Abstract
Cdc45, which binds to the minichromosomal maintenance (Mcm) proteins, has a pivotal role in the initiation and elongation steps of chromosomal DNA replication in eukaryotes. Here we show that throughout the cell cycle in Saccharomyces cerevisiae, Cdc45 forms a complex with a novel factor, Sld3. Consistently, Sld3 and Cdc45 associate simultaneously with replication origins in the chromatin immunoprecipitation assay: both proteins associate with early-firing origins in G1 phase and with late-firing origins in late S phase. Moreover, the origin associations of Sld3 and Cdc45 are mutually dependent. The temperature-sensitive sld3 mutation confers a defect in DNA replication at the restrictive temperature and reduces an interaction not only between Sld3 and Cdc45, but also between Cdc45 and Mcm2. These results suggest that the Sld3–Cdc45 complex associates with replication origins through Mcm proteins. At the restrictive temperature in sld3-5 cells, replication factor A, a single-strand DNA binding protein, does not associate with origins. Therefore, the origin association of Sld3–Cdc45 complex is prerequisite for origin unwinding in the initiation of DNA replication.
Keywords: budding yeast/CDC45/DNA replication/Mcm proteins/SLD3
Introduction
Eukaryotic DNA replication initiates from multiple origins to replicate large genome DNA. Every replication origin fires once and only once during the S phase. This is mainly regulated by the initiation step of chromosomal DNA replication. The initiation of eukaryotic DNA replication is a complex process. In Saccharomyces cerevisiae, chromosomal DNA replication is initiated at a restricted region referred as an autonomously replicating sequence (ARS), which was initially identified as the DNA sequence that promoted the replication of episomal DNA (reviewed in Campbell and Newlon, 1991). ARS is bound by the origin recognition complex (Orc) throughout the cell cycle (Bell and Stillman, 1992; Diffley et al., 1995). The prereplicative complex (pre-RC) is assembled on each origin from late M to G1 phase. The pre-RC contains Orc, Cdc6 and six minichromosomal maintenance (Mcm) proteins (Diffley et al., 1995; Aparicio et al., 1997; Donovan et al., 1997; Liang and Stillman, 1997; Tanaka et al., 1997). Finally, origins fire only upon the subsequent activation of S phase cyclin-dependent kinases (S-Cdks) and Cdc7–Dbf4 kinase (Diffley, 1996; Stillman, 1996; Nougaréde et al., 2000). This activation causes origin unwinding and loading of replication machinery. Although it is not clear what catalyzes origin unwinding, Mcm complex is the most prevailing candidate to unwind origins, because a weak DNA helicase activity associates with a complex composed of human Mcm4, Mcm6 and Mcm7 (Ishimi, 1997).
In the well characterized simian virus 40 (SV40) system, large T antigen is an origin-binding protein and also a helicase. Initiation of DNA replication from SV40 origin begins with local unwinding of the origin by cooperative action of large T antigen and replication factor A (RF-A), a single-strand DNA-binding protein (reviewed in Herendeen and Kelly, 1996; Waga and Stillman, 1998). Subsequently, DNA polymerase (Pol) α-primase is loaded on the unwound origin to synthesize the RNA primer for the initiation of leading strand synthesis. In yeast, RF-A associates with origins at the timing corresponding to the initiation of replication, and thus this association reflects origin unwinding (Tanaka and Nasmyth, 1998).
Cdc45, which is involved in not only the initiation step, but also the elongation step of chromosomal DNA replication, associates with pre-RC at different timing at each origin: Cdc45 association with late origins is delayed relative to early origins (Aparicio et al., 1999; Tercero et al., 2000; Zou and Stillman, 2000). Moreover, the Cdc45 activity is required for Pol α and Pol ε associations with origins (Aparicio et al., 1999). In Xenopus egg extracts, Cdc45 forms a complex with Pol α and is required for chromatin associations of RF-A and Pol α (Mimura and Takisawa, 1998; Mimura et al., 2000; Walter and Newport, 2000). These results strongly suggest that Cdc45 has a pivotal role in the step from origin unwinding to loading of replication machinery.
Dpb11 is also involved in the loading of replication machinery in S.cerevisiae. The DPB11 gene was originally isolated as a multicopy suppressor of mutations in the POL2 and DPB2 genes, which encode the catalytic and second-largest subunits of Pol ε (Araki et al., 1995). The amino acid sequence of Dpb11 has similarity to the sequence of the Cut5 (also known as Rad4) protein of Schizosaccharomyces pombe. This protein is required for the onset of the S phase and cell cycle checkpoint in S.pombe (Saka and Yanagida, 1993; Saka et al., 1994, 1997; McFarlane et al., 1997; Verkade and O’Connell, 1998). Both Dpb11 and Cut5 have four copies of the BRCA1 C-terminus (BRCT) domain, which is thought to be an interaction domain between proteins (Bork et al., 1997; Callebaut and Mornon, 1997; Zhang et al., 1998). Dpb11 is required for DNA replication and the S phase checkpoint. During the S phase of the cell cycle, Dpb11 and Pol ε form a complex and associate preferentially with ARS. Associations of Dpb11 and Pol ε with ARS are mutually dependent and are required for the association of the Pol α-primase complex (Masumoto et al., 2000).
To understand the function of Dpb11, we have identified several factors that interact with Dpb11 by isolating synthetic lethal mutations with dpb11–1 (sld). So far, we have isolated genes SLD1–6. SLD1 is identical to DPB3, which encodes the third-largest subunit of Pol ε, and SLD4 is identical to CDC45. The SLD2 and DPB11 gene products form a complex that is essential for DNA replication (Kamimura et al., 1998). SLD2 is identical to DRC1 (DNA replication and checkpoint 1), which has been independently isolated, and the drc1-8 mutant was found to be defective in the S phase checkpoint (Wang and Elledge, 1999). SLD6 is identical to RAD53, which is required for S phase and damage checkpoints (our unpublished results).
In this paper, we report the characterization of one of the SLD genes, SLD3. The SLD3 gene is a novel gene encoding a 77 kDa protein essential for cell growth. We show that Sld3 and Cdc45 form a complex throughout the cell cycle using a cross-linking agent. Sld3 associates with different replication origins at different timing as does Cdc45, and the origin associations of Sld3 and Cdc45 are mutually dependent. Furthermore, we show that the function of Sld3 is required for the loading of RF-A onto origins. Therefore, we suggest that Sld3 together with Cdc45 functions at the origin unwinding and polymerase-loading steps of chromosomal DNA replication.
Results
The SLD3 gene is essential for cell growth
Previously, we reported the isolation of sld mutants and cloning of SLD genes (Kamimura et al., 1998). One of the SLD genes, SLD3, was later found to be identical to one of the genes whose mutation confers temperature-sensitive growth in spk1-101 background (Sugimoto et al., 1996; K.Sugimoto, personal communication) and corresponds to the YGL113w ORF (Paoluzi et al., 1997). SLD3 encodes a 77 kDa protein and its predicted amino acid sequence has similarities with those of the budding yeast YOR165w ORF and psl3 in S.pombe (R.Nakajima and H.Masukata, personal communication).
One of the copies of the SLD3 gene was disrupted in a diploid strain by replacement with LEU2, and the resultant strain was sporulated and the spores were dissected. All tetrads showed two viable and two lethal spores. All viable spore clones showed Leu–, indicating that SLD3 is essential for cell growth. To confirm that we did not incidentally inactivate any essential gene other than SLD3, the sld3Δ::LEU2/SLD3 diploid was transformed with YEp195SLD3 before sporulation. Many tetrads generated from this diploid strain yielded more than two viable spores, and some of them were Leu+. All Leu+ survivors were also Ura+, showing that the lethality of sld3Δ::LEU2 haploid was rescued by a wild-type (WT) SLD3 on the URA3 plasmid. Consistently, none of the Leu+ spores could survive on a 5-fluoroorotic acid (5-FOA) plate, confirming that the viability of sld3Δ::LEU2 haploid completely relied on the presence of the SLD3 plasmid.
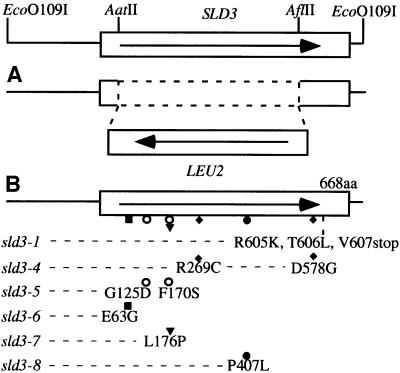
Since sld3-1 isolated by a synthetic lethal screening did not have any phenotype, we isolated five temperature-sensitive alleles (sld3-4, -5, -6, -7 and -8) of the SLD3 gene for the further analysis of Sld3 function, and identified their mutation sites (Figure 1B). After temperature was shifted up to 37°C, all the mutant cells arrested with a dumb-bell shape, with one nucleus adjacent to the isthmus between mother and daughter cells, which is the typical morphology for mutants defective in DNA replication.
Fig. 1. Location of mutation sites in the SLD3 gene. (A) For deletion, the 1476 bp region between AatII and AflII was replaced with LEU2. (B) Mutation sites in the SLD3 gene. The amino acid substitution is shown for each mutant allele. Nucleotide substitution occurring in each mutant is as follows (nucleotide 1 is A in the first ATG of the ORF): deletion of G at nucleotide 1817 in sld3-1; T for C at nucleotide 369, T for C at nucleotide 805 and G for A at nucleotide 1733 in sld3-4; A for G at nucleotide 374 and C for T at nucleotide 509 in sld3-5; G for A at nucleotide 188 in sld3-6; C for T at nucleotide 527 and C for T at nucleotide 1582 in sld3-7; and T for C at nucleotide 1220 in sld3-8.
High-copy CDC45 and SLD3 suppress growth defect caused by sld3 and cdc45 mutations
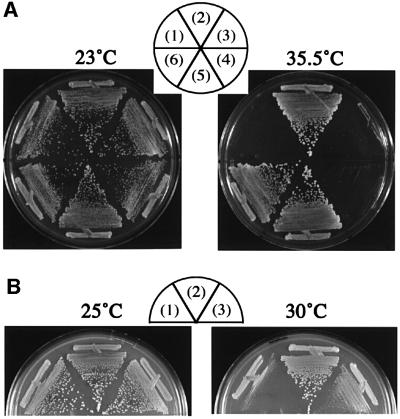
To identify the factors interacting with Sld3, we screened a multicopy suppressor(s) of sld3-5 temperature-sensitive growth. Sld3-5 cells transformed with the YEp24-based library were incubated at 23°C for 1 day, shifted to 35.5°C and incubated at 35.5°C for 3 days. Forty colonies, out of 36 400 transformants, appeared; 36 contained SLD3 and four contained CDC45 (Figure 2A). In fact, high-copy CDC45 also suppressed the growth defect of sld3-4, -7 and -8, but not -6 (data not shown).
Fig. 2. Suppression of temperature-sensitive growth of sld3-5 and cdc45-27. (A) YYK15 (sld3-5) cells harboring YCplac111 (1), YCp111SLD3 (2), YCp111CDC45 (3), YEplac195 (4), YEp195SLD3 (5) and YEp195CDC45 (6) plasmids were streaked onto YPD plates and incubated at the indicated temperature for 2 days. (B) YYK32 (cdc45-27) cells harboring YEplac195 (1), YEp195CDC45 (2) and YEp195SLD3 (3) plasmids were streaked onto YPD plates and incubated at the indicated temperature for 2 days.
We also studied whether high-copy SLD3 suppresses growth defect of cdc45 mutations. We used cold-sensitive cdc45-1 and thermosensitive cdc45-27 mutations. The cdc45-27 mutation was recently isolated by us and its detailed analysis will be described elsewhere. As shown in Figure 2B, cell growth of cdc45-27, but not cdc45-1 (data not shown), was suppressed by high-copy SLD3. These results show that Sld3 and Cdc45 interact genetically.
Physical interaction between Sld3 and Cdc45
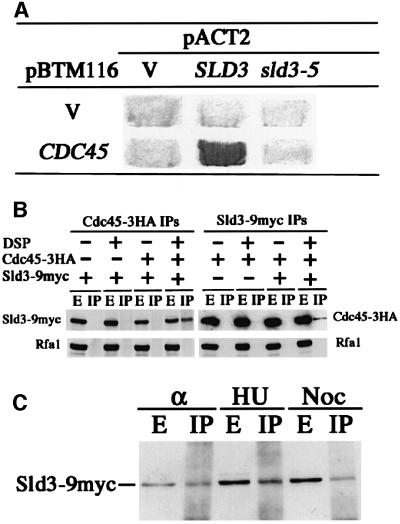
Genetic interactions described above suggested that the Sld3 and Cdc45 proteins interact physically. We therefore employed a two-hybrid assay. The ORF of CDC45 was fused to the LexA-binding domain of pBTM116 (Bartel and Fields, 1995), and the ORF of SLD3 was fused to the Gal4 activation domain of pACT2 (Bai and Elledge, 1997). The two resultant plasmids complemented the thermosensitive growth defect of cdc45-1 or sld3-5, respectively, indicating that the fused genes are functional. They were introduced into L40 cells, and expression of the lacZ gene was measured. As shown in Figure 3A, the lacZ gene was expressed clearly in cells transformed with those two plasmids, suggesting that Sld3 interacts physically with Cdc45. On the other hand, cells transformed with the sld3-5 plasmid instead of SLD3 showed reduced expression of lacZ (Figure 3A), suggesting that the interaction between Sld3-5 and Cdc45 is significantly reduced.
Fig. 3. Physical interaction between Sld3 and Cdc45. (A) V and Cdc45 in pBTM116 denote plasmids that express LexA and LexA–Cdc45, respectively. V, SLD3 and sld3-5 in pACT2 denote plasmids that express Gal4, Gal4–Sld3 and Gal4–Sld3-5, respectively. Transformants of L40 carrying each pair of the plasmids were assayed for β-galacto sidase activity by colony color with X-Gal. (B) Cells of YYK21 (Sld3-9myc), YYK22 (Cdc45-3HA) and YYK20 (Sld3-9myc, Cdc45-3HA) were spheroplasted and treated with DSP (+) or left untreated (–). HA-tagged Cdc45 was immunoprecipitated with anti-HA mouse Mab 12CA5. Myc-tagged Sld3 was immunoprecipitated with anti-myc mouse Mab 9E11. Extracts (E) or immunoprecipitates (IP) were separated in an SDS–8% polyacrylamide gel, and Sld3-myc proteins, Rfa1 proteins and Cdc45-HA proteins were detected by immuno blotting using anti-myc mouse Mab 9E10, anti-Rfa rabbit polyclonal antiserum and anti-HA rat Mab 3F10 (Roche Diagnostics), respectively. No specific Rfa1 signal in IP appeared even after longer exposure. (C) Lysates were prepared from cells arrested by α-factor (α), HU and nocodazole (Noc). Co-immunoprecipitation between Cdc45-HA and Sld3-myc was performed as described in (B).
To obtain the biochemical evidence of interaction between Sld3 and Cdc45, we employed co-immunoprecipitation assay. Cells harboring SLD3-9myc and CDC45-3HA on their chromosomes were spheroplasted, treated with the cross-linking agent, dithiobis (succimidylpropionate) (DSP), and Cdc45 or Sld3 was precipitated from cell lysate by the anti-HA or the anti-myc antibody. The immunoprecipitates were subjected to SDS–PAGE, followed by the detection of Sld3-9myc and Cdc45-3HA using anti-myc and anti-HA antibodies, respectively. As shown in Figure 3B, Sld3 was precipitated with Cdc45 by the anti-HA antibody and Cdc45 was precipitated with Sld3 by the anti-myc antibody. This co-precipitation is dependent on the presence of the cross-linking agent. As a control, we followed the Rfa1 protein, the largest subunit of RF-A complex associating with unwound origins and replication forks (Tanaka and Nasmyth, 1998), using the same membrane and the anti-Rfa antibodies, and found that neither Sld3 nor Cdc45 precipitates with Rfa1. Therefore, Sld3 co-immunoprecipitates specifically with Cdc45.
Next, we also examined the formation of the complex containing Sld3 and Cdc45 (Sld3–Cdc45 complex) during the cell cycle. Cells were arrested with α-factor (G1 phase), hydroxyurea (HU) (S phase) or nocodazole (G2–M phase) and treated with DSP, and Cdc45 was immunoprecipitated as described above. Co-immunoprecipitation of Sld3 with Cdc45 was observed in all stages of cells examined (Figure 3C). We were also able to co-precipitate Sld3 and Cdc45 from extracts prepared from cells with in vivo formaldehyde cross-linking throughout the cell cycle (data not shown). These results indicate that Sld3 and Cdc45 form a fragile complex throughout the cell cycle.
Sld3-5 cells are defective in the DNA replication
Sld3 and Cdc45 form a complex and Cdc45 is involved in the initiation step of chromosomal DNA replication. We therefore examined whether Sld3 is involved in the same step as Cdc45.
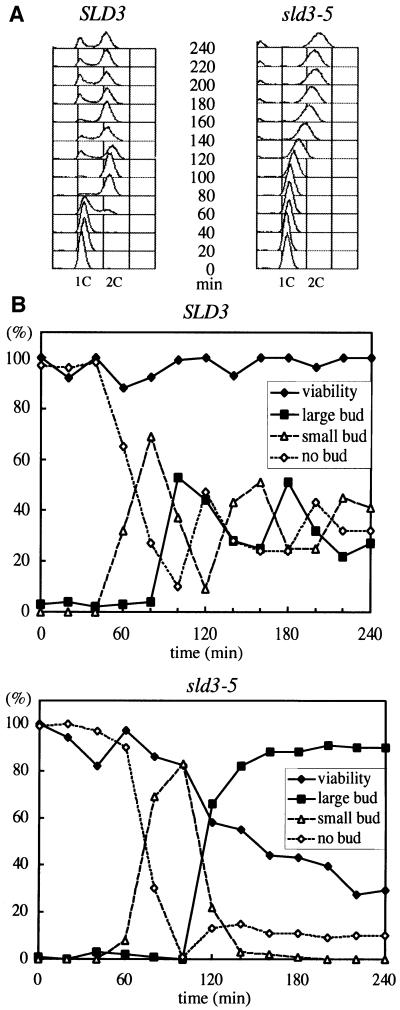
WT and sld3-5 cells were arrested in G1 phase by α-factor, released at 37°C, and DNA content was measured by FACS. The DNA content of WT cells began to increase at 60 min after release from α-factor block and completely shifted to 2C at 80 min. On the other hand, the DNA content of sld3-5 cells began to increase 40 min later than WT and it took at least 100 min to reach 2C, while the bud emerged in these two strains with almost the same timing (Figure 4). This result suggests that sld3-5 cells are defective in DNA replication.
Fig. 4. Delayed entry into S phase in sld3-5 cells. (A) Asynchronous cultures of YYK9 (wild type, SLD3) and YYK19 (sld3-5) cells were arrested in G1 with α-factor at 23°C in YPD. Cells were released from the block at 37°C and samples were collected at the time points indicated and analyzed by FACS. (B) Viability of YYK9 (wild type, SLD3) and YYK19 (sld3-5) cells after release from α-factor arrest at 37°C. Time course was the same as (A). Aliquots were withdrawn at the indicated times to determine cell morphology and cell number. Cells were spread on YPD plates and incubated at 23°C for 3 days. Colonies grown on YPD plates were scored.
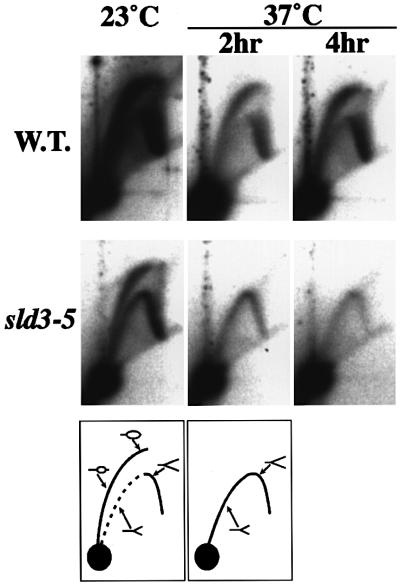
To examine the DNA replication defect in sld3-5 cells in detail, an active origin, ARS1, was analyzed by neutral/neutral two-dimensional gel (N/N 2-D gel) (Brewer and Fangman, 1987). WT and sld3-5 cells were grown to log phase at 23°C and shifted to 37°C for 2 or 4 h before harvest. At both 23 and 37°C, the WT cells gave a clear transition signal from bubble to fork arcs. This result indicates that replication initiates in the ARS1 region. On the contrary, a complete fork in addition to a bubble arc was observed in sld3-5 cells at 23°C. Furthermore, the bubble arc signals completely disappeared after temperature shift-up (Figure 5). This can be explained by the reduced frequency of initiation from ARS1 and the replication forks coming from outside of ARS1 region.
Fig. 5. N/N 2-D gel analysis of the chromosomal ARS1 locus in YYK9 (wild type, SLD3) and YYK19 (sld3-5) cells. Cells were grown and harvested at 23°C or shifted to 37°C for 2 or 4 h prior to harvest. DNA was digested with NcoI and probed with the ARS1 containing fragment. The diagrams below the photographs show the bubble and fork arcs.
Of chromosomal DNA replication, Cdc45 has roles not only in the initiation but also in the elongation step (Tercero et al., 2000). To further examine the function of Sld3 in the progression of S phase after DNA replication initiates, we tested whether Sld3 executes any function after an HU block, because HU is an inhibitor of ribonucleotide reductase and causes replication forks to stall.
WT or sld3-5 cells were arrested in G1 phase with α-factor and released into YPD medium containing 0.2 M HU at 23°C. Cells were then released from the HU block at 37°C, and nuclei were observed under an epifluorescence microscope after DNA was stained with 4′, 6-diamino-2-phenylindole. As shown in Table I, most of the sld3-5 cells showed one nucleus at 80 min after release from HU, while >50% of WT cells had two nuclei. Further incubation of sld3-5 cells did not significantly increase the population of cells having two nuclei. Since sld3-5 cells that were arrested in G2–M by nocodazole and released at the restrictive temperature divided with the same efficiency as did the WT cells (data not shown), the results indicate that Sld3 functions in S phase after the HU block.
Table I. Nuclear division after release from an HU block.
| Genotype | % of cells having two nucleia at |
||
|---|---|---|---|
| 0 min | 80 min | 200 min | |
| SLD3 | 0 | 53 | n.d.b |
| sld3-5 | 0 | 0 | 7 |
aCells were synchronized with α-factor, released to YPD medium containing 0.2 M HU at 23°C for 90 min, and then transferred to 37°C. After 30 min of incubation, cells were washed and released from the HU block at 37°C. At the indicated time after release from HU, nuclei were observed.
bn.d., not determined.
Sld3 associates with different origins in a temporally controlled manner
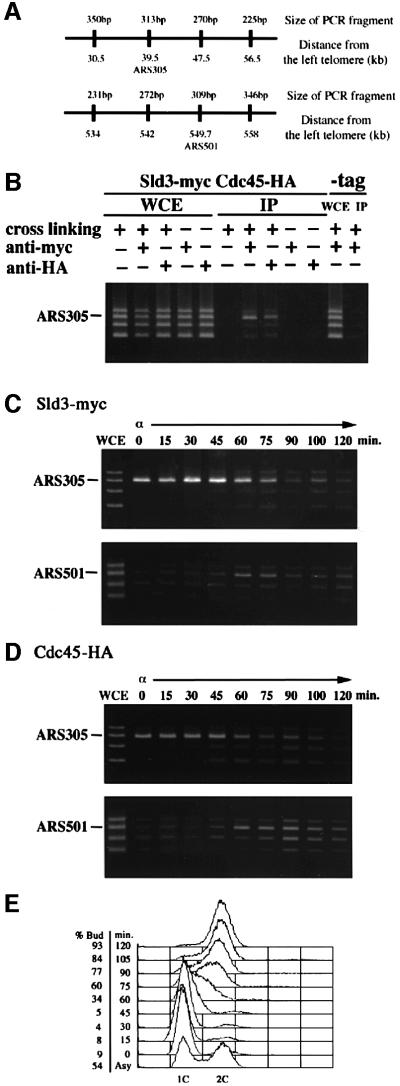
An in vivo cross-linking chromatin immunoprecipitation (CHIP) assay has been adopted successfully to localize several replication proteins on replication origins (Aparicio et al., 1997, 1999; Tanaka et al., 1997; Tanaka and Nasmyth, 1998; Masumoto et al., 2000; Zou and Stillman, 2000). Using this method, we demonstrated the association of Sld3 with origins. The Sld3-myc-tagged strain and the WT strain were treated with formaldehyde to promote cross-linking. The lysates from cells were immunoprecipitated with anti-myc antibody. The presence of ARS-containing fragments in immunoprecipitates was determined by PCR amplification. As shown in Figure 6B, ARS305 fragments were specifically amplified by PCR. This preferential amplification of ARS fragment was dependent on the treatment with formaldehyde and on the inclusion of anti-myc antibody. Furthermore, preferential amplification of ARS1, but not ARS305, was abolished in immunoprecipitates from a point mutant of ARS1-A (Marahrens and Stillman, 1994) that greatly reduces frequency of the initiation from ARS1 (data not shown). Therefore, it was concluded that Sld3-9myc associates with ARS.
Fig. 6. In vivo association of Sld3 protein with ARS-containing fragments. (A) Locations of fragments amplified by PCR. (B) Association of Sld3 or Cdc45 with ARS305 in vivo. YYK20 (SLD3-9myc CDC45-3HA) or YYK9 (-tag) cells were grown in YPR (2% yeast extract, 1% polypeptone, 2% raffinose) medium at 23°C. Immunoprecipitation procedures were performed with or without prior cross-linking, with or without anti-myc antibody or anti-HA antibody. PCR was performed on chromatin fragments isolated after the immunoprecipitation (IP) or on those from the whole-cell extract (WCE). (C) Temporally regulated origin associations of Sld3. YYK20 (SLD3-9myc CDC45-3HA) cells were arrested in G1 by α-factor and released in YPR at 23°C. Cells were withdrawn from the culture every 15 min. Sld3-myc was immunopreciptated from each extract by anti-myc antibody. PCR was performed on immunoprecipitates derived from the same number of cells at each time point. (D) Temporally regulated origin associations of Cdc45. Samples were the same as (C) except that Cdc45-HA was immunoprecipitated by anti-HA antibody. (E) DNA content was measured by FACS from samples collected in (C) and (D).
Next, cells were arrested in G1 phase with α-factor and released into the cell cycle at 23°C. In α-factor-blocked cells in G1 phase, Sld3 associated with an early origin, ARS305 (Figure 6C). This early origin association increased after α-factor release, peaked at 30 min and then gradually decreased. Since Cdc45 associated with ARS305 in G1 phase and this association signal slightly increased after α-factor release, the association mode of Cdc45 was almost the same as that observed for Sld3 (Figure 6D). In contrast to ARS305, Sld3 did not associate significantly with late-firing origin, ARS501, in G1 phase. However, it associated significantly with late origin in late S phase (Figure 6C). This result shows that Sld3 associates with late-firing origins substantially later in the cell cycle than with the early firing origin.
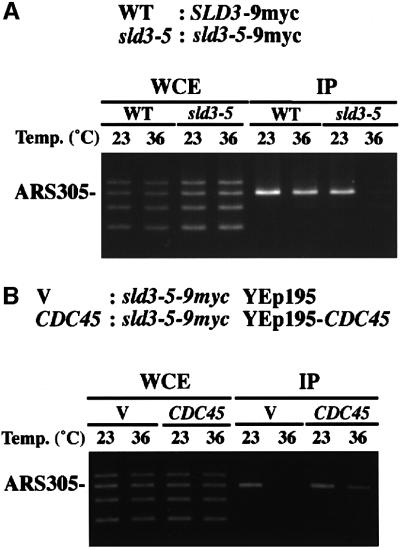
We tested further whether the Sld3-5 protein associates with origins. Cells having Sld3-9myc or Sld3-5-9myc were arrested in G1 phase by α-factor at 23°C. Half of each culture was shifted to 36°C and further incubated for 1 h in the presence of α-factor, while the other half was maintained at 23°C. Sld3-9myc associated with ARS305 at 23°C and 36°C, while Sld3-5 associated with ARS305 at 23°C, but this association was greatly reduced at 36°C (Figure 7A). Since sld3-5 cells were defective in DNA replication, these results suggest that association of Sld3 with origins is important for their role in DNA replication and the sld3-5 mutation reduced this activity at the restrictive temperature.
Fig. 7. The origin association of Sld3-5 is reduced and overproduction of Cdc45 restores the origin association of Sld3-5. (A) YYK21 (SLD3-9myc; WT) and YYK23 (sld3-5-9myc) cells were arrested in G1 phase by α-factor at 23°C for 3 h. Half of each culture was shifted to 36°C while the other half was maintained at 23°C and incubations were continued in the presence of α-factor for 1 h. (B) YYK23 (sld3-5-9myc) cells were introduced by YEplac195 (V) or YEp195-CDC45 plasmids (CDC45). V and CDC45 cells were arrested in G1 phase by α-factor at 23°C for 3 h. Half of each culture was shifted to 36°C while the other half was maintained at 23°C and incubations were continued in the presence of α-factor for 1 h. PCR was performed on chromatin fragments (IP) isolated after the immunoprecipitation or on those from the whole-cell extract (WCE).
As described above, high-copy CDC45 suppresses the thermosensitive growth of sld3-5 cells (Figure 2). We thus examined whether this suppression results from enhanced association of Sld3-5 with origins. Sld3-5-9myc cells harboring multicopy-plasmid (YEplac195) or YEp195CDC45 were subjected to CHIP assay as above. High-copy CDC45 partially restored association of Sld3-5-9myc with ARS305 at 36°C (Figure 7B). This result is consistent with the idea that high-copy CDC45 enhances the association of the Sld3-5 protein with origins, and that consequently sld3-5 cells grow at the restrictive temperature.
The origin associations of Sld3 and Cdc45 are mutually dependent
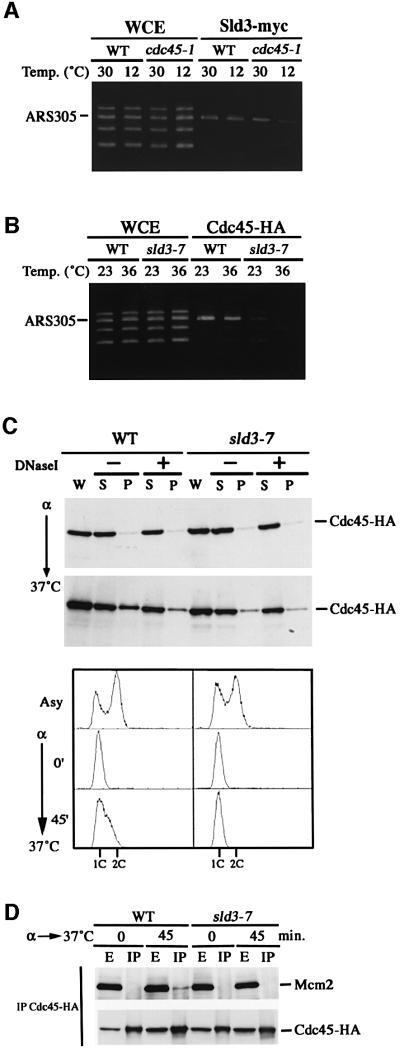
As Sld3 and Cdc45 form a complex, we investigated whether origin associations of Sld3 and Cdc45 are mutually dependent. A cold-sensitive cdc45-1 and the congenic WT cells were arrested in G1 phase by α-factor at 30°C. Half of each culture was shifted to 12°C and incubated further for 1 h in the presence of α-factor while the other half was maintained at 30°C. Then, we used these cells for the CHIP assay with the antibody against myc epitope. As shown in Figure 8A, Sld3-myc associated with ARS305 in both WT and cdc45-1 cells at 30°C. However, the association in cdc45-1 cells at 12°C was greatly reduced, indicating that Sld3 association with origins depends on functional Cdc45.
Fig. 8. Origin associations of Sld3 and Cdc45 are mutually dependent. (A) Origin association of Sld3 requires Cdc45. YYK21 (WT) and YYK29 (cdc45-1) cells were arrested in G1 phase by α-factor at 30°C for 2.5 h. Half of each culture was shifted to 12°C while the other half was maintained at 30°C and incubations were continued in the presence of α-factor for 1 h. PCR was performed on chromatin fragments (Sld3-myc) isolated after the immunoprecipitation or on those from the whole-cell extract (WCE). (B) Origin association of Cdc45 requires Sld3. YYK22 (WT) and YYK31 (sld3-7) cells were arrested in G1 by α-factor at 23°C for 3 h. Half of each culture was shifted to 36°C while the other half was maintained at 23°C and incubations were continued in the presence of α-factor for 1 h. PCR was performed on chromatin fragments (Cdc45-HA) isolated after the immunoprecipitation or on those from the whole-cell extract (WCE). (C) Cdc45 association of chromatin is reduced in the sld3-7 mutant cells. YYK22 (WT) and YYK31 (sld3-7) cells expressing Cdc45-3HA were synchronized in G1 phase by α-factor and released at 37°C. The cells were collected at the α-factor block or 45 min after release. Chromatin-binding experiments were performed as described in Materials and methods. The proteins present in the different fractions of the chromatin purification were examined by immunoblotting of SDS–PAGE: W, whole-cell extract; S, supernatant; P, pellet fraction. Extract was incubated on ice either without (–) or with (+) DNase I as described in Materials and methods. The lower panel was DNA content of the samples used in upper panel. (D) Cdc45–Mcm2 interaction is reduced in sld3-7 mutants at the nonpermissive temperature. Cells were arrested as in (C) and released at 37°C. Whole-cell extracts (E) were prepared from the cells collected at the α-factor block or 45 min after release and subjected to immunoprecipitations (IP) using an anti-HA antibody.
Next, we examined whether Sld3 is required for Cdc45 association with origins. For this purpose, we first attempted to substitute CDC45-3HA for WT CDC45 in sld3-5 cells. This was not successful, probably because CDC45-3HA is not compatible with the sld3-5 mutation. We then tried another SLD3 mutation, sld3-7, which occurred in the site close to the sld3-5 mutation site (Figure 1B) and is suppressed by high-copy CDC45, as is the sld3-5 mutation. Since the sld3-7 cells show higher restrictive temperature than the sld3-5 cells, we expected that even at the permissive temperature the sld3-7 mutation would confer a milder effect on cell growth than the sld3-5 mutation. As expected, we succeeded in replacing CDC45 with CDC45-3HA in sld3-7 cells. The resultant sld3-7 CDC45-3HA cells as well as the isogenic WT cells bearing CDC45-3HA were arrested in G1 phase by α-factor at 23°C. Half of each culture was shifted to 36°C, while the other half was maintained at 23°C, and incubation was continued for 1 h in the presence of α-factor. After incubation, these cells were used for CHIP assay with an antibody against HA epitope. As shown in Figure 8B, the ARS305 fragment was preferentially amplified at 23 and 36°C in WT cells. However, in sld3-7 cells, ARS association signal was faint even at 23°C and abolished at 36°C (Figure 8B). This result suggests that Cdc45 association with origin is dependent on the function of Sld3. Taken together, origin associations of Cdc45 and Sld3 in G1 phase are mutually dependent.
Since we do not have evidence that origin associations of Cdc45 and Sld3 in G1 phase are prerequisite for their origin associations after release from G1 arrest, we examined further whether Cdc45 and Sld3 in mutant cells associated with origins after α-factor release. Cdc45-1 SLD3-9myc and sld3-7 CDC45-3HA cells synchronized in G1 phase with α-factor at permissive temperature (30°C for cdc45-1 and 23°C for sld3-7) were shifted to the restrictive temperature (12°C for cdc45-1 and 36°C for sld3-7) and incubated further for 30 min. Cells were then released from the block at the restrictive temperature and samples were collected at various time. Origin associations of Cdc45 in sld3-7 cells and Sld3 in cdc45-1 cells were reduced in comparison with those in WT cells (data not shown). Hence, the origin associations of Sld3 and Cdc45 were found to be mutually dependent.
Chromatin-binding assays showed that Cdc45 associates with chromatin during S phase (Zou and Stillman, 1998). The sld3-7 and the isogenic WT cells were released from an α-factor block and were incubated further at 37°C to start the cell cycle. The bud emerged in these two strains with almost the same timing (data not shown). At 45 min, Cdc45 on chromatin in sld3-7 cells was less abundant than that in WT cells. Cdc45 in the pellet (P) fraction from the WT cells associated with chromatin because it was reduced by DNase I treatment. On the contrary, DNase I treatment did not reduce the amount of Cdc45 in the pellet (P) fraction from sld3-7 cells (Figure 8C). Furthermore, we did not detect the chromatin binding of Cdc45 in sld3-7 cells until 60 min after release from an α-factor (data not shown). These results showed that Cdc45 in sld3-7 cells hardly binds to chromatin and chromatin association of Cdc45 is dependent on Sld3. Although we can not neglect the possibility that Sld3 function is prerequisite for chromatin binding of Cdc45, Sld3-dependent chromatin binding of Cdc45 is consistent with the previous result obtained using the CHIP assay.
During S phase, Cdc45 on chromatin physically interacts with Mcm proteins, at least one of which, Mcm2, is required for chromatin association of Cdc45 (Zou and Stillman, 1998). Since Cdc45 does not bind to chromatin in sld3-7 cells, we examined the Cdc45–Mcm2 association in sld3-7 cells. WT and mutant cells were first synchronized in G1 phase with α-factor and then released at 37°C. At 45 min after release, cells were collected, disrupted, and Cdc45-3HA was precipitated with anti-HA antibody. While the Mcm protein co-precipitated with Cdc45 in WT cells, we did not observe Mcm2 in the immunoprecipitates from sld3-7 cells (Figure 8D). The reduction of Cdc45–Mcm2 interaction in the sld3-7 mutant cells is consistent with the reduction of chromatin-bound Cdc45 in the mutant cells. These results suggest that Sld3 is prerequisite or required directly for the association of Cdc45 with Mcm2.
Sld3 is required for RF-A association with origins
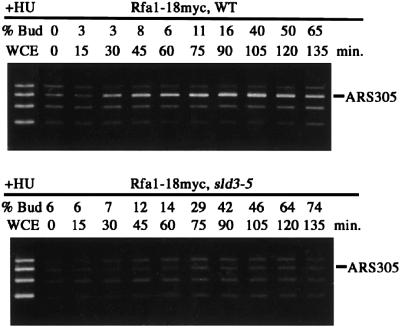
The pre-RC is assembled at ARSs from late M to G1 phase. This complex is then activated by Cdk and Cdc7 protein kinase, and origin DNAs are unwound and stabilized by RF-A (Tanaka and Nasmyth, 1998). For association of RF-A with origin, Cdc45 is required (Mimura et al., 2000; Zou and Stillman, 2000). Moreover, we showed above that Cdc45 does not bind chromatin without functional Sld3. Thus, it was expected that RF-A association with origin does not occur if the Sld3 function is lost. To examine this hypothesis, we analyzed ARS association of RF-A in the temperature-sensitive sld3-5 mutant and WT cells released from α-factor block at 34°C. As shown in Figure 9, Rfa1 associated with ARS305 in WT cells. However, no significant amplification of ARS DNA fragments was detected in the Rfa1 immunoprecipitates from sld3-5. Therefore, this result indicates that Sld3 is required for ARS association of RF-A and further suggests that the Sld3 function is prerequisite for origin unwinding.
Fig. 9. The origin association of Rfa1 depends on Sld3. YHM014 (RFA1-18myc; WT) and YYK32 (RFA1-18myc; sld3-5) cells were arrested in G1 phase by α-factor at 23°C for 3 h and released in YPR containing 0.2 M HU at 34°C. Cells were withdrawn from the culture every 15 min. Rfa1-18myc was immunoprecipitated from each extract by an anti-myc antibody. PCR was performed on immunoprecipitates derived from the same number of cells at each time point.
Discussion
Sequential assembly of replication proteins on origins
To initiate DNA replication, dynamic changes must occur at origins. In S.cerevisiae, DNA replication origins are bound by Orc throughout the cell cycle, and the Mcm complex is recruited by Cdc6 from late M phase to G1 phase to form the pre-RC complex. Cdc45 joins the pre-RC and then replication origins are unwound in cooperation with Cdk and Cdc7 protein kinases. Finally, DNA polymerases are recruited to origins and this step requires Dpb11 (Masumoto et al., 2000).
We showed contradictory observations for Cdc45 association with replication origins. Using CHIP assay, we observed that Cdc45 associates with early-firing origins in G1 and S phase; we did not detect significant association of Cdc45 with total chromatin in G1 phase in chromatin-binding assays. This observation agrees with the results of the CHIP assay described in Aparicio et al. (1997, 1999) and with the results of chromatin binding assay in Zou and Stillman (1998, 2000). Diffley (1998) and Aparicio et al. (1999) explained the discrepancy by reflection of distinct modes of Cdc45–chromatin association and Aparicio et al. (1999) termed these putative binding modes of Cdc45 ‘bound’ (Cdk independent and occurring in G1 phase) and ‘engaged’ (Cdk dependent and occurring only in S phase). That is, bound-mode Cdc45 association may be disrupted during isolation without cross-linking, while engaged-mode Cdc45 association may be stable. In contrast to this dual mode hypothesis, Zou and Stillman (2000) described the association signal of Cdc45, even with early origins in CHIP assay, being detected only in S phase. Although we do not know why the signal was detected only in S phase, we agree that the strongest origin association of Cdc45 occurs in S phase, since we and Aparicio et al. (1997, 1999) observed an increase of the association signal in S phase. Nonetheless, Sld3 protein is required for Cdc45 chromatin association, because the association signal of Cdc45 was not detected in sld3-7 cells after release from G1 arrest or in G1 phase (Figure 8C and D).
Sld3 and Cdc45 form a complex (Figure 3). In thermosensitive sld3 cells, the interaction between Sld3 and Cdc45 is reduced (Figure 3A), Cdc45 does not associate with replication origins (Figure 8B) and chromosomal DNA replication is defective (Figures 4A and 5). Moreover, Sld3 does not associate with origins in cdc45-1 cells (Figure 8A). These facts suggest that Sld3–Cdc45 complex formation is required for the origin associations of Sld3 and Cdc45. Mutations in SLD3 and CDC45 are suppressed by high-copy CDC45 and SLD3 (Figure 2), and the origin association of Sld3-5 was restored by high-copy CDC45 (Figure 7B). This observation can be explained by the fact that formation of Sld3–Cdc45 complex in these mutant cells is restored by high-copy SLD3 and CDC45. In S phase, we detected co-immunoprecipitation of Mcm2 and Cdc45 (Figure 8D) without cross-linking agent, but not that of Mcm2 and Sld3 (our unpublished result). These results suggest that the Sld3–Cdc45 complex associates with origin by the interaction between Cdc45 and Mcm proteins, at least in S phase and probably in G1 phase as well.
After the Sld3–Cdc45 complex associates with origins, origin DNA is unwound and bound by the single-strand DNA-binding protein, RF-A. DNA polymerases are recruited to this unwound region. Although we have not detected physical interaction between Dpb11 and Sld3–Cdc45 complex, an interaction between Sld3 and Dpb11 was observed using two-hybrid assay (our unpublished result). This result, together with synthetic lethality between dpb11, sld3 and cdc45 (Kamimura et al., 1998), strongly suggests that Dpb11 works for polymerase loading in cooperation with Sld3–Cdc45. The recent discovery of a complex consisting of pre-RC, Cdc45 and Pol ε (Zou and Stillman, 2000) further supports this idea.
After initiation, Cdc45 moves with replication fork and functions for the elongation step of DNA replication (Aparicio et al., 1997). This observation is consistent with the fact that Cdc45 is required not only for the initiation step of DNA replication (Hopwood and Dalton, 1996; Zou et al., 1997) but also for the elongation step (Tercero et al., 2000). Sld3 is also required for the step after HU (Table I). Furthermore, we observed the Sld3–Cdc45 complex throughout the cell cycle. Thus, the Sld3–Cdc45 complex seems likely to move together for the step(s) other than the initiation step; for example, the elongation step or recovery of stalled replication fork.
Regulation of Sld3–Cdc45 association with origins
Engaged Cdc45 chromatin association requires S-Cdk activity (Zou and Stillman, 1998; Aparicio et al., 1999). Cdc7/Dbf4 kinase is also required for Cdc45 association with chromatin (engaged mode) (Zou and Stillman, 2000) and this kinase phosphorylates Cdc45 in vitro (Nougaréde et al., 2000). Thus, it is possible that this phosphorylation is required for engaged association. It is also possible that Mcm proteins are phosphorylated by Cdk or Cdc7 kinases, and that this phosphorylation enhances the binding of Mcm to the Sld3–Cdc45 complex. In fact, Mcm2 is a good substrate of Cdc7 kinase in vitro (Lei et al., 1997; Kihara et al., 2000).
Bound mode of the Cdc45–Sld3 complex with early origins, however, is not regulated by Cdk and Cdc7 kinases, because they bind early origins in G1 phase. So far, we do not know whether there is a regulation for bound-mode association of Cdc45–Sld3 with early origins. The Cdc45–Sld3 complex may be loaded onto pre-RC with Mcm proteins in early-firing origins. In late origins there may be structural hindrance to avoid loading of Cdc45–Sld3 when Mcm proteins are loaded.
Molecular function of the Sld3–Cdc45 complex
To initiate DNA replication, replication origins must be unwound. Without Cdc45 or Sld3, origin is not unwound as RF-A does not associate with origins (Figure 9). However, neither Cdc45 nor Sld3 has a helicase motif in their amino acid sequence. Thus, Sld3–Cdc45 may be required for recruitment of helicase to replication origins. As Mcm complex has weak helicase activity (Ishimi, 1997), Sld3–Cdc45 may remodel Mcm proteins to enhance their helicase activity.
Amino acid sequence of Cdc45 is well conserved from yeast to human cells. On the contrary, we can not find a good candidate for Sld3 in higher eukaryotes from its amino acid sequence. Thus, either higher eukaryotic cells do not need Sld3 for Cdc45 loading to Mcm or Sld3 is diversified from organism to organism. Although we can not find a good candidate for Dpb11 in higher eukaryotes by its amino acid sequence, Drosophila Mus101 (Yamamoto et al., 2000) and human TopBP1 (J.Syväoya, personal communication), which have seven and eight BRCT domains, respectively, and play roles in DNA replication and repair, are suggested as functional homologs of Dpb11 and Cut5/Rad4, a counterpart of Dpb11 in S.pombe. Moreover, Sld3 and Psl3 of S.pombe are similar in not only amino acid sequences, but also biological functions (R.Nakajima and H.Masukata, personal communication). These findings suggest that a functional homolog of Sld3 exists in higher eukaryotes and functions with Cdc45 for DNA replication.
Materials and methods
Yeast strains
All the yeast strains used in this study except L40 were originated from W303 and are listed in Table II.
Table II. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R.Rothstein |
| W303-1A/1B | MATa/α diploid, cross of W303-1A and W303-1B | R.Rothstein |
| YYK9 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 | this study |
| YYK10 | MATa dpb11-1 ade2 ade3 leu2 trp1 ura3 sld3-1 [pYK1] | Kamimura et al. (1998) |
| YYK11 | MATa dpb11-1 ade2 ade3 leu2 trp1 ura3 sld4-1 [pYK1] | Kamimura et al. (1998) |
| YYK12 | MATa/α sld3Δ::LEU2/SLD3 W3031A/1B | this study |
| YYK13 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sld3Δ::LEU2 [YEp195SLD3] | this study |
| YYK14 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sld3-4 | this study |
| YYK15 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sld3-5 | this study |
| YYK16 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sld3-6 | this study |
| YYK17 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sld3-7 | this study |
| YYK18 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sld3-8 | this study |
| YYK19 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 sld3-5 | this study |
| YYK20 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 SLD3-9myc::LEU2 CDC45-3HA::TRP1 | this study |
| YYK21 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 SLD3-9myc::LEU2 | this study |
| YYK22 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 CDC45-3HA::TRP1 | this study |
| YYK23 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 sld3-5-9myc::LEU2 | this study |
| YYK24 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 cdc45-1 | this study |
| YYK29 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 cdc45-1 SLD3-9myc::LEU2 | this study |
| YYK30 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 cdc45-1 SLD3-9myc::LEU2 | this study |
| YYK31 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 sld3-7 CDC45-3HA::TRP1 | this study |
| YYK32 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 cdc45-27 bar1 | this study |
| YHM014 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 RFA1-18myc::TRP1 | Masumoto et al. (2000) |
| YYK32 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1 sld3-5 RFA1-18myc::TRP1 | this study |
| L40 | MATa ade2 his3Δ200 leu2-3,112 trp1-901 ura3-1 LYS::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ | Bartel and Fields (1995) |
Plasmids
pBS(SK+)SLD3 was constructed by subcloning the 2.9 kb EcoO109I SLD3 DNA fragment blunted by T4 DNA polymerase into the EcoRV site of pBS(SK+). YCp22SLD3, YCp33SLD3 and YEp195SLD3 were constructed by subcloning the 2.9 kb PstI–SalI SLD3 DNA fragment into PstI–SalI sites of YCplac22, YCplac33 and YEplac195. YCp22CDC45 and YEp195CDC45 were constructed by subcloning the 2.2 kb SpeI–HindIII CDC45 DNA fragment into the XbaI–HindIII sites of YCplac22 or YEplac195. The myc-tagged version of the SLD3 gene was produced as follows. The C–terminal and terminator region of SLD3 were amplified by PCR and cloned into the SmaI site of YIplac128. The resultant plasmid contains a BglII site at the stop codon of SLD3. Nine tandem copies of the myc epitope were cloned into the BglII site. This plasmid was digested with NsiI before yeast transformation. For Cdc45-3HA, p404-Cdc45-HA/C (kindly provided by S.Bell) was digested with ClaI prior to yeast transformation. The plasmids used in the two-hybrid assay were constructed as follows. PCR amplified DNA from CDC45 was cloned into pBTM116 (Bartel and Fields, 1995) to allow production of LexA–Cdc45 protein. PCR amplified DNA from the SLD3 and sld3-5 was cloned into pACT2 (Bai and Elledge, 1997) to allow production of Gal4–Sld3 and Sld3-5 proteins.
Gene disruption of the SLD3 gene
The genomic sequence between the AatII and AflII sites was removed from the pBS(SK+)SLD3 plasmid and replaced by the 1.6 kb LEU2 fragment isolated from YDp-L (Berben et al., 1991) (Figure 1A). The LEU2-disrupted genomic fragment was subsequently removed from the plasmid and introduced into the W303 diploid. Southern blot analysis was performed on the Leu2+ transformants to confirm that one copy of the endogenous SLD3 was successfully disrupted.
Isolation of temperature-sensitive sld3 alleles
The diploid strain containing the disrupted SLD3 gene was transformed with YEp195SLD3, and the resultant Ura+ transformants were sporulated and dissected. One Ura+ Leu+ segregant, YYK13, was used for further study. The SLD3 was amplified using PCR with Taq DNA polymerase, and was used for transformation of YYK13 with PstI- and SalI-cleaved YCplac22 (Muhlrad et al., 1992). Approximately 6400 transformants grown at 23°C on Ura– Trp– plates were replica-plated to 5-FOA plate and grown at 23°C. Subsequently, these 5-FOA plates were replica-plated to one set of YPD plates and incubated at 23 and 37°C, respectively. Five clones showed temperature-sensitive growth. From these thermosensitive clones, plasmid DNAs were recovered. A 2.9 kb PstI–SalI fragment of the plasmids was transferred to YIplac211, which was used for transformation of W303–1A after digestion with AflII. Ura+ transformants were grown at 23°C, spread onto 5-FOA plates and temperature-sensitive colonies were selected. The sld3-5 allele has two alterations occurring at nucleotide 374 and 509 (nucleotide 1 is the A in the ATG of the ORF) (Figure 1). We do not know which mutation in the sld3-5 mutant is responsible for temperature sensitivity. Genomic DNA sequence confirmed that both mutations were present in genome.
Chromatin immunoprecipitation assay
The CHIP assay used in this study is based on the methods described by Tanaka et al. (1997), except that we used Dynabeads protein A (Dynal) for immunoprecipitation. Sequences of PCR primers used are: ARS305–30.5kb, (S) 5′-TGCAAACAGTATTCCGGCAC-3′, (AS) 5′-ACACGATCCACGCTGTCCCA-3′; ARS305–39.5kb, (S) 5′-TTT CAGAGCCTTCTTTGGAG-3′, (AS) 5′-CAAACTCCGTTTTTAGC CCC-3′; ARS305–47.5kb, (S) 5′-GAAGATGCTAAGAAATGCAG-3′, (AS) 5′-AGTTGAGGCGCAGAATCCCA-3′; ARS305–56.5kb, (S) 5′-TTCGCTAGAAAATGAGTAAG-3′, (AS) 5′-CAGCTATGAAA GATGCGTAG-3′; ARS501–558kb, (S) 5′-AACTTTTACGATCCAAC GCC-3′, (AS) 5′-GCCTCTACGGGTATTAGCTG-3′; ARS501–549.7kb, (S) 5′-ATGAAGATGACATTGCTCCT-3′, (AS) 5′-GTATCTGGATAA TGGATCTG-3′; ARS501–542kb, (S) 5′-GGGAAAAGTACGTGAA AGTC-3′, (AS) 5′-CTTCTTAATCTTAGCACCCT-3′; ARS501–534kb, (S) 5′-ACAAGCATCATTCATAGCCT-3′, (AS) 5′-ATCGTGGCTAGG ACATTTTG-3′.
Chromatin-binding assay
Chromatin-binding assay was performed as described by Desdouets et al. (1998). Cell extracts were incubated for 15 min on ice with 0.46 U/µl of DNase I (Worthington Biochemical Corporation). No DNA was observed in the fraction after DNase I treatment when it was run in an agarose gel and stained with ethidium bromide. Lysates were underlayered with 50% volume of 30% sucrose (volume refers to the volume of the spheroplast suspension), and spun at 14 000 r.p.m. in a TMP-11 rotor (Tomy) for 15 min at 4°C.
Other methods
Synchronization of yeast cells, two-hybrid analysis, N/N 2-D gel analysis and immunoprecipitation without cross-linking agent were performed as described in Kamimura et al. (1998). For immunoprecipitation experiment with cross-linking agent, we followed the method developed by Masumoto et al. (2000).
Acknowledgments
Acknowledgements
We thank T.Tanaka, K.Nasmyth, O.M.Aparicio and S.Bell for information about PCR primers, strains and plasmids; K.Sugimoto, H.Masukata and J.Syväoja for information about a mutation in SLD3 and homologs of Sld3 and Dpb11 before publication; and J.F.X.Diffley and H.Masukata for critical reading of the manuscript. This study is partially supported by grants-in-aid from the Ministry of Education, Science, Sports and Culture, Japan, to Y.K., A.S. and H.A.
References
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Stout,A.M. and Bell,S.P. (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl Acad. Sci. USA, 96, 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Leem,S.-H., Phongdara,A. and Sugino,A. (1995) Dpb11, which interacts with DNA polymerase II(ε) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl Acad. Sci. USA, 92, 11791–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C. and Elledge,S.J. (1997) Gene identification using the yeast two-hybrid system. Methods Enzymol., 283, 141–156. [DOI] [PubMed] [Google Scholar]
- Bartel P.L. and Fields,S. (1995) Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol., 254, 241–263. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origin of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Berben G., Dumont,J., Gilliquet,V., Bolle,P.-A. and Hilger,F. (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast, 7, 475–477. [DOI] [PubMed] [Google Scholar]
- Bork P., Hoffman,K., Bucher,P., Neuwald,A.F., Altschul,S.F. and Koonin,E.V. (1997) A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J., 11, 68–76. [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- Callebaut I. and Mornon,J.-P. (1997) From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett., 400, 25–30. [DOI] [PubMed] [Google Scholar]
- Campbell J.L. and Newlon,C.S. (1991) Chromosomal DNA replication. In Broach,J.R., Pringle,J.R. and Jones,E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 41–146.
- Desdouets C., Santocanale,C., Drury,L.S., Perkins,G., Foiani,M., Plevani,P. and Diffley,J.F.X. (1998) Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase α. EMBO J., 17, 4139–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F.X. (1996) Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev., 10, 2819–2830. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X. (1998) Replication control: choreographing replication origins. Curr. Biol., 8, R771–R773. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X., Cocker,J.H., Dowell,S.J., Harwood,J. and Rowley,A. (1995) Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle. J. Cell Sci. Suppl., 19, 67–72. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F.X. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herendeen D. and Kelly,T.J. (1996) SV40 DNA replication. In Blow,J.J. (ed.), Eukaryotic DNA Replication. Oxford University Press, Oxford, UK, pp. 29–65.
- Hopwood B. and Dalton,S. (1996) Cdc45p assembles into a complex with Cdc46/Mcm5p, is required for minichromosome maintenance and is essential for chromosomal DNA replication. Proc. Natl Acad. Sci. USA, 93, 12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Masumoto,H., Sugino,A. and Araki,H. (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol., 18, 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Nakai,W., Asano,S., Suzuki,A., Kitada,K., Kawasaki,Y., Johnston,L.H. and Sugino,A. (2000) Characterization of the yeast Cdc7p/Dbf4p complex purified from insect cells. J. Biol. Chem., 275, 35051–35062. [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki,Y., Young,M.R., Kihara,M., Sugino,A. and Tye,B.K. (1997) Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis. Genes Dev., 11, 3365–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. and Stillman,B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y. and Stillman,B. (1994) Replicator dominance in a eukaryotic chromosome. EMBO J., 13, 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Sugino,A. and Araki,H. (2000) Dpb11 controls the association between DNA polymerase α and ε and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol., 20, 2809–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane R.J., Carr,A.M. and Price,C. (1997) Characterization of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol. Gen. Genet., 255, 332–340. [DOI] [PubMed] [Google Scholar]
- Mimura S. and Takisawa,H. (1998) Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J., 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S., Masuda,T., Matsui,T. and Takisawa,H. (2000) Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells, 5, 439–452. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Nougaréde R., Seta,F.D., Zarzov,P. and Schwob,E. (2000) Hierarchy of S-phase-promoting factors: yeast Dbf4–Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol., 20, 3795–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoluzi S., Minenkova,O. and Castagnoli,L. (1997) The genes encoding the transcription factor YTAF (II) 60, the G4p1 protein and putative glucose transporter are combined in a 12.3 kb DNA fragment on the left arm of Saccharomyces cerevisiae chromosome VII. Yeast, 13, 85–91. [DOI] [PubMed] [Google Scholar]
- Saka Y. and Yanagida,M. (1993) Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell, 74, 383–393. [DOI] [PubMed] [Google Scholar]
- Saka Y., Fantes,P., Sutani,T., McInerny,C., Creanor,J. and Yanagida,M. (1994) Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J., 13, 5319–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Esashi,F., Matsusaka,T., Mochida,S. and Yanagida,M. (1997) Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev., 11, 3387–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. (1996) Cell cycle control of DNA replication. Science, 274, 1659–1664. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Shimomura,T., Hashimoto,K., Araki,H., Sugino,A. and Matsumoto,K. (1996) Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc. Natl Acad. Sci. USA, 93, 7048–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. and Nasmyth,K. (1998) Association of RPA with chromosomal replication origins requires an Mcm protein and is regulated by Rad53 and cyclin- and Dbf4-dependent kinases. EMBO J., 17, 5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tercero J.A., Labib,K. and Diffley,J.F.X. (2000) DNA synthesis at individual replication forks requires the essential initiation factor Cdc45. EMBO J., 19, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade H.M. and O’Connell,M.J. (1998) Cut5 is a component of the UV-responsive DNA damage checkpoint in fission yeast. Mol. Gen. Genet., 260, 426–433. [DOI] [PubMed] [Google Scholar]
- Waga S. and Stillman,B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- Walter J. and Newport,J. (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA and DNA polymerase α. Mol. Cell, 5, 617–627. [DOI] [PubMed] [Google Scholar]
- Wang H. and Elledge,S.J. (1999) DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R.R., Axton,J.M., Yamamoto,Y., Saunders,R.D.C., Glover,D.M. and Henderson,D.S. (2000) The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encoded a protein with seven BRCA1 C-terminus domains. Genetics, 156, 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. et al. (1998) Structure of an XRCC1 BRCT domain: a new protein–protein interaction module. EMBO J., 17, 6404–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (1998) Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (2000) Assembly of a complex containing Cdc45p, Replication protein A and Mcm2 at replication origins controlled by S-phase cyclin dependent kinases and Cdc7p–Dbf4p kinase. Mol. Cell. Biol., 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Mitchell,J. and Stillman,B. (1997) CDC45, a novel yeast gene that functions with the origin recognition complex and proteins in initiation of DNA replication. Mol. Cell. Biol., 17, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]