hydrogen-bonded to a sulfate ion, which originates from the crystallization buffer and is conserved in the two crystal structures presented here. The remainder of the AdoHcy molecule is stabilized by hydrogen bonds to two well ordered water molecules, the side chain oxygen of Ser56 and Tyr219, and the main-chain nitrogens of Gly86 and Trp87 (residues from helices X and A).
Fig. 3. The AdoMet/AdoHcy-binding site. (A) General view of the position of AdoHcy within NS5MTaseDV. The solvent-accessible surface of NS5MTaseDV was calculated and is displayed using GRASP (Nicholls et al., 1991). It has been coloured according to electrostatic potential. Potential values range from –25 kT (red) to 0 (white) to +25kT (blue), where k is the Boltzman constant and T is the temperature. The AdoHcy molecule is shown in stick representation. The area indicated by the red dotted square delineates the sector shown in detail later in Figure 7B. (B) Stereo diagram of the AdoMet/AdoHcy-binding site in NS5MTaseDV. Carbons are displayed in yellow, oxygens in red, nitrogens in cyan and sulfur in green. The Fo – Fc difference map, contoured at 3σ, was calculated at 2.4 Å resolution from a model in which the ligand was omitted. It is clear from the electron density map that the molecule is a co-product of the methyltransfer, i.e. the AdoHcy molecule. Main hydrogen bonds between NS5MTaseDV and AdoHcy are indicated by black dotted lines. Also shown is a sulfate ion, originating from the crystallization conditions. This sulfate ion is hydrogen-bonded to the 2′- and 3′-oxygen of the AdoHcy ribose. For clarity, two water molecules and Tyr219, which are also interacting via hydrogen bonds, are not represented.
Methyltransferase activity assay
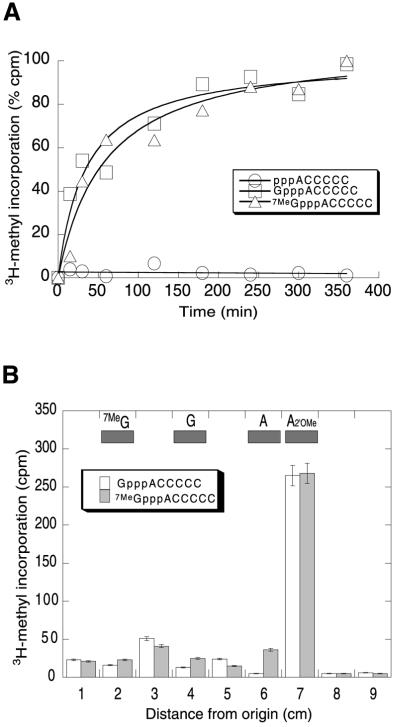
The enzymatic MTase activity of NS5MTaseDV was assayed by following the transfer of a radiolabelled methyl group from AdoMet to various RNA substrates using a filter-binding assay. Capped and non-capped short RNA substrates (GpppACCCCC, 7MeGpppACCCCC and ppp ACCCCC) were used as methyl acceptors. As shown in Figure 4A, the protein is able to transfer a methyl group from AdoMet to the capped RNA subtrates GpppACCCCC and 7MeGpppACCCCC, but not to the non-capped substrate pppACCCCC. Methyltransfer to capped RNA occurs even when the N7 position of the guanine is already methylated.
Fig. 4. MTase activity. (A) Assay of MTase activity. The extent of methyltransfer from Ado[methyl-3H]Met to three different RNA substrates (pppACCCCC, GpppACCCCC and 7MeGpppACCCCC) by 5 µg of NS5MTaseDV is plotted as a function of time. Data points represent the averages of three independent experiments and are presented as percentage of [methyl-3H] incorporation. The plateau of 100% incorporation represents a concentration of 1.5 µM transferred methyl groups in the reaction at the final reaction time. (B) Identification of the nucleoside methylated by NS5MTaseDV. RNAs incubated in the presence of Ado[methyl-3H]Met and purified recombinant NS5MTaseDV were treated with phosphodiesterase and alkaline phosphatase, and analysed using thin-layer chromatography. The experiment was performed independently twice. The figure shows a qualitative analysis of one chromatogram. Indicated positions of marker nucleosides [N7-methylated guanosine, (7MeG), guanosine (G), adenosine (A) and 2′-O-methylated adenosine (A2′OMe)] were determined under UV light. The radiolabelled products were analysed as described in Materials and methods.
In order to characterize the methylated nucleoside(s), the reaction mixture was treated with phosphodiesterase, which cleaves both RNA and cap structure, and alkaline phosphatase to render the nucleoside components. Separation of the reaction products using thin-layer chromatography (Figure 4B) shows that most of the radioactivity co-migrates with 2′-O-methylated adenosine (A2′OMe), and not with N7-methylated guanosine (7MeG). These results demonstrate that, under our experimental conditions, methylation occurs exclusively at the 2′-O position of the second nucleotide. They do not exclude, however, that the N7 position of the guanine would be methylated by NS5MTaseDV under conditions found in the replication complex in vivo. We conclude that NS5MTaseDV is the 2′OMTase of the Dengue virus. The physical coupling of this domain to the polymerase domain should be relevant to the coordination of the initiation of genomic (+) RNA synthesis and RNA capping.
Comparative analysis of NS5MTaseDV in relation to AdoMet-dependent MTases
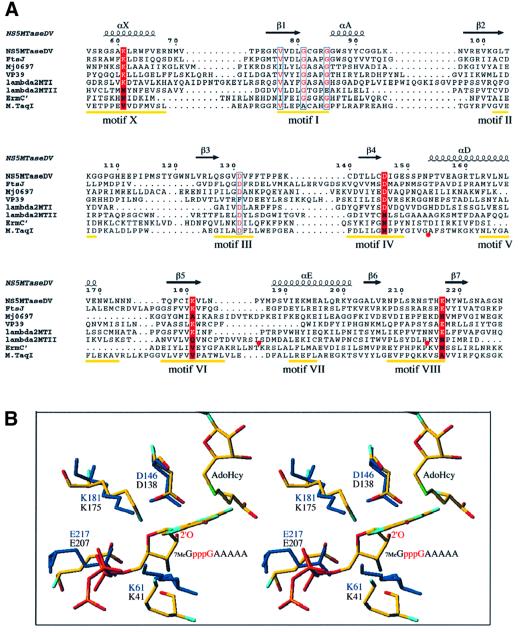
The large family of AdoMet-dependent MTases shows a high degree of structural homology that is not reflected at the amino-acid sequence level. It is thus difficult to establish relationships between existing AdoMet-dependent MTases (Fauman et al., 1999). For DNA MTases of different origins and specificities (methyltransfer to C5-cytosine, N6-adenosine and N4-cytosine) it has been possible to define nine conserved motifs that are involved in AdoMet binding and in catalysis of methyltransfer (Malone et al., 1995). Within the group of RNA MTases, comparable sequence conservation can only be found in specific subfamilies. For example, cellular and double-stranded DNA virus N7MTases show conservation at the sequence level that is not shared by RNA viruses (Bujnicki et al., 2001). Nevertheless, the existence of three-dimensional structures of RNA MTases allows a comparative analysis of these enzymes even if they present a high degree of diversity with respect to origin and specificity. Figure 5A shows a structure-based sequence alignment of the MTase core domain of NS5MTaseDV with rRNA MTase FtsJ (Bugl et al., 2000), the C-terminal rRNA MTase domain of Mj0697 (Wang et al., 2000), mRNA MTase VP39 (Hodel et al., 1996), mRNA MTase domains I and II of Reovirus protein λ2 (Reinisch et al., 2000) and the rRNA MTase ErmC′ (Bussiere et al., 1998). M.TaqI (Schluckebier et al., 1997) was included as a representative of the DNA MTases and a close structural neighbour of NS5MTaseDV. The positions of conserved DNA MTase motifs I–X (except for motif IX, which is conserved for C5-cytosine DNA MTases only; Posfai et al., 1989) are indicated. As expected, calculating the homology within the secondary structural elements present in NS5MTaseDV (134 residues spanning the β-strands and α-helices), we could not detect significant sequence conservation within the group of RNA MTases. NS5MTaseDV shares between 10 and 19% identity with the seven MTases listed in Figure 5A. After searching for conserved residues between all listed MTases within the nine motifs defined for DNA MTases (Figure 5A), sequence conservation was found in motif I, the universal AdoMet-binding motif (Koonin, 1993), and in motif III, which is also involved in AdoMet binding (Malone et al., 1995). Within the group of RNA MTases, we detected conservation of residues K61, D146, K181 and E217 (NS5MTaseDV numbering, marked in red in Figure 5A) for NS5MTaseDV, and FtsJ, VP39 and MTaseI of the λ2 protein of Reovirus. These residues are within motifs X, IV, VI and VIII defined for DNA MTases and, with the exception of motif X, are primarily involved in catalysis (Malone et al., 1995).
Fig. 5. Comparative analysis of NS5MTaseDV. (A) Structure-based sequence alignment of the MTase core domain of NS5MTaseDV with rRNA MTase FtsJ (Bugl et al., 2000), the C-terminal rRNA MTase domain of Mj0697 (Wang et al., 2000), mRNA MTase VP39 (Hodel et al., 1996), mRNA MTase domains I and II of Reovirus protein λ2 (Reinisch et al., 2000), the rRNA MTase ErmC′ (Bussiere et al., 1998) and the DNA MTase M.TaqI (Schluckebier et al., 1997). The positions of conserved DNA MTase motifs I to X [except for motif IX, which is only conserved for (cytosine-5-) DNA MTases; Posfai et al., 1989] are indicated in yellow (Malone et al., 1995). In order to simplify the alignment, additional structural elements in the sequences of λ2 MTase II (between residues Ile936 and Ser961 as well as Arg989 and Lys1014) and in M.TaqI (between residues Gly112 and Ala132) were omitted. The omissions are indicated by red circles below the sequences. The alignment was done manually using Seaview (Galtier et al., 1996) according to the superposition of the structures using TURBO (Roussel and Cambillau, 1991), and the final form of the alignment was generated with ESPript (Gouet et al., 1999). The secondary structure elements of the NS5MTaseDV core are given above the alignment and are named as in Figure 1B. The residues within DNA MTase motifs I and III, presenting similarity as detected by ESPript, are shown in red with blue outlines. Positions conserved between NS5MTaseDV, FtsJ, VP39 and λ2 MTase I, which coincide with motifs X, IV, VI and VIII, are marked by a red background where the conserved active-site residues K-D-K-E of 2′OMTases are in white. (B) Stereo view of the superposition of active site residues K-D-K-E of VP39 and NS5MTaseDV. Both structures are shown as sticks. The VP39 structure (PDB accession code 1AV6) is depicted with a RNA substrate (7MeG-capped RNA hexamer) and AdoHcy. VP39 residues K41-D138-K175-E207, AdoHcy and the RNA substrate are coloured according to atom type (oxygen in red, nitrogen in blue, carbon in yellow, sulfur in green and phosphorous in orange; hydrogens are not shown). The target 2′-OH group of the second nucleotide of the substrate is annotated (only a pppG fragment of the substrate 7MeGpppGAAAAA is depicted). NS5MTaseDV residues K61-D146-K181-E217 are shown in dark blue, AdoHcy was omitted.
VP39 has been demonstrated to be a 2′OMTase (Barbosa and Moss, 1978), and there is strong evidence that FtsJ exhibits the same substrate specificity (Caldas et al., 2000). The conservation of the previously mentioned K-D-K-E residues and their spacial position in the active sites of both VP39 and FtsJ was recently proposed as a characteristic of specific RNA 2′OMTases, leading to the re-assignment of the MTase domain I of λ2 protein as the Reovirus 2′OMTase (Bujnicki and Rychlewski, 2001). Figure 5B shows the position of the four conserved residues within the active site of VP39, the sole 2′OMTase for which a complex of the protein with its substrate and AdoHcy has been structurally characterized (Hodel et al., 1998). The complex did not allow determination of the exact catalytic mechanism of the transfer reaction. Nevertheless, the four conserved residues K41, D138, K175 and E207 (VP39 numbering) are the nearest to the active 2′-OH group of the substrate and may contribute to its deprotonation, which is required for the nucleophilic attack on the carbon atom of the AdoMet methyl group. The superposition of VP39 and NS5MTaseDV in Figure 5B shows the active site residues and their structural conservation in NS5MTaseDV. This structural conservation of the active site residues between NS5MTaseDV and other specific 2′OMTases is in agreement with our enzymatic data showing that NS5MTaseDV is the Dengue virus 2′OMTase. NS5MTaseDV does not show any structural resemblance in its active site region to the putative N7MTase of Reovirus (MTase domain II of λ2) (not shown). As no dual-specificity MTases have yet been characterized, from a structural viewpoint it is difficult to imagine how NS5MTaseDV would catalyse methylation of the N7-guanine of the RNA cap.
The binding of GTP and RNA cap analogues to NS5MTaseDV
The general description of the structure and the comparison of NS5MTaseDV to the consensus MTase fold showed the presence of additional structural elements at the N-terminus (subdomain 1) and the C-terminus (subdomain 3) of the core structure. Accessory substrate-recognizing domains following β-strand 7 and located in front of the β-sheet have been described for DNA and rRNA MTases (Fauman et al., 1999). In contrast, the mRNA cap 2′OMTase VP39 does not contain an additional domain, but N- and C-terminal ‘appendages’ of the MTase consensus fold (Hodel et al., 1999), and so does NS5MTaseDV. Nevertheless they are structurally different, as illustrated in Figure 2. In NS5MTaseDV, the N-terminal appendage (subdomain 1) consists of a helix–turn–helix motif involving helices A1 and A2, followed by β-strand B1 and helix A3. The C-terminal extension (subdomain 3) of the MTase core packs against the N-terminal subdomain and stabilizes it. It consists of helix A4, which is close and nearly parallel to helix A1, followed by a loop, and β-strand B2, which interacts via hydrogen bonding with B1 (Figure 1B). In VP39, the appendage provides a 7MeG recognition site, accommodating the first nucleotide of the mRNA cap 0. It was therefore of interest to test whether the NS5MTaseDV appendage could have a similar function.
To test the putative existence of a cap-recognition site in NS5MTaseDV we conducted binding experiments using purified NS5MTaseDV and various guanosine analogues. When [α-32P]GTP is incubated in the presence of NS5MTaseDV, no radiolabel remains bound to the protein upon denaturing gel electrophoresis (data not shown). UV irradiation allows a Mg2+-independent attachment of [α-32P]GTP to NS5MTaseDV that is stable enough to resist boiling and SDS–PAGE (Figure 6A, lanes 1 and 2). [α-32P]ATP is unable to label NS5MTaseDV significantly under similar conditions (Figure 6A, lane 3). Figure 6B shows the saturation curve of GTP binding to NS5MTaseDV, from which a dissociation constant (Kd) of 58 µM (±14 µM) was determined. Several nucleotides, such as CTP, UTP or 2′-deoxynucleoside 5′-triphosphates, were tested as possible ligands to NS5MTaseDV, but none competed significantly with GTP (not shown). The binding of radiolabelled GTP provided a convenient assay to test the binding of guanosine analogues, such as 7MeGTP and cap analogues GpppA and 7MeGpppA, to the same GTP-binding site. 7MeGTP and the cap analogue GpppA showed dissociation constants similar to that of GTP (64 ± 20 and 65 ± 1 µM, respectively), whereas 7MeGpppA showed a dissociation constant of 255 ± 5 µM, 4-fold higher than that of GTP (Figure 6C). These results indicate that NS5MTaseDV might bind the N7-unmethylated RNA cap preferentially to perform the 2′-O-methyltransfer reaction. The structural determinants of guanosine analogue binding were then investigated.
Fig. 6. The binding of GTP by NS5MTaseDV. (A) Cross-linking of [α-32P]GTP to NS5MTaseDV. NS5MTaseDV was incubated with 50 µM (1 µCi) of either [α-32P]GTP (lanes 1 and 2) or [α-32P]ATP (lane 3) in the presence (lanes 1 and 3) or absence (lane 2) of 5 mM Mg2+. Bacteriophage T4 DNA ligase (Lig) served as a positive control (lane 4). The upper panel shows the Coomassie Blue-stained gel, and the lower panel the corresponding autoradiographic analysis. (B) Determination of the dissociation constant Kd of [α-32P]GTP for NS5MTaseDV. Increasing concentrations of [α-32P]GTP were incubated with NS5MTaseDV, then UV irradiated to form a covalent complex and finally analysed using SDS–PAGE (upper panel) and autoradiography (lower panel). Background radioactivity in lane 0 results from cross-contamination from the adjacent well (not shown). Data points represent bound radioactivity and are the averages of three independent experiments. The Kd was determined using hyperbolic curve fitting. (C) Competition assay for the binding of cap analogues 7MeGTP, 7MeGpppA and GpppA, and ATP to wild-type NS5MTaseDV. NS5MTaseDV was incubated with 50 µM radiolabelled GTP and increasing concentrations of competitors. Data points represent the percentage of [α-32P]GTP-binding to NS5MTaseDV in the presence of a given competitor and are the averages of three independent experiments. Dissociation constants were determined using a descending hyperbolic curve fit.
Structural determinants of GTP binding to NS5MTaseDV
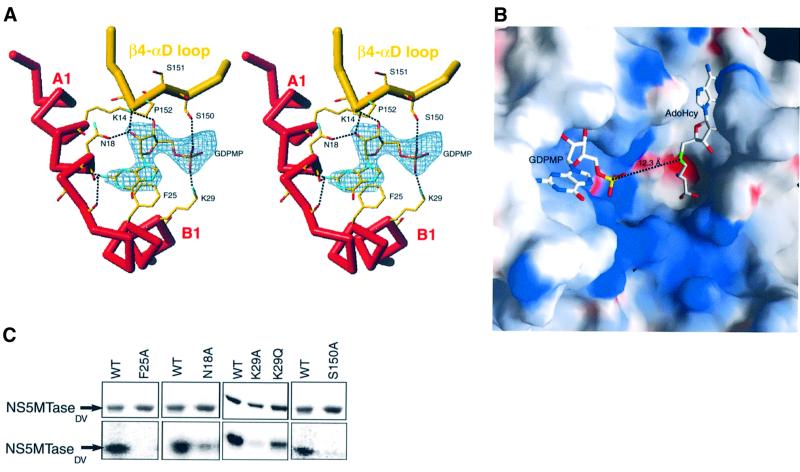
When crystals were soaked in a solution containing β,γ-methylene GTP (GDPMP), a non-hydrolysable GTP analogue, no rearrangement occurred in the crystal structure of the protein. A calculated difference Fourier map showed an additional density corresponding to the GDPMP molecule bound to subdomain 1 (Figure 7A). The base, ribose hydroxyl and α-phosphate moieties of GDPMP are well defined in the electron density map. The density was weaker and discontinuous for β- and γ-phosphates of GDPMP, suggesting mobility in the crystal; as a result they are not shown in Figure 7A and B. The phosphates of GDPMP point towards the MTase active site cleft, suggesting that the position of GTP might mimick that of an RNA cap. Support for this proposition comes from a comparison of VP39 with NS5MTaseDV. The relative position of the cap-binding site to the methyltransferase active site is comparable between the two proteins. The distance of 12.3 Å between the α-phosphorous atom of GDPMP and the sulfur atom of AdoHcy in NS5MTaseDV is similar to the 13.2 Å distance between the corresponding atoms of VP39 in complex with the RNA substrate and AdoHcy [Protein Data Bank (PDB) accession code 1AV6]. In addition, the positively charged region between GDPMP and AdoHcy could bind the negatively charged backbone of the RNA substrate (Figure 7B).
Fig. 7. The GTP-binding site of NS5MTaseDV. (A) Stereo view of the experimental (Fo – Fc) difference map (2.9 Å) contoured at 3σ in the vicinity of Phe25 in a GDPMP-soaked crystal. The Cα trace is shown in bold sticks and coloured as in Figure 1B and C. Side chains are shown in stick representation and coloured according to atom type. For clarity, non-interacting side-chains of residues 17, 19 and 20 are not shown. Dotted lines indicate hydrogen bonds between GDPMP and NS5MTaseDV. (B) Electrostatic surface representation of the MTase active site cleft and of the positively charged GDPMP binding site (framed area of Figure 3A). The figure was generated with GRASP (Nicholls et al., 1991) and coloured as in Figure 3A. (C) Mutational analysis of NS5MTaseDV. Purified NS5MTaseDV mutants (2 µg) were tested for their ability to bind [α-32P]GTP, as described for wild-type NS5MTaseDV in the legend to Figure 6A.
Detailed interactions with amino acids are indicated in Figure 7A. The amino acid side chains closest to the α-phosphate are those of Lys29 and Ser150. Ser150 and Pro152 belong to a loop connecting β4 to helix αD. The conformation of this loop seems to be important for GTP fixation. Interestingly, West Nile and Yellow Fever NS5MTases have a serine residue at position 152, compared with Pro152 in the case of Dengue (Figure 1C). One may anticipate that GTP or ribose-modified GTP analogues would bind to NS5MTaseWNV and NS5MTaseYFV with a different affinity to that observed for the Dengue enzyme. The ribose pucker is of Northern (3′-endo) configuration. The specificity for ribonucleotides (2′-OH) is provided by two specific hydrogen bonds involving Lys14 and Asn18 side chains, which are conserved amongst flaviviruses. The purine base of the nucleotide stacks against the aromatic ring of Phe25. The similarity in affinity for NS5MTaseDV in GTP and 7MeGTP, as observed in our cross-linking studies, is consistent with the lack of specific contacts at the N7 position. There is enough space to accomodate a methyl group at the N7 position without steric hindrance. Consequently, the difference in affinity between GpppA and 7MeGpppA is intriguing and indicates that GpppA and 7MeGpppA do not bind in an identical manner to NS5MTaseDV. Specificity for guanine is achieved via three interactions of main-chain carbonyl groups with the 2-amino group of guanine. None of these interactions would be possible either with adenine or with any other nucleobase, a finding consistent with our cross-linking studies.
A series of NS5MTaseDV mutants was constructed, and the corresponding proteins purified and tested for their ability to bind GTP (Figure 7C). The amino acid substitution F25A eliminates GTP binding, confirming the essential role of Phe25 in guanine binding. The K29A substitution nearly abolishes GTP binding, whereas the K29Q substitution retains subsequent GTP binding (∼50%). The S150A substitution nearly abolishes GTP binding in either wild-type or K29Q (not shown) background. This suggests that both a hydrogen bond from the serine hydroxyl group and a contact from residue 29 to the GTP α-phosphate are essential. N18A reduces GTP binding to ∼20% of the value found with the wild-type protein, indicating a possible permissivity for substitution at the 2′-OH position. These biochemical results are thus in agreement with our structural data.
An original feature of the putative cap-binding site of NS5MTaseDV lies in its high specificity for guanine (see Figure 6C). VP39 preferentially binds 7MeGTP and 7MeRNA cap analogues. Nevertheless it is able to bind other methylated nucleobases, although with 10- to 20-fold less affinity relative to methylated guanosine (Hu et al., 1999). In NS5MTaseDV, the specific contacts to the guanine base are provided exclusively by main-chain atoms to the 2-amino group. Examination of crystal structures of guanine-binding proteins deposited in the PDB failed to reveal the same type of specific interactions. For example, in two guanylyltransferase structures, the mRNA capping enzyme Paramecium bursaria Chlorella virus PBCV-1 guanylyltransferase (Hakansson et al., 1997) and the adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium (Thompson et al., 1999), and in the guanylate kinase structure from yeast (Stehle and Schulz, 1990), the specifity for guanine versus adenine is achieved at two guanine positions, the 6-oxo and the 2-amino group. In addition, this nucleotide-binding subdomain has no known homologue apart from that found in other flaviviruses, nor does it share any common structural feature with any protein structure deposited in the PDB as determined using the DALI server (Holm and Sander, 1993). Thus, the structural basis of both binding and selectivity for guanine found in NS5MTaseDV appears to be novel.
Conclusion
We have determined the crystal structure of an active MTase domain from a single-stranded RNA virus belonging to a family of emerging human pathogens. Enzymatic analysis and structural conservation of characteristic residues in the active site demonstrate that NS5MTaseDV functions as a 2′OMTase. A ternary complex made of GDPMP, AdoHcy and NS5MTaseDV suggests how cap-binding and 2′-O-methyltransfer are spatially organized. The GTP-binding site located on the N-terminal appendage of the typical MTase domain shows a previously unreported fold, and a structurally novel way of promoting specific binding of guanine. These structural features provide a unique basis for rational drug design against the emerging flaviviruses.
Materials and methods
Expression and purification of NS5MTaseDV
The full-length NS5 DNA of Dengue virus type 2 (New Guinea) was obtained from genomic RNA using RT–PCR. The N-terminal part of the NS5 DNA comprised amino acids 1–296 and was expressed as a hexahistidine recombinant protein using expression vector pQE30 (Qiagen) in E.coli strain BL21[pREP4]. NS5MTaseDV was obtained as a partially soluble protein after expression at 30°C for 5 h and sonication in 50 mM Bicine buffer, pH 7.5, 300 mM NaCl. It was adsorbed onto Ni-NTA–Sepharose (Pharmacia) and eluted with 50 mM EDTA. A second purification step consisted of cation-exchange chromatography (SP-Sepharose; Pharmacia) in 50 mM Bicine buffer, pH 7.5, 1 mM dithiothreitol (DTT), 10% glycerol, applying a salt gradient from 0.15 to 1.5 M NaCl. NS5MTaseDV eluted around 1 M NaCl. A size-exclusion column (Superdex-200 10/30; Pharmacia) was used to verify that the protein was homogeneous and monomeric, but was omitted from the standard purification protocol. The procedure described above typically yielded 3 mg of purified NS5MTaseDV from 1 l of bacterial culture. Final purity was >95% based on SDS–PAGE and mass spectrometry analysis. The protein was concentrated up to 22 mg/ml in 50 mM Bicine, pH 7.5, 0.8 M NaCl, 1 mM DTT and 10% glycerol using a Microsep 10 concentrator, and used for crystallization and biochemical assays. Mutant NS5MTaseDV genes were produced using commercially available kits from Stratagene (CA, USA). All gene constructs were verified by nucleotide sequencing. The variant proteins were purified following the same procedure used for wild-type NS5MTaseDV. They exhibited a similar behaviour during purification and an identical profile upon analytical gel filtration.
AdoMet-dependent methyltransferase assay
The methyltransferase assay was performed in 100 µl final volume containing 40 mM Tris–HCl pH 7.1, 100 µM AdoMet with 10 µCi of Ado[methyl-3H]Met (74 Ci/mmol; Amersham), purified NS5MTaseDV (5 µg) and 70 µl RNA substrate. The RNA substrate mix was produced using purified T7 DNA primase in conjunction with the synthetic DNA oligonucleotide T10CTG5 (10 µM), 300 µM CTP, and either 200 µM ATP to obtain pppAC5 or 200 µM cap analogue to obtain capped AC5 as described in Matsuo et al. (2000). The final RNA concentration in the primase reaction was ∼20 µM, as judged by analytical gel electrophoresis. The plasmid encoding T7 DNA primase was a kind gift from Charles C.Richardson (Harvard Medical School, Boston). The primase was purified as described previously (Frick et al., 1998). The methyltransfer reaction was monitored as follows: during 6 h of incubation at 30°C, 8-µl samples were spotted onto a DEAE-81 filter (Whatman) at given time points and washed with 20 mM sodium formate (pH 8) to remove the remaining AdoMet. Radioactively labelled RNA was quantified using liquid scintillation counting.
Analysis of the methylated cap nucleoside
Capped and non-capped RNA substrates were methylated in the presence of Ado[methyl-3H]Met and purified recombinant NS5MTaseDV for 3 h as described above. To identify the methylated nucleoside, RNAs were digested for 3 h at 37°C by adding 3 U of nucleotide pyrophosphat ase, type III (Sigma) containing 0.1 U phosphodiesterase, and 6 U calf intestinal alkaline phosphatase (New England Biolabs) in 15 µl of 50 mM Tris–HCl, pH 8.5, 5 mM MgCl2. Digests were applied directly onto a silica thin-layer sheet (Merck Eurolab Polylabo). Ascending chromatography was performed in ethylacetate/2-propanol/7.5 M NH4OH/1-butanol (3:2:2:1 v/v). Strips corresponding to individual migration lanes were cut into 1 cm2 pieces, which had been positioned relative to the origin of migration. The [methyl-3H]-labelled nucleosides present in individual squares were quantitated using scintillation counting, and were identified by comparison with authentic standards spotted on the same plate and visible under UV light (see Figure 4 legend). Background radioactivity, measured in triplicate (27 ± 3 c.p.m.), was substracted from each square.
Crystallization
Crystals were grown at room temperature in hanging drops. The protein solution (1 µl at 22 mg/ml) was mixed with 1 µl of a reservoir solution containing 0.1 M sodium citrate pH 5.8, 1.2 M lithium sulfate, 0.5 M ammonium sulfate, and was allowed to equilibrate by vapour diffusion for 1 week. Crystals belonging to space group P3121 (a = 111.5 Å, c = 56.3 Å) were cryoprotected in an artificial mother liquor supplemented with 20% glycerol, and flash frozen in a nitrogen stream. The asymmetric unit contained one protein molecule and 55% solvent. The complex with GDPMP (GTP analogue) was obtained by soaking crystals for 4–6 h in the artificial mother liquor, to which 2 mM GDPMP had been added. Data were collected at the European Synchrotron Radiation Facility (ESRF) on beamlines ID14-2, ID14-3 and BM14 using charge-coupled device detectors (ADSC Q4 or MAR 165). Images were processed using DENZO (Otwinowski and Minor, 1997) and intensities were merged with SCALA (CCP4, 1994).
Structure determination and refinement
The addition of selenomethionine to the bacterial growth medium was toxic to E.coli[p NS5MTaseDV]. Thus, crystals were soaked for 5 h in 0.5 mM Hg(CN)2. Difference isomorphous and anomalous Patterson functions were calculated using the CCP4 program RSPS (CCP4, 1994); they revealed one heavily occupied heavy-atom binding site. The structure was solved using the MAD method, taking advantage of the anomalous scattering properties of mercury. Three data sets were collected: (i) at the anticipated peak of the absorption edge; (ii) at its presumed inflection point; and (iii) at a high energy remote wavelength. Phases between 30 and 2.8 Å were calculated using MLPHARE (CCP4, 1994), and solvent flattening and phase extension to 2.4 Å were performed using DM (Cowtan, 1994). Despite the presence of only one anomalous scatterer, the initial electron density map was of good quality, and residues 10–261 were built and assigned unambiguously. A random 5% of the data was omitted from refinement for Rfree calculation (Brünger, 1992). Refinement was carried out using CNS (Brünger et al., 1998). It consisted of multiple cycles of torsion angle simulated annealing and B-factor refinement, including bulk solvent correction for data at <6 Å resolution. A maximum likelihood target was used and maps were calculated using SIGMAA weighting. Manual rebuilding between refinement cycles was performed using TURBO (Roussel and Cambillau, 1991). Stereochemistry was evaluated using PROCHECK (Laskowski et al., 1993) and indicated that the structure was within the idealized targets considering its resolution. The final model contained 261 residues (7–267), 65 water molecules, five sulfate ions, one mercury ion and one AdoHcy molecule. No density was observed for the N-terminal His6-tag and the following six residues, or for the 29 C-terminal residues.
Crystals of NS5MTaseDV in complex with GDPMP diffracted to 2.9 Å. The unit cell dimensions of this crystal were slightly different to those of the NS5MTaseDV with AdoHcy alone (Table I). The structure was solved by molecular replacement using the polypeptidic chain of the NS5MTaseDV–AdoHcy complex as a model. Both Fo – Fc and 3Fo – 2Fc SIGMAA-weighted electron density maps clearly indicated the presence of AdoHcy, five sulfate ions, and of the nucleoside and α-phosphate of GDPMP. The electron density for the β- and γ-phosphates was weak and therefore they were not modelled. As in the case of the NS5MTaseDV–AdoHcy complex, no density was observed for the first six and the last 31 residues. Refinement statistics are given in Table I. Atomic coordinates have been deposited in the PDB (accession code 1L9K).
Nucleotide binding experiments
The NTP binding assay was performed in the presence or absence of 5 mM MgCl2, in a 10 µl reaction volume containing 50 mM Tris pH 7.6, 5 mM DTT, [α-32P]NTP as indicated, and 2 µg of recombinant protein. The mixture was UV-irradiated for 3 min using a UV lamp (40 W, λ = 254 nm) at a 12 mm distance. The cross-linking reaction was stopped by the addition of gel-loading buffer, and samples were boiled for 5 min then subjected to SDS–PAGE using a 10% polyacrylamide gel. The gel was stained using Coomassie Blue dye, and radioactive products were visualized using photostimulatable plates and a FujiImager.
The dissociation constant Kd of GTP for NS5MTaseDV was determined using increasing concentrations of [α-32P]GTP (10, 25, 50, 100, 200 and 300 µM) cross-linked to 2 µg of NS5MTaseDV as described above. The bound radioactivity was quantified after SDS–PAGE using photostimulatable plates and a FujiImager. Data were fit to a hyperbolic function from which the Kd was determined. Dissociation constants for analogues were determined using a competitive binding assay based on the addition of increasing amounts of unlabelled analogues to a fixed concentration of NS5MTaseDV and radiolabelled GTP (58 µM, i.e. the Kd value of GTP for NS5MTaseDV).
Acknowledgments
Acknowledgements
We thank Christian Cambillau, Sylvie Doublié, Tom Ellenberger, Karine Alvarez, François Ferron, Hélène Dutartre and Sonia Longhi for helpful suggestions and critical reading of the manuscript, and Hugues Tolou and Boris Pastorino for help in the initial phase of the project. We thank Charles C.Richardson for the T7 DNA primase expression clone, and David Karlin for primase purification. We thank the staff from the JSBG beamlines for data collection at the ESRF Grenoble. This work was supported by a grant from the French Ministery of Research and Technology (programme ‘Maladies Infectieuses’). B.S. and D.B. are the recipients of post- and pre-doctoral fellowships from the Centre National de la Recherche Scientifique and the Direction Générale des Armées, respectively.
References
- Ahola T. and Kaariainen,L. (1995) Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl Acad. Sci. USA, 92, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.F., Andreadis,T.G., Vossbrinck,C.R., Tirrell,S., Wakem,E.M., French,R.A., Garmendia,A.E. and Van Kruiningen,H.J. (1999) Isolation of West Nile virus from mosquitoes, crows and a Cooper’s hawk in Connecticut. Science, 286, 2331–2333. [DOI] [PubMed] [Google Scholar]
- Barbosa E. and Moss,B. (1978) mRNA(nucleoside-2′-)-methyltrans ferase from vaccinia virus. Characteristics and substrate specificity. J. Biol. Chem., 253, 7698–7702. [PubMed] [Google Scholar]
- Bisaillon M. and Lemay,G. (1997) Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology, 236, 1–7. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–474. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed]
- Bugl H., Fauman,E.B., Staker,B.L., Zheng,F., Kushner,S.R., Saper,M.A., Bardwell,J.C. and Jakob,U. (2000) RNA methylation under heat shock control. Mol. Cell, 6, 349–360. [DOI] [PubMed] [Google Scholar]
- Bujnicki J.M. and Rychlewski,L. (2001) Reassignment of specificities of two cap methyltransferase domains in the reovirus lambda 2 protein. Genome Biol., 2, RESEARCH0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki J.M., Feder,M., Radlinska,M. and Rychlewski,L. (2001) mRNA:guanine-N7 cap methyltransferases: identification of novel members of the family, evolutionary analysis, homology modeling and analysis of sequence–structure–function relationships. BMC Bioinformatics, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussiere D.E. et al. (1998) Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry, 37, 7103–7112. [DOI] [PubMed] [Google Scholar]
- Caldas T., Binet,E., Bouloc,P., Costa,A., Desgres,J. and Richarme,G. (2000) The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem., 275, 16414–16419. [DOI] [PubMed] [Google Scholar]
- CCP 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chambers T.J., Hans,C.S., Galler,R. and Rice,C.M. (1990) Flavivirus genome organization, expression and replication. Annu. Rev. Microbiol., 44, 649–688. [DOI] [PubMed] [Google Scholar]
- Changela A., Ho,C.K., Martins,A., Shuman,S. and Mondragon,A. (2001) Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J., 20, 2575–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. (1994) DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsl. Protein Crystallogr., 31, 34–38. [Google Scholar]
- Djordjevic S. and Stock,A.M. (1997) Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure, 5, 545–558. [DOI] [PubMed] [Google Scholar]
- Enserink M. (2001) West Nile Watch. Science, 293, 1571. [Google Scholar]
- Fauman E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and evolution of AdoMet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyl transferases: Structures and Funtions. World Scientific Publishing, Singapore, pp. 1–38.
- Forwood J.K., Brooks,A., Briggs,L.J., Xiao,C.Y., Jans,D.A. and Vasudevan,S.G. (1999) The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem. Biophys. Res. Commun., 257, 731–737. [DOI] [PubMed] [Google Scholar]
- Frick D.N., Baradaran,K. and Richardson,C.C. (1998) An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc. Natl Acad. Sci. USA, 95, 7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Hu,Y., Konishi,K., Takata,Y., Ogawa,H., Gomi,T., Fujioka,M. and Takusagawa,F. (1996) Crystal structure of glycine N-methyl transferase from rat liver. Biochemistry, 35, 11985–11993. [DOI] [PubMed] [Google Scholar]
- Furuichi Y. and Shatkin,A.J. (2000) Viral and cellular mRNA capping: past and prospects. Adv. Virus Res., 55, 135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Gouy,M. and Gautier,C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Hakansson K., Doherty,A.J., Shuman,S. and Wigley,D.B. (1997) X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell, 89, 545–553. [DOI] [PubMed] [Google Scholar]
- Hodel A.E., Gershon,P.D., Shi,X. and Quiocho,F.A. (1996) The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell, 85, 247–256. [DOI] [PubMed] [Google Scholar]
- Hodel A.E., Gershon,P.D. and Quiocho,F.A. (1998) Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell, 1, 443–447. [DOI] [PubMed] [Google Scholar]
- Hodel A.E., Quiocho,F.A., Gershon,P.D. (1999) VP39—an mRNA cap-specific 2′-O-methyltransferase. In Cheng,X., Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltranferases: Structures and Functions. World Scientific Publishing, Singapore, pp. 255–282.
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hu G., Gershon,P.D., Hodel,A.E. and Quiocho,F.A. (1999) mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl Acad. Sci. USA, 96, 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubalek Z. and Halouzka,J. (1999) West Nile fever—a reemerging mosquito-borne viral disease in Europe. Emerg. Infect. Dis., 5, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingrosso D., Fowler,A.V., Bleibaum,J. and Clarke,S. (1989) Sequence of the d-aspartyl/l-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA and small molecule S-adenosylmethionine-dependent methyltransferases. J. Biol. Chem., 264, 20131–20139. [PubMed] [Google Scholar]
- Isturiz R.E., Gubler,D.J. and Brea del Castillo,J. (2000) Dengue and dengue hemorrhagic fever in Latin America and the Caribbean. Infect. Dis. Clin. North Am., 14, 121–140. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. (1991) The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol., 72, 2197–2206. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. (1993) Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and λ2 protein of reovirus. J. Gen. Virol., 74, 733–740. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of a protein structure. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lima C.D., Wang,L.K. and Shuman,S. (1999) Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell, 99, 533–543. [DOI] [PubMed] [Google Scholar]
- Lvov D.K. et al. (2000) Isolation of two strains of West Nile virus during an outbreak in southern Russia, 1999. Emerg. Infect. Dis., 6, 373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- Matsuo H., Moriguchi,T., Takagi,T., Kusakabe,T., Buratowski,S., Sekine,M., Kyogoku,Y. and Wagner,G. (2000) Efficient synthesis of 13C, 15N-labeled RNA containing the cap structure m7GpppA. J. Am. Chem. Soc., 122, 2417–2421. [Google Scholar]
- Merrit E.A. and Murphy,M.E.P. (1994) Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr., 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamics properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- O’Gara M., Roberts,R.J. and Cheng,X. (1996) A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase. J. Mol. Biol., 263, 597–606. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget,I., Delarue,M. and Tordo,N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J., 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai J., Bhagwat,A.S., Posfai,G. and Roberts,R.J. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch K.M., Nibert,M.L. and Harrison,S.C. (2000) Structure of the reovirus core at 3.6 Å resolution. Nature, 404, 960–967. [DOI] [PubMed] [Google Scholar]
- Rigau-Perez J.G., Clark,G.G., Gubler,D.J., Reiter,P., Sanders,E.J. and Vorndam,A.V. (1998) Dengue and dengue haemorrhagic fever. Lancet, 352, 971–977. [DOI] [PubMed] [Google Scholar]
- Roussel A. and Cambillau,C. (1991) In Graphics,S. (ed.), Silicon Graphics Directory. Mountain View, CA, p. 97.
- Schluckebier G., Kozak,M., Bleimling,N., Weinhold,E. and Saenger,W. (1997) Differential binding of S-adenosylmethionine S-adenosyl homocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI. J. Mol. Biol., 265, 56–67. [DOI] [PubMed] [Google Scholar]
- Shuman S. (2001) Structure, mechanism and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol., 66, 1–40. [DOI] [PubMed] [Google Scholar]
- Stehle T. and Schulz,G.E. (1990) Three-dimensional structure of the complex of guanylate kinase from yeast with its substrate GMP. J. Mol. Biol., 211, 249–254. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Clustal_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T.B., Thomas,M.G., Escalante-Semerena,J.C. and Rayment,I. (1999) Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU) complexed with GMP: evidence for a substrate-induced transferase active site. Biochemistry, 38, 12995–13005. [DOI] [PubMed] [Google Scholar]
- Wang H., Boisvert,D., Kim,K.K., Kim,R. and Kim,S.H. (2000) Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J., 19, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G. (1993) The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology, 197, 265–273. [DOI] [PubMed] [Google Scholar]