Abstract
The 5′ capping of mammalian pre-mRNAs is initiated by RNA triphosphatase, a member of the cysteine phosphatase superfamily. Here we report the 1.65 Å crystal structure of mouse RNA triphosphatase, which reveals a deep, positively charged active site pocket that can fit a 5′ triphosphate end. Structural, biochemical and mutational results show that despite sharing an HCxxxxxR(S/T) motif, a phosphoenzyme intermediate and a core α/β-fold with other cysteine phosphatases, the mechanism of phosphoanhydride cleavage by mammalian capping enzyme differs from that used by protein phosphatases to hydrolyze phosphomonoesters. The most significant difference is the absence of a carboxylate general acid catalyst in RNA triphosphatase. Residues conserved uniquely among the RNA phosphatase subfamily are important for function in cap formation and are likely to play a role in substrate recognition.
Keywords: crystal structure/cysteine phosphatase/mRNA capping/RNA processing/RNA triphosphatase
Introduction
RNA triphosphatase is an essential mRNA processing enzyme that catalyzes the first step in 5′ cap formation: the hydrolysis of the γ-phosphate of nascent pre-mRNA to form a diphosphate end, which is then capped with GMP by the enzyme RNA guanylyltransferase (Shuman, 2000). In yeast, the triphosphatase and guanylyltransferase activities reside in separate polypeptides (Cet1 and Ceg1, respectively) encoded by different genes, whereas mammalian species contain a single bifunctional polypeptide (Mce1; 597 amino acids) composed of an N-terminal triphosphatase domain (amino acids 1–210) and a C-terminal guanylyltransferase domain (amino acids 211–597). The RNA triphosphatases of fungi are strictly dependent on a divalent cation, whereas the RNA triphosphatases of mammals and other metazoa do not require a metal for activity and are instead inhibited by divalent cations.
The fungal RNA triphosphatases belong to a new family of metal-dependent phosphohydrolases that includes the poxvirus, baculovirus and phycodnavirus RNA capping enzymes. The crystal structure of yeast Cet1 revealed a unique active site architecture in which an eight-stranded β-barrel forms a topologically closed tunnel (Lima et al., 1999). The proposed mechanism of phosphoanhydride cleavage by fungal and viral RNA triphosphatases entails the direct attack of water on the γ-phosphate with no formation of a covalent intermediate.
The triphosphatase domain of metazoan and plant capping enzymes contains an HCxxxxxR(S/T) motif that defines the cysteine phosphatase superfamily, which includes protein tyrosine phosphatases, dual-specificity phosphatases and phosphoinositide phosphatases. The tyrosine-specific and dual-specificity protein phosphatases catalyze a two-step ping-pong phosphoryl-transfer reaction (Denu and Dixon, 1998). First, the conserved cysteine of the signature motif attacks the phosphoamino acid substrate to form a covalent protein-cysteinyl-S-phosphate intermediate and expel the hydroxyamino acid product (Tyr, Ser or Thr). Secondly, the covalent phosphoenzyme intermediate is hydrolyzed to liberate inorganic phosphate.
The finding of the HCxxxxxR(S/T) motif in metazoan capping enzymes and the demonstration that the conserved cysteine is required for RNA triphosphatase activity (Takagi et al., 1997; Ho et al., 1998b; Wen et al., 1998) challenged the presumption that polypeptides containing the signature sequence necessarily act as protein phosphatases. The functional diversity of the cysteine phosphatase superfamily was underscored further by the discovery that the tumor suppressor PTEN dephosphorylates phospholipid substrates (Maehama and Dixon, 1998).
Phylogenetic comparisons and biochemical studies have now delineated a family of RNA-specific phosphatases within the cysteine phosphatase superfamily. The RNA phosphatases are divisible into two subgroups: (i) the triphosphatase domains of the bifunctional metazoan and plant mRNA capping enzymes, which exclusively hydrolyze the γ-phosphate of RNA; and (ii) monofunctional enzymes that convert RNA 5′ triphosphate ends to 5′ diphosphates and 5′ monophosphates. The latter subgroup includes the baculovirus phosphatase BVP and the human phosphatase PIR1 (Gross and Shuman, 1998; Takagi et al., 1998; Deshpande et al., 1999). The amino acid sequences of the RNA-specific phosphatases show extensive similarity, but bear little resemblance to the other cysteine phosphatases outside of the HCxxxxxR(S/T) motif.
X-ray crystallography has illuminated three structural classes of cysteine phosphatases. The first class includes protein tyrosine phosphatases, dual-specificity protein phosphatases and PTEN (Barford et al., 1994; Stuckey et al., 1994; Yuvaniyama et al., 1996; Lee et al., 1999); the second class encompasses the low-molecular-weight phosphatases (Su et al., 1994); and the third class includes the Cdk-activating phosphatase Cdc25 (Fauman et al., 1998). All three classes require the presence of a general acid for catalysis.
How did the RNA triphosphatases evolve to catalyze the hydrolysis of phosphoanhydrides? Are RNA triphosphatases related structurally to any of the three known branches of the cysteine phosphatase superfamily? Do they employ the same catalytic mechanism? To address these questions, we determined the crystal structure of the triphosphatase domain of Mce1, the mouse mRNA capping enzyme. We report that mouse RNA triphosphatase is a member of the first branch of cysteine phosphatases and is especially similar to PTEN, which, like Mce1, acts on substrates containing multiple phosphates. Structural and biochemical analyses of Mce1 confirm the catalytic role of the conserved cysteine of Mce1 and the formation of a phosphoenzyme, while indicating that the mammalian capping enzyme cleaves phosphoanhydrides via a mechanism distinct from that of the cysteine phosphomonoesterases.
Results and discussion
Structure determination
The structure of the mammalian RNA triphosphatase domain Mce1(1–210) was solved using data from a selenomethionine multiwavelength anomalous dispersion (MAD) experiment. MAD phases calculated to 2.05 Å yielded an excellent electron density map, which showed unambiguous density for ∼90% of the polypeptide chain. The resolution of the structure was extended to 1.65 Å during refinement. There was no density for the first four N-terminal residues or the last 12 residues at the C-terminus, and a short surface loop (amino acids 114–118) was also disordered. The final model had an R-factor of 19.6% and an Rfree of 22.5%, with excellent stereochemistry.
Mce1(1–210) was also crystallized in the presence of sodium tungstate in an attempt to bind a phosphate analog in the active site. Although there was no evidence of a tungstate molecule in the resulting electron density map, the map did reveal that Cys126 had been oxidized to cysteine sulfenic acid (see Materials and methods). Four N-terminal and 16 C-terminal amino acids were disordered in the model of the oxidized enzyme. The surface loop (amino acids 114–118) was ordered when the enzyme was crystallized in the presence of tungstate. No other differences from the native structure were apparent in the structure of the oxidized enzyme; the root mean square deviation (r.m.s.d.) between the native and oxidized enzyme structures was 0.31 Å for all atoms. The structure of the oxidized enzyme was refined to 1.7 Å with an R-factor of 19.3% and an Rfree of 21.7%. Data collection, phasing and refinement statistics for both models are listed in Table I.
Table I. Data collection and refinement statistics.
| Native I | Native IIa | Se-Met λ1 | Se-Met λ2b | Se-Met λ3 | Se-Met λ4 | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| detector type/source | MAR-345/SSRL | MAR-CCD/APS | MAR-CCD/APS | MAR-CCD/APS | MAR-CCD/APS | MAR-CCD/APS |
| wavelength (Å) | 0.97 | 1.20374 | 0.97912 | 0.97922 | 0.96390 | 0.99484 |
| resolution (Å) | 1.65 | 1.7 | 2.05 | 2.05 | 2.05 | 2.05 |
| measured reflections | 143 292 | 145 070 | 120 097 | 120 156 | 120 001 | 119 083 |
| unique reflections | 26 506 | 24 075 | 14 150 | 14 159 | 14 159 | 14 150 |
| completeness (%)c | 99.4 (98.6) | 98.0 (96.5) | 99.4 (100) | 99.4 (100) | 99.5 (100) | 99.4 (99.7) |
| Rsym (%)c,d | 8.4 (25.1) | 6.0 (12.0) | 6.8 (15.9) | 7.0 (16.9) | 7.1 (17.5) | 7.6 (32.6) |
| Rmeas (%)c,e | 9.3 (28.2) | 7.0 (15.5) | 7.8 (18.0) | 7.9 (18.2) | 8.1 (19.8) | 8.6 (38.7) |
| Phasing (27–2.05 Å) | ||||||
| phasing power (acentric/centric)f | 0.41/0.30 | 1.59/1.14 | 1.31/0.94 | |||
| Rcullis (acentric/centric)g | 0.96/0.96 | 0.73/0.67 | 0.79/0.75 | |||
| figure of merit | 0.62 | |||||
| Refinement | ||||||
| resolution (Å) | 27–1.65 | 27–1.7 | ||||
| no. of reflections: working set/test set | 25 162/1334 | 22 841/1228 | ||||
| R-factorh | 19.6 | 19.3 | ||||
| Rfreei | 22.5 | 21.7 | ||||
| protein atoms | 1536 | 1544 | ||||
| water molecules | 210 | 209 | ||||
| other atoms | 23 | 23 | ||||
| r.m.s.d. | ||||||
| bond lengths (Å) | 0.01 | 0.01 | ||||
| bond angles (°) | 1.6 | 1.7 | ||||
| average B-factor (Å2)j | ||||||
| main chain | 15.6 (6.4) | 16.6 (7.4) | ||||
| side chain | 20.1 (8.9) | 21.1 (8.7) | ||||
| solvent | 35.8 (12.9) | 34.7 (11.3) |
aNative II refers to data set collected on a crystal grown in the presence of tungstate.
bData set used as reference or native set for phasing.
cNumbers in parentheses represent values in the highest resolution shell.
dRsym = Σ|I – <I>|/ΣI, where I = observed intensity and <I> = average intensity obtained from multiple measurements.
eRmeas as defined by Diederichs and Karplus (1997).
fPhasing power = r.m.s.d. (|Fh|/E), where |Fh| = heavy atom structure factor amplitude and E = residual lack of closure error.
gRcullis = Σ||Fh(obs)| – |Fh(calc)||/Σ|Fh(obs)|, where |Fh(obs)| = observed heavy atom structure factor amplitude and |Fh(calc)| = calculated heavy atom structure factor amplitude.
hR-factor = Σ||Fo| – |Fc||/Σ|Fo|, where |Fo| = observed structure factor amplitude and |Fc| = calculated structure factor amplitude.
iRfree, R-factor based on 5% of the data excluded from refinement.
jNumbers in parentheses represent r.m.s.d.
Overview of the RNA triphosphatase structure
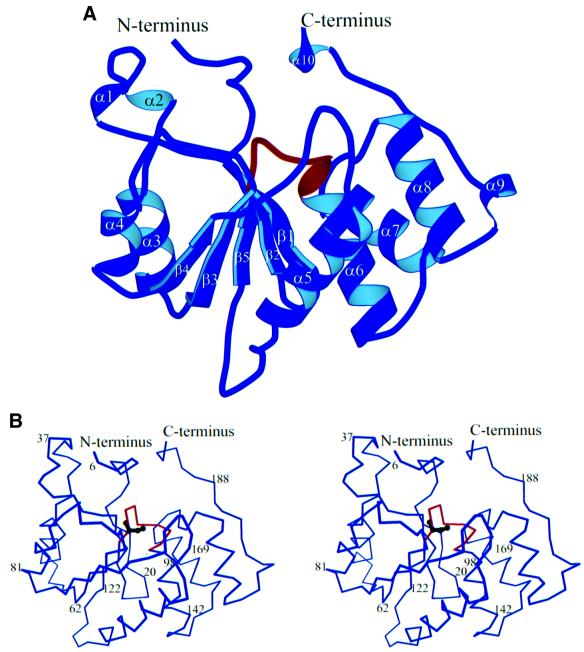
The RNA triphosphatase domain of Mce1 is an α/β-fold that consists of five β-strands, which form a central twisted sheet, flanked by two α-helices on one side and four α-helices on the other (Figure 1). The C-terminus travels back towards the N-terminus, where it would ultimately lead into the guanylyltransferase domain of the bifunctional mRNA capping enzyme. The HCxxxxxR(S/T) signature motif of the cysteine phosphatase superfamily comprises a phosphate binding loop (P-loop) located between the β5 strand and the α6 helix in Mce1(1–210). The P-loop contains the active site cysteine (Cys126), which is situated in a deep pocket formed by residues from loops joining several of the secondary structural elements and also from residues in the N- and C-terminal regions. As expected, Mce1(1–210) displays no structural similarity to the yeast RNA triphosphatase Cet1 (Lima et al., 1999).
Fig. 1. Overall structure of the Mce1 RNA triphosphatase domain. (A) Schematic representation showing the α/β-fold of the RNA triphosphatase domain (amino acids 5–194). The phosphate binding loop (P-loop) is shown in red. Secondary structural elements are labeled as α# for α-helices and β# for the β-strands. (B) Stereo view of a Cα trace of the RNA triphosphatase domain. The orientation is the same as in (A) with the P-loop colored red. The side chain of the active site cysteine, Cys126, is depicted in black.
Comparison with protein phosphatases and phosphoinositide phosphatases
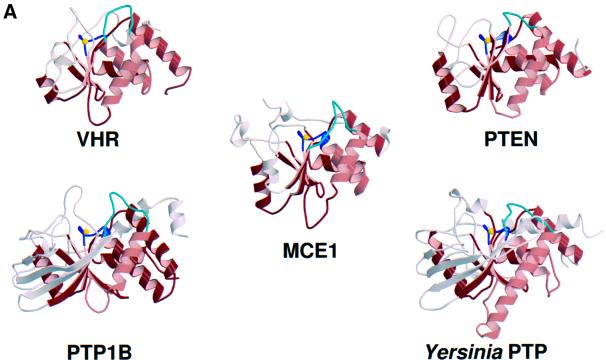
A search of the protein structure database using DALI (Holm and Sander, 1993) found significant similarities between mammalian RNA triphosphatase and other members of the cysteine phosphatase superfamily, including the protein tyrosine phosphatases PTP1B (Barford et al., 1994), SHP-2 (Hof et al., 1998), LAR (Nam et al., 1999) and Yersinia PTP (Stuckey et al., 1994), the dual-specificity protein phosphatase VHR (Yuvaniyama et al., 1996) and the phosphoinositide phosphatase PTEN (Lee et al., 1999). A comparison of the crystal structures of Mce1(1–210), PTP1B, Yersinia PTP, VHR and PTEN highlights a conserved fold of five twisted β-strands packed against four α-helices on one side and an α-helix on the other (Figure 2A). In pairwise comparisons of Mce1(1–210) to PTEN, VHR, Yersinia PTP and PTP1B, the r.m.s.d. values for Cα atoms of the core elements were 1.96, 2.07, 2.20 and 2.21 Å, respectively. The structures diverge outside of the core region. Overall, Mce1 is most similar to VHR and PTEN. All three proteins have a compact fold with few additional secondary structural elements.
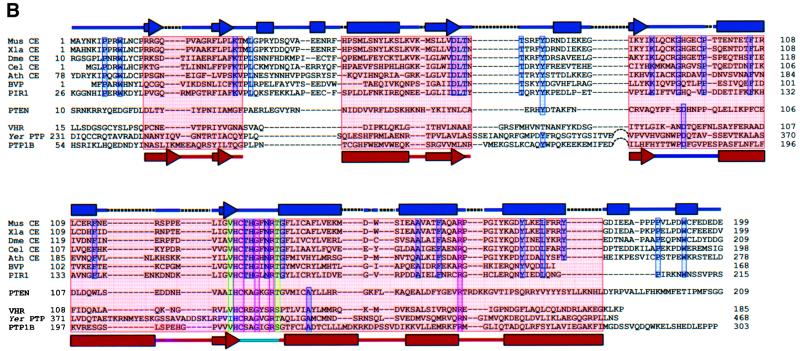
Fig. 2. Comparison of the Mce1 RNA triphosphatase with other phosphatases. (A) Structural comparison of the Mce1 RNA triphosphatase domain with the dual-specificity protein phosphatase VHR, the phosphatase domain of PTEN tumor suppressor, human PTP1B and Yersinia protein tyrosine phosphatase. The P-loop is colored blue, with the active site cysteine depicted as a yellow sphere; the general acid loop is colored cyan. Other common core elements are shaded red; these consist of four twisted β-strands surrounded by four α-helices on one side and one helix on the other. (B) Structure-based sequence alignment of the Mce1 RNA triphosphatase domain with other members of the metazoan RNA triphosphatase and protein phosphatase families. For the RNA triphosphatases, the sequences were aligned based on sequence similarity, but whenever possible, insertions and deletions were confined to loop regions. To align the sequences of the protein phosphatases, the structure of the Mce1 RNA triphosphatase domain was superimposed onto the structures of the phosphatases. The common core region is highlighted in pink. Identical residues found in both families are highlighted in red, while highly conserved residues in the P-loop are in green and conserved residues within each family are blue. Structural elements of the Mce1 RNA triphosphatase are shown at the top with α-helices depicted as boxes, β-strands as arrows, insertions as dotted lines, and loops as solid lines. Structural elements representing the common core region are shown at the bottom colored the same way as in (A). The RNA triphosphatases correspond to the capping enzyme triphosphatase domains of mouse (Mus CE), Xenopus laevis (Xla CE), Drosophila melanogaster (Dme CE), Caenorhabditis elegans (Cel CE) and Arabidopsis thaliana (Ath CE), along with the baculovirus-encoded triphosphatase (BVP) and the human triphosphatase (PIR1).
A structure-based amino acid sequence alignment of the cysteine phosphatases shows that the conserved residues, HCxxGxxR(T/S), are located in the P-loop (Figure 2B). The only other residue common to all cysteine phosphatases is an arginine (Arg160 in Mce1) that hydrogen bonds to main chain groups of the P-loop.
Active site
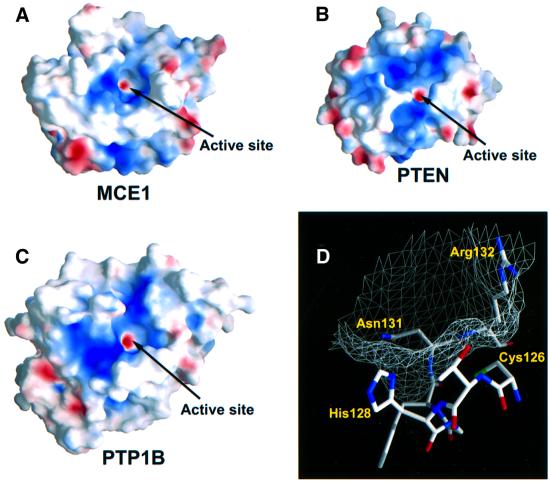
The active site of the Mce1 triphosphatase domain is found in a deep, positively charged pocket created primarily by residues of the P-loop (Figure 3). The Cys126 nucleophile is located at the base of the cavity and displays unusual φ,ϕ angles (φ = –117°, ϕ = –150°). This deviation has been noted in other phosphatases and is postulated to help in positioning the nucleophile for catalysis (Jia et al., 1999). The Mce1 crevice is ∼8 × 10 Å wide and ∼8 Å deep, similar in depth to the catalytic pockets of human PTP1B, Yersinia PTP and PTEN. Modeling suggests that a 5′ triphosphate moiety of RNA can fit into the active site without the need for a conformational rearrangement.
Fig. 3. Surface representation of the Mce1 RNA triphosphatase reveals a deep, positively charged pocket similar to that found in other phosphatases. Surface electrostatic potentials of (A) Mce1 RNA triphosphatase, (B) PTEN phosphatase and (C) human PTP1B. Blue and red correspond to positively and negatively charged areas, respectively. The active site cysteine was assigned a –1 charge to reflect its presumed existence as a thiolate ion at physiological pH. The active site pocket is indicated for all the proteins. (D) Surface mesh rendering of the Mce1 catalytic region shows the overall size and depth of the active site pocket. Residues 126–132 of the P-loop are also shown.
The hydrogen bonding network between the P-loop residues in Mce1 resembles that seen in other cysteine phosphatases (Figure 4). For the protein tyrosine phosphatases and dual-specificity phosphatases, it has been determined that the pKa of the active site cysteine is low (∼4.7–5.6) such that it exists as a thiolate in the ground state (Zhang and Dixon, 1993; Denu and Dixon, 1995; Lohse et al., 1997). Multiple hydrogen bonding interactions are available to stabilize a cysteine thiolate in Mce1. The main chain amide groups of seven residues in the P-loop of Mce1 are oriented towards Cys126, with N to S distances of 3.2–4.3 Å. The Sγ of Cys126 is also within hydrogen bonding distance of Oδ of Thr133, which is conserved as threonine or serine in all of the other cysteine phosphatases (Figure 2B). Tyr74, which is conserved in the RNA triphosphatase family, makes a hydrogen bond with Nε of His125, while Nδ of His125 hydrogen bonds to the main chain carbonyl of Cys126 (Figure 4). Arg160 forms hydrogen bonds with the carbonyl groups of His128 and Gly129 of the P-loop. These interactions with the P-loop may help to maintain the P-loop in a catalytically competent conformation.
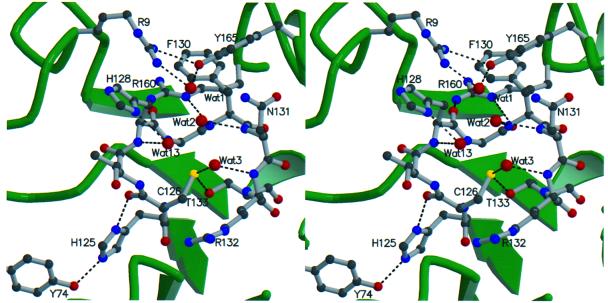
Fig. 4. Stereo view of the active site region in the Mce1 RNA triphosphatase domain. The highly conserved residues of the P-loop are shown along with other conserved residues from nearby loops. Hydrogen bonding interactions within the active site are depicted by dotted lines. Water molecules are shown as red spheres.
Arg9, Tyr165 and Lys166 line the rim of the active site pocket. Arg9, Arg72, His128, Arg132 and Lys166 confer a positive surface potential for interaction with the negatively charged 5′ phosphates of the substrate. A positive surface potential surrounding the active site is also present in the protein tyrosine phosphatases and in PTEN (Figure 3). The invariant arginine of the P-loop (Arg132 in Mce1) is predicted to stabilize the transition state of the γ-phosphate during catalysis (Denu et al., 1996). In the Mce1 structure, the side chain of Arg132 is ∼6 Å from Cys126 and is held there by hydrogen bonds to the main chain carbonyls of Leu67 and Lys92. This position of Arg132 prevents it from blocking access to the cysteine at the base of the pocket. Arg132 is likely to move towards the γ-phosphate upon binding of the triphosphate within the active site.
Three well ordered waters are present near Cys126 (Wat2, Wat3 and Wat13 in Figure 4), and their positions appear to mimic the tetrahedral geometry of the oxygens of a phosphate. Wat2 and Wat3 form hydrogen bonds with backbone amides of the P-loop, while Wat13 is coordinated to a main chain amide and Nδ of His128. This interaction suggests a role for His128 in substrate binding and/or transition state stabilization.
The hydrolysis of phosphomonoesters by most cysteine phosphatases depends on a conserved aspartate general acid that is located on a loop connecting a central β-strand and an adjacent α-helix. In several phosphatases, the general acid loop has been observed to undergo a conformational change upon substrate binding that closes off the active site pocket and brings the aspartate (which is protonated in the ground state) into the active site, where it serves as a proton donor to expel the hydroxyl amino acid leaving group (Stuckey et al., 1994; Jia et al., 1995). The structure of Mce1 reveals a similarly positioned loop (amino acids 88–99) (colored cyan in Figure 2). The conformation of this loop in Mce1 resembles that of the general acid loop in the unliganded forms of the protein tyrosine phosphatases. However, the structural alignment shows that there is no conserved acidic residue within this loop region of Mce1 to serve as a candidate general acid.
Formation of a phosphoenzyme during catalysis
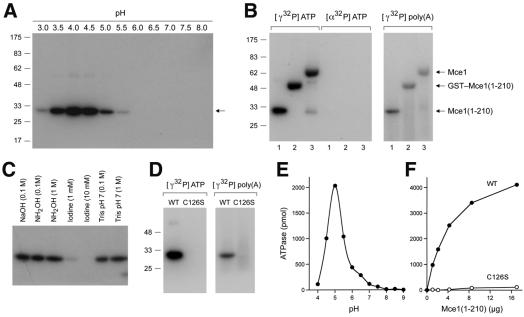
Although the P-loop cysteine is required for the RNA triphosphatase activity of metazoan capping enzyme, the phosphoenzyme intermediate has never been demonstrated directly. Here we found that a phosphoenzyme could be captured during a 15 s reaction of Mce1(1–210) with 10 µM [γ-32P]ATP at 22°C. Label transfer from [γ-32P]ATP to Mce1(1–210) was detected only at acidic pH, with optimal trapping of the phosphoenzyme at pH 4.0–4.5 (Figure 5A). Mce1(1–210) also catalyzed the release of 32Pi from [γ-32P]ATP. The ATPase activity was optimal at pH 5.0 and there was little activity detected above pH 7.0 (Figure 5E). The extent of ATP hydrolysis was proportional to Mce1(1–210) concentration (Figure 5F) and we calculated a turnover number of 1.6 ATP molecules hydrolyzed per minute per enzyme. Phosphoenzyme formation and ATP hydrolysis coincided with the distribution of Mce1(1–210) when the enzyme preparation was sedimented in a glycerol gradient (not shown), suggesting that both activities are intrinsic to the mammalian RNA triphosphatase.
Fig. 5. Demonstration of a cysteinyl phosphoenzyme. (A) pH dependence of phosphoenzyme formation. Reaction mixtures (10 µl) containing 4 µg of Mce1(1–210), 10 µM [γ-32P]ATP, 5 mM DTT and 50 mM Tris buffer [either Tris–formate (pH 3.0, 3.5), Tris–acetate (pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5) or Tris–HCl (pH 7.0, 7.5, 8.0)] were incubated at 22°C for 15 s and then quenched with 2% SDS. The samples were analyzed by SDS–PAGE. An autoradiograph of the gel is shown. The positions and sizes (kDa) of marker proteins are indicated on the left. (B) Phosphoenzyme formation by Mce1 and the isolated RNA triphosphatase domain. Reaction mixtures (10 µl) containing 50 mM Tris–acetate pH 4.0, 5 mM DTT, either 10 µM [γ-32P]ATP or [α-32P]ATP (at equivalent specific radioactivity) or 5.3 µM [γ-32P]poly(A), and 4 µg of Mce1(1–210) (lanes 1), GST–Mce1(1–210) (lanes 2) or Mce1 (lanes 3) were incubated at 22°C for 15 s. The positions of the 32P-labeled enzymes are indicated on the right. (C) Chemical stability of the phosphoenzyme. [32P]Mce1(1–210) was denatured with SDS and exposed to NaOH, hydroxylamine or iodine for 30 min at 37°C. Control samples were exposed to Tris buffer pH 7.0. The treated samples were analyzed by SDS–PAGE. An autoradiograph of the gel is shown. (D) Phosphoenzyme formation requires Cys126. Reaction mixtures (10 µl) containing 50 mM Tris–acetate pH 4.0, 5 mM DTT, either 10 µM [γ-32P]ATP or 5.3 µM [γ-32P]poly(A), and 4 µg of wild type (WT), Mce1(1–210) or Mce1(1–210)-C126S were incubated at 22°C for 15 s, then denatured and analyzed by SDS–PAGE. (E) pH dependence of ATP hydrolysis by Mce1. Reaction mixtures (10 µl) containing 4 µg of Mce1(1–210), 0.5 mM [γ-32P]ATP, 1 mM DTT and 50 mM Tris buffer [either Tris–acetate (pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5) or Tris–HCl (pH 7.0, 7.5, 8.0, 8.5, 9.0)] were incubated at 37°C for 15 min. The reactions were quenched by adding 2.5 µl of 5 M formic acid. Aliquots of the reaction mixtures were applied to polyethyleneimine cellulose TLC plates, which were developed in 0.5 M LiCl, 1 M formic acid. [γ-32P]ATP and 32Pi were quantitated by scanning the TLC plate with a phosphorimager. (F) ATP hydrolysis requires Cys126. Reaction mixtures (10 µl) containing 50 mM Tris–acetate pH 5.0, 5 mM DTT, 0.5 mM [γ-32P]ATP and wild type (WT), Mce1(1–210) or Mce1(1–210)-C126S were incubated at 37°C for 15 min. ATP hydrolysis is plotted as a function of input protein.
To prove that the ∼30 kDa 32P-labeled polypeptide was indeed the recombinant mammalian RNA triphosphatase, we purified full-length Mce1 and a glutathione S-trans ferase (GST)–Mce1(1–210) fusion protein, and incubated them with 10 µM [γ-32P]ATP in parallel with Mce1(1– 210). The apparent sizes of the 32P-labeled reaction products reflected those of the input recombinant proteins (Figure 5B). No label transfer was observed when Mce1(1–210), GST–Mce1(1–210) or Mce1 was incubated with 10 µM [α-32P]ATP (Figure 5B). Purified GST– Mce1(1–210) and Mce1 catalyzed the release of 32Pi from [γ-32P]ATP (data not shown), and their respective turnover numbers (2.1 and 2.0 min–1) were similar to that of Mce1(1–210).
Mce1(1–210), GST–Mce1(1–210) and Mce1 all formed phosphoenzyme adducts during a 15 s reaction with 5 µM γ-32P-labeled poly(A) (Figure 5B). Thin layer chromatography (TLC) analysis of the RNA preparation verified that it was exclusively polynucleotide and devoid of residual [γ-32P]ATP. Label transfer from γ-32P-labeled poly(A) to Mce1(1–210) was optimal at pH 4.0 (data not shown). Mce1(1–210) catalyzed the release of 32Pi from [γ-32P]poly(A), and the RNA triphosphatase activity was proportional to enzyme concentration at either pH 5.0 or 7.5. We calculated turnover numbers of 1.7 min–1 at pH 5.0 and 15 min–1 at pH 7.5 (not shown). Thus, mammalian RNA triphosphatase displays equivalent activity to RNA and ATP substrates at pH 5, but specifically hydrolyzes triphosphate-terminated RNA at pH 7.5.
The SDS-denatured [32P]Mce1(1–210) adduct was stable during a 30 min exposure at 37°C to 0.1 M NaOH, or to 0.1 or 1 M hydroxylamine at pH 7.0, but the 32P was released from Mce1(1–210) by treatment with 1–10 mM iodine at pH 7.0 (Figure 5C). Stability to neutral hydroxylamine argues against an acylphosphate linkage, whereas hydrolysis by iodine is strongly indicative of a thiophosphate linkage, presumably to Cys126.
Phosphoenzyme formation during the reaction of Mce1(1–210) with ATP or triphosphate-terminated RNA was abrogated by mutating Cys126 to serine (Figure 5D), and the hydrolysis of ATP by the C126S mutant was <1% of the wild-type activity (Figure 5E). These results provide evidence of a cysteinyl phosphoenzyme intermediate in the catalysis of γ-phosphate cleavage, and they show that the phosphohydrolase activity is not limited to polynucleotide substrates.
Mutational analysis
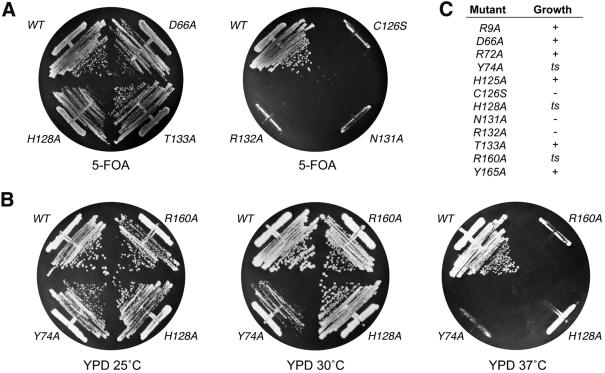
The functional relevance of individual amino acids high lighted by the Mce1(1–210) structure was gauged by alanine scanning in the full-length Mce1. The mutated capping enzymes were tested for their ability to function in vivo in yeast, in lieu of the endogenous RNA triphosphatase Cet1. MCE1 alleles were cloned into a yeast CEN TRP1 plasmid so as to place their expression under the control of a constitutive promoter. The plasmids were transformed into a Saccharomyces cerevisiae cet1Δ strain in which the chromosomal CET1 gene was deleted and replaced by LEU2. Growth of cet1Δ is contingent on maintenance of a wild-type CET1 allele on a CEN URA3 plasmid. Therefore, the cet1Δ strain is unable to grow on agar medium containing 5-fluoroorotic acid (5-FOA) unless it is first transformed with a biologically active RNA triphosphatase gene (Ho et al., 1998a). Expression of wild-type Mce1 in cet1Δ cells permitted their growth on 5-FOA, whereas expression of the catalytically inactive C126S mutant did not (Figure 6A).
Fig. 6. Mutational effects on Mce1 RNA triphosphatase activity in vivo. (A) Complementation in yeast. Yeast strain YBS20 (cet1Δ p360-CET1 [CEN URA3 CET1]) was transformed with CEN TRP1 plasmids containing either wild-type MCE1 or the indicated mutant alleles under the control of the yeast TPI1 promoter. Individual Trp+ transformants were selected and then patched on agar medium lacking tryptophan. Cells from single patches were then streaked on agar medium containing 0.75 mg/ml 5-FOA. The plates were photographed after incubation for 4 days at 30°C. (B) Temperature-sensitive mutants. Trp+ FOA-resistant derivatives of YBS20 containing the indicated alleles of MCE1 on CEN TRP1 plasmids were streaked on YPD agar. The plates were photographed after incubation for 4 days at 25, 30 or 37°C as specified. (C) Summary of mutational effects. Lethal mutants C126S, N131A and R132A (– growth) yielded no 5-FOA-resistant colonies after incubation for 4 days at either 25 or 30°C. All of the other mutants supported growth on 5-FOA at 25 and 30°C; these were then tested for growth on YPD. Mutants R9A, D66A, H125A and T133A grew as well as wild-type MCE1 cells at 25, 30 and 37°C (+ growth). ts mutants Y74A, H128A and R160A either failed to grow or formed only pinpoint colonies at 37°C.
The plasmid shuffle assay was used to test the effects of other alanine mutations within the P-loop. Replacement of Asn131 or Arg132 with alanine abolished in vivo activity of Mce1 (Figure 6A). In contrast, the expression of Mce1 mutants H125A, H128A and T133A supported growth of cet1Δ cells on 5-FOA during selection at either 25 or 30°C. The viable MCE1-Ala strains were then tested for growth on rich medium (YPD) at 25, 30 and 37°C. H128A cells displayed a temperature-sensitive phenotype, growing well at 25 and 30°C, but not at 37°C (Figure 6B). H125A and T133A cells grew at all temperatures and their colony sizes were similar to that of wild-type MCE1 cells (data not shown).
Arg160 is the only residue outside of the P-loop that is conserved in the cysteine phosphatase structures aligned in Figure 2B. Tyr74, conserved in the RNA triphosphatases, forms a hydrogen bond to His125 of the P-loop (Figure 4). Yeast cells expressing the R160A or Y74A mutant enzymes were viable at 25 and 30°C, but failed to grow at 37°C (Figure 6B). We infer that the direct interactions of Arg160 with the P-loop serve to stabilize the triphosphatase active site. However, the favorable effects of Tyr74 on in vivo function of Mce1 do not seem dependent on direct bonding to His125 of the P-loop, since the elimination of the His125 side chain did not phenocopy Y74A.
We extended the alanine scan to selected residues outside of the P-loop. Asp66 is the only acidic residue in Mce1(1–210) that is conserved in all known metazoan-type RNA triphosphatases. Asp66 has been invoked as a general acid catalyst (Wen et al., 1998), but the structure reveals that Asp66 is not located in a position corresponding to the essential aspartate-containing general acid loop described for the protein phosphatases and PTEN. Instead, the carboxylate of Asp66 hydrogen bonds to the hydroxyls of Tyr86 and Thr68. Replacement of Asp66 by alanine had no effect on the ability of the mutant to complement growth of cet1Δ cells on 5-FOA; indeed, D66A cells grew as well as MCE1 cells on rich medium at all temperatures tested (Figure 6C). To evaluate the contributions of Asp66 further, we introduced the D66A mutation into the triphosphatase domain. The RNA triphosphatase specific activity of the D66A mutant (measured at pH 7.5) was 60% of the wild-type activity (not shown). Thus, the conserved aspartate is not essential for Mce1 activity in vivo or in vitro.
Residues Arg9 and Tyr165 are located at the rim of the active site pocket, where they hydrogen bond to each other and coordinate a water molecule (Figure 4). Arg72 is also located on the protein surface at the opposite edge of the pocket, and this residue is conserved in all of the RNA triphosphatases and PTEN (Figure 2). Yet, replacement of either Arg9, Tyr165 or Arg72 by alanine had no discernible effect on Mce1 function in vivo in yeast (Figure 6C).
Mechanistic implications
The studies presented here provide insight into the catalytic mechanism of mammalian RNA triphosphatase and emphasize the marked differences between the triphosphatase components of the metazoan and fungal capping apparatus. Moreover, the results suggest that, despite sharing the P-loop motif, a phosphoenzyme intermediate and a core tertiary structure with other cysteine phosphatases, the mechanism of phosphoanhydride cleavage by mammalian capping enzyme is different in key respects from the mechanisms used by the phosphomonoesterases of the cysteine phosphatase superfamily. The most significant difference is the apparent absence of a carboxylate general acid catalyst in Mce1. Mutation of the only acidic side chain conserved in all members of the metazoan RNA triphosphatase subfamily indicates that this residue is not essential for Mce1 activity in vivo or in vitro. Elimination of the equivalent aspartate in BVP had no effect on its catalytic activity in vitro (Martins and Shuman, 2000). It was proposed for BVP that the low pKa of the nucleoside diphosphate leaving group circumvents the need for a general acid catalyst (Martins and Shuman, 2000). This hypothesis is consistent with the Mce1 structure, which shows that no acidic residue is located in a position suited for proton donation. Furthermore, the loop in Mce1 that is equivalent to the general acid loop in the protein and phosphoinositide phosphatases contains no conserved acidic residue. The Mce1 loop does contain a non-conserved histidine (His94), which could, in principle, serve as a proton donor. However, we consider it unlikely that His94 acts as a general acid in light of the finding that Mce1 mutant Q90L/H94A supports the growth of yeast cet1Δ cells on 5-FOA (not shown). Although a catalytic acidic residue is also lacking in Cdc25, its activity is still dependent on a general acid catalyst, which is postulated to be located on its substrate (Chen et al., 2000). In contrast, Mce1 and other metazoan RNA triphosphatases seem to cleave the β–γ phosphoanhydride bond of RNA without the aid of a general acid catalyst.
The present study highlights functionally important P-loop residues, His128 and Asn131, which are unique to the RNA triphosphatase subfamily and may therefore determine the specificity of these enzymes for the hydrolysis of phosphoanhydrides. Asn131 of the P-loop is required for Mce1 RNA triphosphatase activity in vivo. Mutation of the equivalent Asn residue in BVP resulted in the loss of phosphohydrolase activity and the ability of BVP to form a phosphoenzyme intermediate (Martins and Shuman, 2000). The position of Asn131 and the orientation of Nδ into the active site suggest that it forms a hydrogen bond to a phosphate of the substrate. Asn131 is situated in the active site at a position similar to that of residues Gln262 of PTP1B and Gln446 of Yersinia PTP. These glutamines orient the activated water that attacks the phosphoenzyme intermediate in the second step of the PTPase reaction. An attractive hypothesis is that Asn131 interacts with the β-phosphate of the 5′ triphosphate substrate during formation of the phosphoenzyme intermediate (this contact being inherently unique to the RNA 5′ phosphatase subfamily) and then with a water during subsequent hydrolysis of the intermediate. The Mce1 structure suggests that His128 of the P-loop, which hydrogen bonds to Wat13 (Figure 4), may also interact with one of the 5′ phosphates of the substrate. Mutation of His128 to alanine confers a temperature-sensitive phenotype in vivo.
Two in vivo structure–activity relationships for the P-loop of Mce1 differed significantly from the in vitro results obtained for other members of the cysteine phosphatase family, including the RNA triphosphatase BVP. Thr133 was not essential for Mce1 function in vivo, and Wen et al. (1998) found that the T133A mutation elicited only a 2-fold reduction in Mce1 RNA triphosphatase activity in vitro. The 40–50% residual activity of T133A sufficed for Mce1 function in vivo in yeast. In contrast, replacing the equivalent Thr or Ser residues of protein tyrosine phosphatases, dual-specificity protein phosphatases and BVP by alanine lowered kcat by at least two orders of magnitude (Denu and Dixon, 1995; Evans et al., 1996; Lohse et al., 1997; Martins and Shuman, 2000). The structures of multiple cysteine phosphatases, including Mce1, show that the hydroxyl donates a hydrogen bond to the active site cysteine. This interaction is proposed to stabilize the cysteine thiolate. We infer that in Mce1 the interactions of the backbone amides of the P-loop with Cys126 are sufficient to stabilize it as the thiolate in vivo in the absence of Thr133.
His125 is dispensable for Mce1 function in vivo. Wen et al. (1998) reported that the H125A mutant protein was 40% as active as the wild-type RNA triphosphatase in vitro. However, substitution of the equivalent histidines of Yersinia protein tyrosine phosphatase, human PTP1B and BVP by alanine reduced kcat by two or three orders of magnitude (Zhang and Dixon, 1993; Flint et al., 1997; Martins and Shuman, 2000). The histidine in Mce1 makes a hydrogen bond with the main chain carbonyl of the P-loop cysteine, just as it does in Yersinia PTP (Stuckey et al., 1994). Apparently, Mce1 does not depend acutely on this interaction to hold its P-loop in a catalytically active conformation. The finding that Arg132 is essential for Mce1 function is the only instance (other than the P-loop cysteine) where there is complete agreement with available mutational data for Mce1 and all other cysteine phosphatases.
Structural comparisons of protein tyrosine phosphatases and dual-specificity phosphatases suggest that the depth of the active site pocket is a key determinant of phosphoamino acid substrate specificity. Similar considerations may apply to the RNA triphosphatases. Mammalian and Caenorhabditis elegans capping enzymes hydrolyze only the β–γ phosphoanhydride bond of triphosphate-terminated substrates. The active site pocket in the Mce1(1–210) structure can accommodate a triphosphate group linked to a nucleoside, but it may be too deep to allow diphosphate-terminated substrates to reach the active site cysteine. The active sites of BVP and PIR1, which hydrolyze 5′ triphosphates and 5′ diphosphates, are likely to be shallower than that of Mce1.
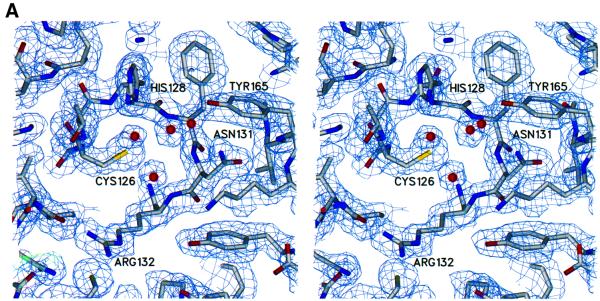
Finally, it has been suggested that cysteine phosphatase activity may be regulated in vivo by reversible oxidation of the active site cysteine. Denu and Tanner (1998) have shown that the inactivation of protein phosphatases by low concentrations of hydrogen peroxide correlates with conversion of the cysteine thiolate to cysteine sulfenic acid. Oxidation of the Mce1 Cys126 to cysteine sulfenic acid was observed from a crystal grown in the presence of tungstate (Figure 7B). Thus, stable cysteine sulfenic acid intermediates exist in two different subgroups of the cysteine phosphatase superfamily. Cysteine oxidation was also observed when the human Cdc25A protein phosphatase was crystallized in the presence of vanadate, although no vanadate was bound at the active site (Fauman et al., 1998). We speculate in light of the present findings for Mce1 that vanadate elicited the formation of a sulfenic acid intermediate in Cdc25, which then reacted with a second cysteine thiol near the active site to form the observed disulfide (Denu and Tanner, 1998). The finding of cysteine sulfenic acid in the mammalian capping enzyme raises the interesting prospect that mRNA processing may be regulated via changes in the redox state of the cell or the microenvironment of the nucleus in which transcription occurs.
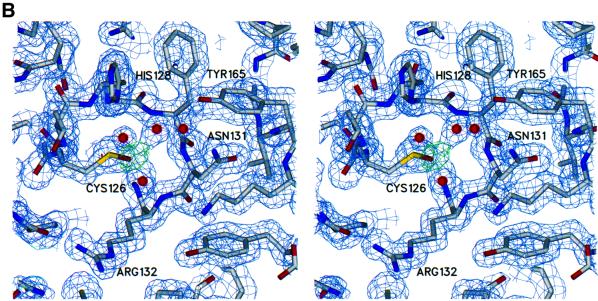
Fig. 7. Electron density of the active site in native and oxidized Mce1 RNA triphosphatase. (A) Stereo view of the experimental electron density of the active site region in the native enzyme. The map is calculated at 2.05 Å from density-modified MAD phases and is contoured at 1.2 σ. (B) Stereo view of the refined electron density at 1.7 Å of the active site region in the oxidized enzyme. The structure of the oxidized enzyme was obtained from data collected on a crystal grown in the presence of tungstate. The 2Fo – Fc map is colored in blue and is contoured at 1.2 σ. The green Fo – Fc map was calculated without the oxygen atom of the oxidized cysteine and is contoured at 4 σ. Cys126 is shown as a cysteine sulfenic acid.
Conclusions
Metazoan RNA triphosphatases share a catalytic P-loop motif with the protein tyrosine/dual-specificity and phosphoinositide phosphatases, which has led to the prediction of structural and mechanistic conservation between RNA triphosphatase and other members of the cysteine phosphatase superfamily. The crystal structure of the mammalian RNA triphosphatase domain, together with biochemical demonstration of a cysteinyl phosphoenzyme and structure-based mutational analysis, reveal structural similarity to one of three classes of the cysteine phosphatase superfamily, while highlighting unique features of the catalytic mechanism of the capping enzyme. Comparative mutagenesis of Mce1 and BVP illuminates distinct structure–activity relationships even within the RNA triphosphatase family. The differences in substrate specificity and structural requirements for catalysis by various families of cysteine phosphatases augur well for the identification of inhibitors that selectively target individual members or subgroups of the superfamily.
Materials and methods
RNA triphosphatase domain purification
His-tagged Mce1(1–210) was produced in Escherichia coli and purified as described (Ho et al., 1998b). For crystallization, the N-terminal histidine tag was removed using limited proteolysis by trypsin. Following trypsin cleavage, the protein was passed through a Ni2+–agarose column to remove uncleaved protein and then purified further using anion exchange chromatography. N-terminal sequencing and mass spectrometry analysis confirmed that trypsin cleavage had removed the histidine tag. Purified Mce1(1–210) was dialyzed into 20 mM Tris pH 8.0, 200 mM NaCl and 1 mM dithiothreitol (DTT) for crystallization. Se-Met-substituted protein was produced using the methionine pathway inhibition method (Van Duyne et al., 1993) and purified in the same manner as for wild-type Mce1(1–210). Incorporation of Se-Met was confirmed by mass spectrometry analysis.
Crystallization and data collection
Crystals were grown at 10°C by hanging drop vapor diffusion. Mce1(1–210) (5 mg/ml) was mixed with equal volumes of 15% isopropanol, 50 mM sodium cacodylate pH 6.5, 25 mM MgCl2 and 25 mM (NH4)2SO4. Crystals appeared overnight and grew as needles of ∼0.05 × 0.05 × 0.70 mm3 in 3–4 days. Streak seeding techniques were used to improve crystal size and reproducibility (Stura and Wilson, 1991). Se-Met Mce1(1–210) crystals were grown under similar conditions by streak seeding them from native crystals. For crystals grown in the presence of tungstate, Mce1(1–210) was incubated with 1 mM sodium tungstate for ∼1 h before setting up the crystallization experiment. Prior to data collection, Mce1(1–210) crystals were transferred in a single step to crystallization solution containing 30% 2,4-methylpentanediol for 2–3 min, and then flash cooled in liquid nitrogen. In order to decrease crystal mosaicity, the method of macromolecular crystal annealing (Harp et al., 1998) was used.
All data were collected at 100 K using synchrotron radiation. Data collection statistics are listed in Table I. Se-Met MAD data (Hendrickson, 1991) were collected at four wavelengths in 15° segments using inverse beam techniques. Data for the crystal grown in the presence of tungstate were collected near the absorption edge peak for tungsten in order to maximize any anomalous signal present. All data were processed using MOSFLM (Leslie, 1990) and scaled using SCALA (CCP4, 1994). Native Mce1(1–210), Se-Met Mce1(1–210) and Mce1(1–210) crystals grown in the presence of tungstate belong to space group C2221 with unit cell dimensions of a = 62.2 Å, b = 98.9 Å and c = 71.8 Å, and have one molecule in the asymmetric unit.
Structure determination and refinement
Four out of five possible selenium sites were identified with CNS (Brünger et al., 1998) using data collected at the selenium absorption peak (λ1). The sites were refined and phases to 2.05 Å were calculated by treating the MAD data as a special case of multiple isomorphous replacement using MLPHARE (CCP4, 1994). Phases were improved by density modification using DM (CCP4, 1994). The resulting electron density map clearly showed elements of protein secondary structure, and even solvent molecules could be identified (Figure 7A). ARP/wARP (Perrakis et al., 1999) automatically traced ∼90% of the polypeptide chain. Additional model building was carried out in O (Jones et al., 1991). The model was refined with REFMAC (CCP4, 1994) using data to 1.65 Å. ARP (CCP4, 1994) was used to place water molecules into Fo – Fc difference density peaks >3σ and that were within hydrogen bonding distances. No electron density was seen for the first four N-terminal residues or the last 12 C-terminal residues, and a small loop region was also found to be disordered. The final model contains residues 5–113, 119–198, 212 water molecules, one sulfate molecule, one octahedrally hydrated magnesium, two isopropanol molecules, and a cacodylate ion attached to Cys193. All residues are found within the most favored or allowed regions in the Ramachandran plot. Refinement statistics are listed in Table I.
Data were also collected from a crystal grown in the presence of tungstate. The structure was solved by rigid body refinement of the native model using REFMAC. The resulting 2Fo – Fc and Fo – Fc electron density maps showed no evidence of tungstate. Furthermore, an anomalous difference map showed peaks for the S atoms and the As atom of the cacodylate molecule, but no peak corresponding to a W atom was detected. However, additional density was seen extending from the active site cysteine (Figure 7B). This density was interpreted as oxidation of the cysteine, and thus, Cys126 was refined as a cysteine sulfenic acid (Cys126-SOH). The sulfur to oxygen distance of Cys126-SOH refined to 1.5 Å, which is consistent with S–O bond lengths in cysteine sulfenic acids found in other crystal structures (Yeh et al., 1996; Becker et al., 1998). Three well ordered water molecules located near Cys126, which were present in the native structure, are also seen. One of these waters was refined at 60% occupancy to account for its close proximity to the oxygen of the oxidized cysteine, which was refined at 40% occupancy. Density was also seen for a small loop that was disordered in the native structure. This model contains residues 4–194, 209 water molecules, one sulfate, one octahedrally hydrated magnesium, two isopropanol molecules, and a cacodylate molecule attached to Cys193. Atomic coordinates and structure factor amplitudes for native and oxidized Mce1(1–210) have been submitted to the RCSB PDB (1I9S, 1I9T).
Figures were prepared with GRASP (Nicholls et al., 1991), MOLSCRIPT (Kraulis, 1991), RASTER3D (Merritt and Murphy, 1994) and SETOR (Evans, 1993).
Mutational analysis of MCE1
Plasmid pYX1-MCE1 encodes full-length 597-amino-acid Mce1 under the control of constitutive yeast TPI1 promoter (Ho et al., 1998b). Missense mutations in the triphosphatase domain were introduced into the MCE1 gene by using the two-stage PCR overlap extension method (Ho et al., 1989). pYX1-MCE1 served as the template for the first round of amplification. The second-stage PCR products were digested with NdeI and BglII, and then ligated into the corresponding sites in pYX1-MCE1 in place of the wild-type fragment. The presence of the desired mutations was confirmed by dideoxy sequencing. We sequenced the entire restriction fragment insert in each pYX1 plasmid to exclude the introduction of unwanted mutations during amplification and cloning. The in vivo RNA capping activities of the wild-type and mutated MCE1 alleles were tested by plasmid shuffle in a yeast cet1Δ strain (Ho et al., 1998a).
The C126S mutation was introduced into a pET16-based expression vector encoding the autonomous RNA triphosphatase domain Mce1(1–210). The His-tagged C126S protein was produced in E.coli BL21(DE3) by a modification of the procedure of Ho et al. (1998b). A 1 l culture was grown at 37°C until the A600 reached 0.4–0.6, then chilled on ice for 30 min and adjusted to 2% ethanol. The culture was then incubated at 17°C for 20 h. The C126S protein was purified from a soluble bacterial lysate by Ni2+–agarose chromatography (Ho et al., 1998b). The wild-type Mce1(1–210) and Mce1 proteins were produced and purified by the same method. A GST–Mce1(1–210) fusion protein was produced by the same method, except that the culture was adjusted to 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) prior to incubation at 17°C. GST– Mce1(1–1210) was purified from a soluble bacterial lysate by adsorption to glutathione–Sepharose and elution with 10 mM reduced glutathione. The affinity-purified protein preparations were then dialyzed against buffer containing 50 mM Tris–HCl pH 8.0, 50 mM NaCl, 2 mM EDTA, 2 mM DTT, 0.1% Triton X-100, 10% glycerol. The concentrations of the recombinant triphosphatase polypeptides were determined as described (Martins and Shuman, 2000).
Acknowledgments
Acknowledgements
We thank A.Rosenzweig and E.Sontheimer for their comments. Research was supported by NIH grants GM52470 (S.S.) and GM51350 (A.Mondragón). A.C. was supported by an NRSA Institutional Training Grant in Molecular Biophysics (GM08382). Portions of this work were performed at the DuPont–Northwestern–Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source and at the Stanford Synchrotron Radiation Laboratory. DND-CAT is supported by DuPont, Dow and the NSF. Use of the APS is supported by the DOE. SSRL is operated by the DOE, Office of Basic Energy Sciences. The SSRL Biotechnology Program is supported by the NIH and the DOE.
References
- Barford D., Flint,A.J. and Tonks,N.K. (1994) Crystal structure of human protein tyrosine phosphatase 1B. Science, 263, 1397–1404. [PubMed] [Google Scholar]
- Becker K., Savvides,S.N., Keese,M., Schirmer,R.H. and Karplus,P.A. (1998) Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers. Nature Struct. Biol., 5, 267–271. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chen W., Wilborn,M. and Rudolph,J. (2000) Dual-specific Cdc25B phosphatase: in search of the catalytic acid. Biochemistry, 39, 10781–10789. [DOI] [PubMed] [Google Scholar]
- Denu J.M. and Dixon,J.E. (1995) A catalytic mechanism for the dual-specific phosphatases. Proc. Natl Acad. Sci. USA, 92, 5910–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu J.M. and Dixon,J.E. (1998) Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol., 2, 633–641. [DOI] [PubMed] [Google Scholar]
- Denu J.M. and Tanner,K.G. (1998) Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry, 37, 5633–5642. [DOI] [PubMed] [Google Scholar]
- Denu J.M., Lohse,D.L., Vijayalakshmi,J., Saper,M.A. and Dixon,J.E. (1996) Visualization of intermediate and transition-state structures in protein-tyrosine phosphatase catalysis. Proc. Natl Acad. Sci. USA, 93, 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande T., Takagi,T., Hao,L., Buratowski,S. and Charbonneau,H. (1999) Human PIR1 of the protein-tyrosine phosphatase superfamily has RNA 5′-triphosphatase and diphosphatase activities. J. Biol. Chem., 274, 16590–16594. [DOI] [PubMed] [Google Scholar]
- Diederichs K. and Karplus,P.A. (1997) Improved R-factors for diffraction data analysis in macromolecular crystallography. Nature Struct. Biol., 4, 269–275. [DOI] [PubMed] [Google Scholar]
- Evans B., Tishmack,P.A., Pokalsky,C., Zhang,M. and Van Etten,R.L. (1996) Site-directed mutagenesis, kinetic, and spectroscopic studies of the P-loop residues in a low molecular weight protein tyrosine phosphatase. Biochemistry, 35, 13609–13617. [DOI] [PubMed] [Google Scholar]
- Evans S.V. (1993) SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Fauman E.B., Cogswell,J.P., Lovejoy,B., Rocque,W.J., Holmes,W., Montana,V.G., Piwnica-Worms,H., Rink,M.J. and Saper,M.A. (1998) Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A. Cell, 93, 617–625. [DOI] [PubMed] [Google Scholar]
- Flint A.J., Tiganis,T., Barford,D. and Tonks,N.K. (1997) Development of ‘substrate-trapping’ mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl Acad. Sci. USA, 94, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.H. and Shuman,S. (1998) Characterization of a baculovirus-encoded RNA 5′-triphosphatase. J. Virol., 72, 7057–7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harp J.M., Timm,D.E. and Bunick,G.J. (1998) Macromolecular crystal annealing: overcoming increased mosaicity associated with cryocrystallography. Acta Crystallogr. D, 54, 622–628. [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A. (1991) Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science, 254, 51–58. [DOI] [PubMed] [Google Scholar]
- Ho C.K., Schwer,B. and Shuman,S. (1998a) Genetic, physical, and functional interactions between the triphosphatase and guanylyl transferase components of the yeast mRNA capping apparatus. Mol. Cell. Biol., 18, 5189–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.K., Sriskanda,V., McCracken,S., Bentley,D., Schwer,B. and Shuman,S. (1998b) The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem., 273, 9577–9585. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hof P., Pluskey,S., Dhe-Paganon,S., Eck,M.J. and Shoelson,S.E. (1998) Crystal structure of the tyrosine phosphatase SHP-2. Cell, 92, 441–450. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jia Z., Barford,D., Flint,A.J. and Tonks,N.K. (1995) Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science, 268, 1754–1758. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of proteins. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lee J.O., Yang,H., Georgescu,M.M., Di Cristofano,A., Maehama,T., Shi,Y., Dixon,J.E., Pandolfi,P. and Pavletich,N.P. (1999) Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell, 99, 323–334. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1990) Crystallographic Computing. Oxford University Press, Oxford, UK.
- Lima C.D., Wang,L.K. and Shuman,S. (1999) Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell, 99, 533–543. [DOI] [PubMed] [Google Scholar]
- Lohse D.L., Denu,J.M., Santoro,N. and Dixon,J.E. (1997) Roles of aspartic acid-181 and serine-222 in intermediate formation and hydrolysis of the mammalian protein-tyrosine-phosphatase PTP1. Biochemistry, 36, 4568–4575. [DOI] [PubMed] [Google Scholar]
- Maehama T. and Dixon,J.E. (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidyl inositol 3,4,5-trisphosphate. J. Biol. Chem., 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- Martins A. and Shuman,S. (2000) Mechanism of phosphoanhydride cleavage by baculovirus phosphatase. J. Biol. Chem., 275, 35070–35076. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Nam H.J., Poy,F., Krueger,N.X., Saito,H. and Frederick,C.A. (1999) Crystal structure of the tandem phosphatase domains of RPTP LAR. Cell, 97, 449–457. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B.H. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–286. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Shuman S. (2000) Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol., 66, 1–40. [DOI] [PubMed] [Google Scholar]
- Stuckey J.A., Schubert,H.L., Fauman,E.B., Zhang,Z.Y., Dixon,J.E. and Saper,M.A. (1994) Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 Å and the complex with tungstate. Nature, 370, 571–575. [DOI] [PubMed] [Google Scholar]
- Stura E.A. and Wilson,I.A. (1991) Applications of the streak seeding technique in protein crystallization. J. Cryst. Growth, 110, 270–282. [Google Scholar]
- Su X.D., Taddei,N., Stefani,M., Ramponi,G. and Nordlund,P. (1994) The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase. Nature, 370, 575–578. [DOI] [PubMed] [Google Scholar]
- Takagi T., Moore,C.R., Diehn,F. and Buratowski,S. (1997) An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell, 89, 867–873. [DOI] [PubMed] [Google Scholar]
- Takagi T., Taylor,G.S., Kusakabe,T., Charbonneau,H. and Buratowski,S. (1998) A protein tyrosine phosphatase-like protein from baculovirus has RNA 5′-triphosphatase and diphosphatase activities. Proc. Natl Acad. Sci. USA, 95, 9808–9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Wen Y., Yue,Z. and Shatkin,A.J. (1998) Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc. Natl Acad. Sci. USA, 95, 12226–12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J.I., Claiborne,A. and Hol,W.G. (1996) Structure of the native cysteine-sulfenic acid redox center of enterococcal NADH peroxidase refined at 2.8 Å resolution. Biochemistry, 35, 9951–9957. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama J., Denu,J.M., Dixon,J.E. and Saper,M.A. (1996) Crystal structure of the dual specificity protein phosphatase VHR. Science, 272, 1328–1331. [DOI] [PubMed] [Google Scholar]
- Zhang Z.Y. and Dixon,J.E. (1993) Active site labeling of the Yersinia protein tyrosine phosphatase: the determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry, 32, 9340–9345. [DOI] [PubMed] [Google Scholar]