Abstract
Yeast prions are protein-based genetic elements that produce phenotypes through self-perpetuating changes in protein conformation. For the prion [PSI+] this protein is Sup35, which is comprised of a prion-determining region (NM) fused to a translational termination region. [PSI+] strains (variants) with different heritable translational termination defects (weak or strong) can exist in the same genetic background. [PSI+] variants are reminiscent of mammalian prion strains, which can be passaged in the same mouse strain yet have different disease latencies and brain pathologies. We found that [PSI+] variants contain different ratios of Sup35 in the prion and non-prion state that correlate with different translation termination efficiencies. Indeed, the partially purified prion form of Sup35 from a strong [PSI+] variant converted purified NM much more efficiently than that of several weak variants. However, this difference was lost in a second round of conversion in vitro. Thus, [PSI+] variants result from differences in the efficiency of prion-mediated conversion, and the maintenance of [PSI+] variants involves more than nucleated conformational conversion (templating) to NM alone.
Keywords: amyloid/conformational conversion/epigenetics/[PSI+]/Sup35
Introduction
The Saccharomyces cerevisiae prion [PSI+] causes translational read-through of stop codons and is detected by the suppression of nonsense mutations (Cox, 1965; Wickner, 1994; Patino et al., 1996; Paushkin et al., 1996). The protein determinant of [PSI+] is the translational termination factor Sup35 (reviewed in Serio and Lindquist, 1999). [PSI+] appears when some Sup35 undergoes a conformational change that renders it insoluble and less active (Patino et al., 1996; Paushkin et al., 1996). Importantly, the insoluble prion form of the protein Sup35PSI+ promotes conversion of newly synthesized Sup35 to the prion form. Thus, the Sup35PSI+ state, as well as its associated phenotype, is heritable, indepen dent of any nucleic acid change (Kushnirov and Ter-Avanesyan, 1998; Liebman and Derkatch, 1999; Wickner and Chernoff, 1999).
Because [PSI+] is a protein-based genetic element, it has peculiar genetic properties. [PSI+] is dominant (Cox, 1965), cytoplasmically inherited (Cox, 1965; Cox et al., 1980) and in [psi–] strains it arises at a low spontaneous rate (Cox, 1965; Lund and Cox, 1981). The frequency with which it appears can be increased by transiently over-producing Sup35 or various fragments containing its N-terminus (Chernoff et al., 1993; Derkatch et al., 1996). [PSI+] is also lost spontaneously at low frequency (Cox, 1965; Cox et al., 1980) and loss is accelerated by growth in low concentrations of guanidinium hydrochloride (GuHCl) (Tuite et al., 1981) or changes in the level of the protein remodeling factor Hsp104 (Chernoff et al., 1995).
These unusual genetic properties reflect its protein-based mode of inheritance, as do its biochemical and cell biological properties. In [psi–] cells, Sup35 is mostly soluble; in [PSI+] cells, it is largely insoluble (Patino et al., 1996; Paushkin et al., 1996). Insight into the molecular mechanism underlying this switch came from biochemical characterization of Sup35 and an N-terminal fragment essential for [PSI+], NM. Like Sup35 in vivo, NM can exist in a soluble or insoluble state in vitro under physiological conditions. The insoluble form is highly organized, rich in β-sheet structure and exhibits an X-ray diffraction pattern characteristic of amyloid fibers (Glover et al., 1997; Serio et al., 2000). Minute amounts of pre-existing NM amyloid greatly accelerate the rate of assembly of soluble NM into amyloid (Glover et al., 1997). [PSI+] may or may not be equivalent to the amyloid observed in vitro, but the conformational changes that produce [PSI+] and amyloid are certainly very closely related. Addition of [PSI+] but not [psi–] lysate greatly accelerates conversion of NM to the prion form (Glover et al., 1997; Paushkin et al., 1997). NM mutations that increase or decrease [PSI+] induction frequency in vivo correspondingly alter the rate of amyloid formation in vitro (Doel et al., 1994; DePace et al., 1998; Liu and Lindquist, 1999). Thus, NM amyloid formation in vitro closely models the induction and propagation of Sup35PSI+ in vivo.
[PSI+] shares many characteristics of mammalian prions, including the existence of heritable variants with distinct biological properties that can be passaged in the same genetic background (Caughey, 2000). Mammalian prion variants, called strains, are distinguished by different rates of disease progression and brain pathology (Bruce and Fraser, 1991; Bruce, 1993). The existence of these strains is the most commonly cited evidence against a protein-based mechanism and in favor of a viral mechanism of prion propagation (Dickinson and Outram, 1979; Bruce and Dickinson, 1987; Scott et al., 1999). Prion proponents posit, however, that such strains result from transmissible conformational differences in the mammalian prion protein (PrP) (Prusiner, 1998; Scott et al., 1999; Caughey et al., 2001). Certainly conformational variation exists (reviewed in Caughey et al., 2001; Collinge, 2001), but whether it is the molecular basis of strains is unproven.
[PSI+] strains are classified by the extent of their translational termination defects and by how stable they are. Here, we refer to them as [PSI+] variants to avoid confusion with the many different yeast genetic backgrounds in which they can exist, which are also referred to as strains. Weak [PSI+] variants suppress nonsense mutations less efficiently and they are typically less stable than strong [PSI+] variants in that they lose [PSI+] more often (Derkatch et al., 1996). Once established, however, variants do not generally switch from weak to strong or vice versa, although an example of this has recently been reported (Kochneva-Pervukhova et al., 2001). [PSI+] variants appear spontaneously or when the prion is induced de novo by overproduction of Sup35 or specific N-terminal fragments (Derkatch et al., 1996; Kochneva-Pervukhova et al., 2001). They are not caused by differences in the genome: when variants from the same genetic background are cured of [PSI+] and new [PSI+] elements are induced, the same broad spectrum of variants arises in each (Derkatch et al., 1996). Thus, the phenomenon is not due to new mutations arising in Sup35 or elsewhere in the genome; it is epigenetic.
Little is known about the molecular basis of [PSI+] variants. Two studies of NM amyloid formation in vitro suggest they might represent conformational differences in Sup35PSI+. Purified NM forms amyloid fibers in vitro that vary in structure (wavy or straight) and these differences are perpetuated throughout the length of the fibers (Glover et al., 1997). Similarly, a chimeric NM mutant can adopt distinct physical states in vitro, perhaps due to quaternary structural differences (Chien and Weissman, 2001). However, there is no direct evidence linking wild-type [PSI+] variants with different Sup35PSI+ conformational states. [PSI+] variants could also be due to stochastic, self-perpetuating differences in the associa tion of Sup35 with other proteins (Schirmer and Lindquist, 1997; Czaplinski et al., 1998; Bailleul et al., 1999; Honey et al., 2001; Wang et al., 2001). Indeed, one such protein, Sup45, associates with Sup35PSI+ in some but not all genetic backgrounds (Patino et al., 1996; Paushkin et al., 1996; Czaplinski et al., 1998; Eaglestone et al., 1999).
One piece to the puzzle is that in the few cases examined, weak [PSI+] variants have more soluble Sup35 than strong [PSI+] variants. An especially weak [PSI+] variant, [ETA+], retains ∼50-fold more soluble Sup35 than a strong [PSI+] variant (Zhou et al., 1999). A different weak variant, in an unrelated genetic background, has 4-fold more soluble Sup35 than an isogenic strong [PSI+] variant (Kochneva-Pervukhova et al., 2001). The solubility of chimeric Sup35 formed by a fusion of NM from two yeast species can also differ (Kushnirov et al., 2000a). Variation in the amount of soluble Sup35 seems linked to the molecular mechanism that underlies these variant prion forms. Here, we test the possibility that Sup35PSI+ protein variants have inherent differences in their capacities to convert Sup35 into the prion form.
Results
Sup35 solubility and nonsense suppression phenotypes in [PSI+] variants
To characterize further the relationship between translational termination defects and Sup35 solubility in [PSI+] variants, we examined one [psi–] and three isogenic [PSI+] variants in the yeast strain 74-D694 (Derkatch et al., 1996, 1998) and one [psi–] and two isogenic [PSI+] variants in the strain SL1010-1A (Zhou et al., 1999). One of the SL1010-1A variants, [ETA+], is an especially weak variant of [PSI+] (Zhou et al., 1999). Both genetic backgrounds contain ade1-14, a stop codon mutation in a gene involved in adenine biosynthesis. In [psi–] strains, ribosomes do not read through this stop codon. Cells can not grow on media lacking adenine (SC–Ade), and on rich media (YPD) they form red colonies because they accumulate a red-pigmented metabolic byproduct. The greater the level of read through (nonsense suppression) in [PSI+] variants, the better they grow on SC–Ade and the whiter the colonies on YPD (Figure 1A). To provide an independent, more quantitative assay of stop codon read through, we measured the β-galactosidase (β-gal) activity of strains carrying plasmids encoding a translational fusion of phosphoglycerate kinase and β-gal with or without an intervening UGA, UAG or UAA stop codon (Figure 1B; Stansfield et al., 1995). Neither of the two [psi–] isolates grew on SC–Ade media and they produced very little β-gal.
Fig. 1. Analysis of the amount of nonsense suppression and soluble Sup35 in [PSI+] variants. (A) Five-fold serial dilutions and growth of [PSI+] variants on rich media (YPD, 30°C, 4 days) and media lacking adenine (SC–Ade, 30°C, 14 days). (B) Quantitation of translational read through of [PSI+] variants. (C) Dot-blot analysis of Sup35 solubility in [PSI+] variants. Proteins from the top fraction of each sucrose cushion were normalized, serially diluted in 2-fold increments and applied to a PVDF membrane. Top, reactivity with a Sup35-specific antibody (Patino et al., 1996). Bottom, membrane stained with a non-specific protein stain, Ponceau-S. The amount of total protein per column is indicated at the top.
By these criteria, one [PSI+] variant in the 74-D694 background and another in the SL1010-1A background were classified as strong; they grew better on adenine-deficient media and produced more β-gal than the other variants (Figure 1A and B). Two 74-D694 variants, 13 and 21, were classified as weak; they grew equally poorly on SC–Ade and their pink colony color was indistinguishable on YPD. By the plasmid-based assay, the weak [PSI+] variant 13 seemed to suppress nonsense codons slightly better than variant 21, although these differences were within the range of experimental error (Figure 1B). By all criteria, [ETA+] was the weakest [PSI+] variant.
By immunoblot analysis of total cell lysates, all of the 74-D694 [PSI+] variants had the same quantities of total Sup35 (data not shown). To determine whether they contained different amounts of Sup35 in the soluble state, lysates were sedimented through a sucrose cushion (Figure 1C; Zhou et al., 1999). Top fractions, which contained the soluble proteins, were serially diluted and spotted onto a membrane. Ponceau S staining confirmed equal protein loading for each dilution series (Figure 1C, bottom).
Within a given genetic background, [psi–] lysates always had ∼100- to 200-fold more soluble Sup35 than the lysates from strong [PSI+] variants (Figure 1C, top; Zhou et al., 1999). Lysates from [psi–] cells had ∼15- to 30-fold more soluble Sup35 than lysates of the two weak variants. The very weak variant [ETA+] from the SL1010-1A strain had ∼2- to 4-fold more soluble Sup35 than either of the weak [PSI+] variants from the 74-D694 strain (Figure 1C; Zhou et al., 1999). In contrast with the plasmid-based suppression assay, we could unambiguously distinguish between the two 74-D694 weak variants with this method; variant 13 consistently had 2- to 3-fold more soluble Sup35 than variant 21. Thus, assaying the amount of soluble Sup35 in this manner provided a very sensitive means of distinguishing between [PSI+] variants.
Sup35 from [PSI+] but not [psi–] lysates stimulated NM conversion
Next, we investigated whether the prion proteins from different [PSI+] variants have intrinsically different capacities to convert the soluble prion-determining region of Sup35 into the prion conformation. We fractionated lysates and quantified how efficiently fractions containing Sup35PSI+ stimulated conversion of soluble NM into in soluble amyloid fibers. A previous method demonstrating that proteins from [PSI+] cells could convert Sup35 to a prion-like state used a C-terminally truncated form of Sup35PSI+ purified in the presence of 2.5 M GuHCl (Paushkin et al., 1997). We used gentler conditions to avoid possible conformational rearrangements or the removal of additional proteins that might be necessary to maintain [PSI+] variants (see Materials and methods). To determine whether other components that altered the kinetics of NM conversion might co-purify with Sup35PSI+, we subjected a lysate isolated from a [psi–] NM-deletion strain (ΔNMSup35) to the same protocol. This strain can not become [PSI+] because the NM region of Sup35 was deleted from the chromosome.
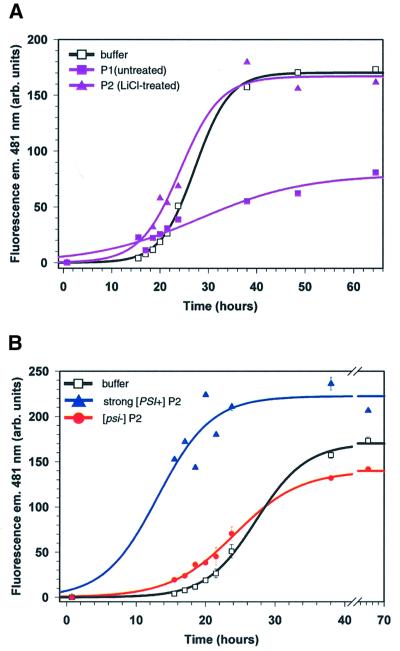
Indeed, adding small amounts of crude fractions from [psi–] lysates often inhibited NM conversion. For example, proteins from a [psi–] strain that pelleted upon high-speed sedimentation [see Materials and methods, preparation 1 (P1)] slowed NM conversion considerably, compared with buffer alone (Figure 2A, ΔNMSup35 strain; data not shown for the wild-type strain). Inhibition was not due to contaminating proteases (data not shown; performed as in Figure 3B) nor was it due to an alternative state of the NM region of Sup35 since the results from a ΔNMSup35 or wild-type fraction were indistinguishable. Although we did not identify the inhibitor, subsequent fractionation eliminated it. The proteins that pelleted after the P1 fraction was treated with 1 M LiCl and sedimented through a sucrose cushion [preparation 2 (P2)] did not inhibit conversion (Figure 2A). As discussed below, this protocol did not disrupt differences between weak and strong Sup35PSI+. Other fractionation approaches also eliminated the inhibitor (below). Here, we report only the results of experiments lacking the inhibitor(s).
Fig. 2. Sucrose cushion pellet proteins from [PSI+] but not [psi–] lysates increased the rate of NM conversion. (A) Kinetics of NM fiber formation with or without proteins pelleted from the ΔNMSup35 strain. NM fiber formation was monitored by fluorescence emission of thioflavin-T (ThT) at 481 nm. Two fractionation methods were used, preparation 1 (untreated, see Materials and methods) or 2 (LiCl treated). The black curve (open squares) represents the progress of NM conversion in the presence of buffer diluted 25-fold; the purple curve with filled triangles represents [psi–] ΔNMSup35 pelleted P2 proteins diluted 25-fold and the purple curve with filled sqaures represents [psi–] strain ΔNMSup35 pelleted P1 proteins diluted 100-fold. (B) Effect of different salt-treated P2 pellets on the kinetics of NM conversion. Conversion was monitored by fluorescence emission of ThT at 481 nm. Sucrose cushion P2 pellets or buffer were diluted 25-fold. Curves are drawn to indicate the likely data trends and are not correlated sigmoidal fits.
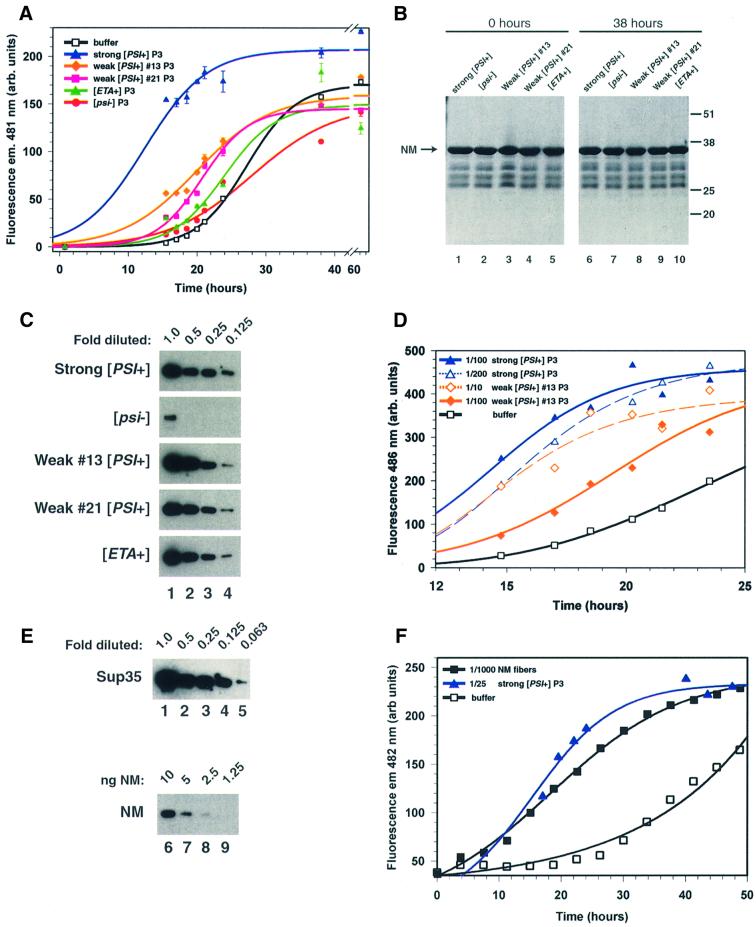
Fig. 3. Sup35PSI+ from different [PSI+] variants differ in NM conversion efficiency. (A) Effect of sucrose cushion P3 proteins isolated from several [PSI+] variants on NM conversion. Sucrose pellet P3 was used at a 100-fold dilution. Curves are drawn to indicate the likely data trends and are not correlated sigmoidal fits. (B) Extent of NM degradation during the conversion in the presence of different P3 pellets as shown in (A). Coomassie Blue R-250 staining of proteins separated by SDS–PAGE. The arrow shows migration of full-length NM. The positions and molecular weights of pre-stained marker proteins are shown on the right. Minor faster migrating bands present in all lanes are NM degradation products that co-purified from E.coli (data not shown). (C) Comparison of the amount of Sup35 in each P3 preparation. Pellet fractions were serially diluted in 2-fold increments as indicated and separated by PAGE. Sup35 was detected with NM-specific antiserum (Patino et al., 1996). (D) Effect of different dilutions of P3 preparations on the conversion efficiency of Sup35PSI+ from a strong and weak [PSI+] variant. P3 from strong [PSI+] was diluted either 100- or 200-fold and P3 from weak variant 13 was diluted 10- or 100-fold as indicated. (E) Estimation of the amount of Sup35 present in each P3 pellet fraction. Purified NM, which was serially diluted as indicated and subjected to PAGE, served as a protein standard. NM and Sup35 were detected by immunoblotting with NM-specific antiserum (Patino et al., 1996). Since P3 preparations from all [PSI+] variants have similar amounts of Sup35 (C), only the strong [PSI+] P3 pellet fraction is shown (upper blot). A darker exposure of the strong [PSI+] samples of (C) is shown here, because the signals were not visible without a longer exposure. (F) Comparison of the conversion efficiency of Sup35PSI+ from a strong [PSI+] variant with that of spontaneously formed NM fibers. P3 from strong [PSI+] was diluted 25-fold and spontaneously formed NM fibers were diluted 1000-fold; thus, the ratio of Sup35PSI+ or NM in fibers to soluble NM was ∼1:1000.
Next, we tested inhibitor-free pellet proteins (P2) from a strong [PSI+] variant for their ability to stimulate NM conversion. We confirmed the presence of Sup35 in the pellet by immunoblot analysis using Sup35-specific antibodies (data not shown). To ensure that conversion was dependent upon Sup35PSI+ we employed the same fractionation procedures with proteins from an isogenic [psi–] strain. The pellet fraction of the [psi–] strain had a small effect on NM conversion (Figure 2B). Pellet proteins from a strong [PSI+] variant, however, dramatically stimulated NM conversion when samples were diluted into a vast excess of NM. Fiber formation was also stimulated by fractions enriched for strong Sup35PSI+ obtained by several other methods, including by sedimentation through a sucrose cushion [preparation 3 (P3), compare with Figures 2B and 3A]. Treatment with 1 M LiCl (Figure 2B), RNases, EDTA or 0.5% (w/v) sodium deoxycholate did not disrupt strong Sup35PSI+ activity (data not shown).
Weak Sup35PSI+ converted NM less efficiently than strong Sup35PSI+
Fractionated proteins from other [PSI+] variants converted NM with strikingly different efficiencies (Figure 3A). The effectiveness of Sup35PSI+ from a given variant correlated with the severity of its termination defect and with the amount of soluble Sup35 it contained in vivo. Proteins of the strong variant were always much more effective than those from the weak [PSI+] variants and proteins from the weak variants were always more effective than those from the very weak variant, [ETA+]. These differences were robust and highly reproducible. Indeed, we repeatedly differentiated between phenotypically indistinguishable variants by how effectively Sup35PSI+ stimulated NM conversion. Sup35PSI+ from weak variant 13 always converted NM more rapidly than Sup35PSI+ from weak variant 21. Moreover, this efficiency difference correlated with differences in Sup35 solubility between these variants in vivo: weak variant 13 had less soluble Sup35 than weak variant 21 (Figure 1C).
A trivial reason for the differences in conversion efficiency is that the Sup35PSI+ fractions were contaminated with different amounts of proteases that degraded NM during fiber formation in vitro. To test this, we compared the total amount of NM remaining after incubation with pellet fractions with the initial amount of NM by SDS–PAGE. For all reactions, proteolytic digestion of NM by any cellular proteases that may have been present in the Sup35PSI+ fractions was negligible (Figure 3B).
Apparent differences in conversion efficiencies could also be produced by differences in the final product’s ability to bind Thioflavin-T (ThT), the dye used to detect fiber formation. To test this, we monitored fiber formation by Congo Red binding, another dye commonly used to detect amyloid (Serio et al., 1999). We also assayed the amount of NM that remained detergent soluble and visualized fibers using electron microscopy (EM) throughout the time course (Serio et al., 1999). ThT fluorescence always coincided with loss of detergent-soluble NM, onset of Congo Red binding and EM-visible fibers (data not shown), thereby eliminating this explanation.
It is possible that yeast factors in addition to Sup35PSI+ modulate conversion of NM in vitro and that these factors were present in different amounts in the various Sup35PSI+ preparations. Glutamine- and asparagine-rich proteins, like the yeast prion Rnq1, are likely candidates for such accessory factors because they can influence the frequency of [PSI+] induction (Derkatch et al., 2001; Osherovich and Weissman, 2001). Molecular chaperones are a second class of likely candidates since they can modulate [PSI+] and can exert variant-specific effects on [PSI+] in vivo (Chernoff et al., 1995, 1999; Newnam et al., 1999; Jung et al., 2000; Kushnirov et al., 2000b). We tested each Sup35PSI+ preparation for the presence of Rnq1, as well as Hsp104, Ssa and Ssb (cytosolic Hsp70 homologs) and Sis1 and Ydj1 (two Hsp40 homologs) by immunoblot analysis. All of these proteins were found in our preparations and the signals obtained for each were different. However, the signal for any particular one was the same in Sup35PSI+ preparations from all the weak and strong variants, and even from preparations from [psi–] strains (data not shown).
Strong Sup35PSI+ converted NM at least 20-fold more efficiently than weak Sup35PSI+
The pellet fractions from weak variants may have converted NM at a slower rate because they contained less Sup35 protein than the pellet fraction from the strong variant. To test this, we compared the relative amounts of Sup35 in each pellet fraction after serially diluting them in 2-fold increments (Figure 3C). As expected, [psi–] pellets had much less Sup35 than [PSI+] pellets; however, the amount of Sup35 in the pellet fractions from all [PSI+] variants differed by <2-fold. Consistent with our observation that only a small fraction of Sup35 remained in the soluble state in different [PSI+] variants (Figure 1C), the quantity of Sup35 in the pellet fractions ranged between 75 and 99% of the total amount present in the cell.
It seems highly unlikely that such small differences in the amount of Sup35 in pellet fractions could account for the differences in NM conversion. To test this possibility more rigorously, we compared the effectiveness of several dilutions of Sup35PSI+ from weak and strong 74-D694 variants. As expected, the more Sup35PSI+ that was added, the more quickly conversion occurred (Figure 3D). The effect of concentration, however, was not proportional; adding 10-fold more of weak Sup35PSI+ increased the rate of conversion by much less than 10-fold. Sonicating NM amyloid fibers fractures them and enhances nucleating activity (Serio et al., 2000) but the conversion efficiency of weak or strong Sup35PSI+ did not change after Sup35PSI+ preparations were sonicated (data not shown). We do not yet understand why there is not a simple linear relationship between the amount of Sup35PSI+ added and the rate of fiber formation. Nevertheless, slight differences in the amount of Sup35 per se can not explain the differences in NM conversion by Sup35PSI+ preparations from different variants.
Using different dilutions enabled us to quantify how much less efficiently weak Sup35PSI+ induced conversion of NM than strong Sup35PSI+. When Sup35PSI+ from weak variant 13 was diluted 10-fold, it accelerated NM conversion to a rate that was nearly equivalent to a 200-fold dilution of strong Sup35PSI+ but slower than a 100-fold dilution (Figure 3D). Thus, the Sup35PSI+ preparation from weak variant 13 contained nearly the same amount of Sup35 protein as Sup35PSI+ preparations from a strong [PSI+] variant yet it converted NM at least 20-fold less efficiently.
Next, we compared the conversion efficiencies of strong Sup35PSI+ with those of NM fibers that were never exposed to Sup35PSI+ but instead were formed by spontaneous nucleation in solutions of soluble NM. To do this, we first determined the amount of Sup35 in our various preparations by immunoblot analysis. Each pellet fraction was serially diluted and the amount of Sup35 per fraction was estimated by comparing it with known amounts of purified NM (Figure 3E). Using this method, we estimated that the molar ratio of Sup35 to NM ranged from 1:1000 to 1:5000, depending on the pellet preparation. Pre-formed NM fibers diluted to this same range stimulated NM polymerization to about the same extent (Figure 3F). Thus, the conversion efficiency of NM fibers formed spontaneously without agitation is comparable to that of strong Sup35PSI+.
Efficiency differences of Sup35PSI+ were not maintained in a second round of NM conversion
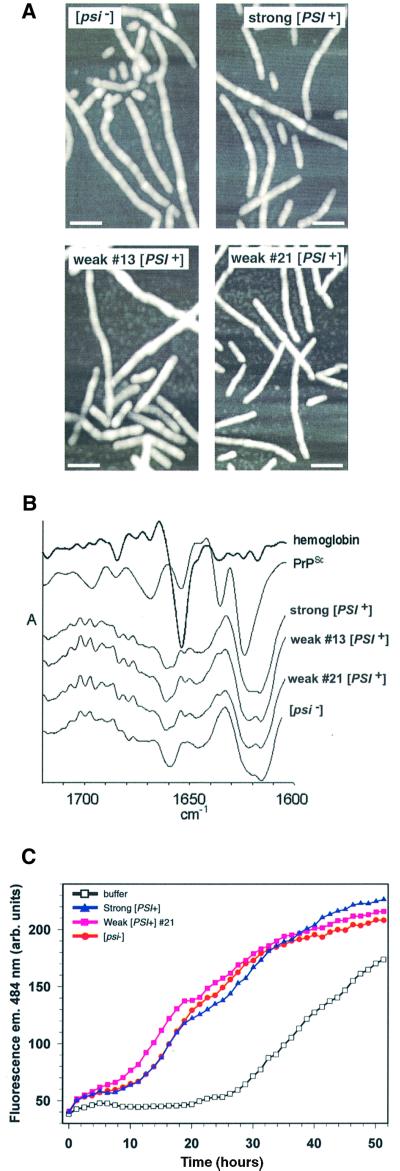
We used several different spectroscopic and biochemical techniques to see whether structural differences between fibers nucleated by weak or strong Sup35PSI+ could be detected. We examined numerous samples of NM fibers formed in the presence of Sup35PSI+ isolated from variants by EM (A.Kowal, S.M.Uptain and S.Lindquist, unpublished data) and atomic force microscopy (AFM; Figure 4A). They were indistinguishable from fibers formed in the presence of buffer. Those same fibers also appeared identical by Fourier transform infrared (FTIR) spectroscopy (Figure 4B) and by digestion with proteinase K (data not shown), two techniques that are used to detect structural differences in PrP derived from different mammalian prion strains (Bessen and Marsh, 1992a; Caughey et al., 1998).
Fig. 4. Structural and biochemical characterization of fibers converted in the presence of Sup35PSI+ from different variants. (A) AFM images taken in tapping mode of fibers formed in the presence of Sup35PSI+ from strong or weak variants or with an identically prepared fraction from a [psi–] strain. The white bar represents 100 µm. (B) Second derivative attenuated total reflectance infrared spectra of NM fibers converted with strong [PSI+], weak [PSI+] (13 and 21) and [psi–] lysates. Second derivative spectra show negative deflections that correspond to positive absorbance maxima in primary spectra and provide a means of better resolving small differences between infrared spectra. For comparison, the spectra of hemoglobin (predominantlyα-helix) and PrPSc (mixed α-helix and β-sheet) are included. The amide I region is shown, which is sensitive primarily to the secondary structure of the polypeptide backbone. No consistent differences were observed between the four types of NM fiber in either the primary (not shown) or second derivative spectra. All of the NM fiber spectra showed predominant negative bands in the region dominated by β-sheet absorbances (∼1616–1640 cm–1); however, the pattern of these probable β-sheet bands was distinct from that of PrPSc. The other major NM fiber band was centered at ∼1660 cm–1, a wavenumber that is inconsistent with either β-sheet or α-helix but consistent with the presence of turn structure(s). (C) Determining whether the characteristic conversion efficiencies of Sup35PSI+of variants can be serially passaged to a second NM preparation. Sucrose P2 pellets from [psi–] and two [PSI+] variants (strong or weak 21) were used to prepare NM fibers that were subsequently diluted 1000-fold into another preparation of 10 µM soluble NM.
Since we could not find any physical difference between them, we tested whether fibers converted by Sup35PSI+ retained the remarkable differences in conversion efficiency characteristic of the initial Sup35PSI+ preparations. We passaged fibers formed in the presence of Sup35PSI+ from variants by diluting them into a second batch of soluble NM. At a 1000-fold dilution, all stimulated NM conversion similarly (Figure 4C). We also tested the effect of using 100-, 10 000- or 100 000-fold dilutions of each seed type. Although the extent that NM conversion was stimulated changed with the dilution, no reproducible differences in the rate of NM conversion were observed, regardless of whether the fibers had been initially converted under the influence of weak or strong Sup35PSI+ preparations (Figure 4C and data not shown) or whether they had formed spontaneously in the presence of [psi–] preparations (Figure 4C and data not shown) or with NM alone (data not shown).
Discussion
Our data provide a molecular explanation for the different nonsense suppression phenotypes and stabilities of genetically identical [PSI+] variants (Derkatch et al., 1996; Zhou et al., 1999). Sup35PSI+ isolated from weak and strong [PSI+] variants converted NM, the prion-determining region of Sup35, with dramatically different efficiencies. Importantly, the conversion efficiency of Sup35PSI+ from a given variant correlated with the amount of soluble Sup35 and the nonsense suppression phenotype characteristic of that variant. Sup35PSI+ preparations from a strong variant that had little soluble Sup35 and a substantial translational termination defect converted NM into amyloid at least 20-fold more efficiently than Sup35PSI+ from two weak variants. Conversion mediated by Sup35PSI+ from [ETA+], a very weak variant, was even less efficient.
These data strongly indicate that in yeast cells, weak and strong Sup35PSI+ convert soluble Sup35 into the prion conformation with a wide range of efficiencies and that this dictates the type of variant that is propagated. In a strong [PSI+] variant, conversion is efficient and consequently the amount of soluble Sup35 is low. Weak Sup35PSI+ converts soluble Sup35 less efficiently; thus weak variants contain more soluble Sup35 and have a weaker termination defect. The poorer conversion efficiency of weak Sup35PSI+ can also explain the reduced genetic stability of weaker variants compared with stronger ones. Most importantly, the differences in conversion rates are likely to be inherent to the prion form of Sup35, since these differences required the presence of Sup35 from a [PSI+] variant and co-purified with it.
Our data also resolve the apparently paradoxical differences in how well Sup35PSI+ mediates conversion in vivo and in vitro. In a less purified cell-free system, Sup35PSI+ from a strong variant was reported to convert twice the amount of NM to an aggregated, proteinase K-resistant state than Sup35PSI+ from a weak variant (Kochneva-Pervukhova et al., 2001). In these experiments, the ratio of Sup35PSI+ to NM was 1:4, yet the majority of NM remained soluble, even when strong Sup35PSI+ was used. This result was perplexing because conversion of Sup35 to the prion state is very efficient in vivo and because the vast majority of Sup35 is insoluble in [PSI+] cells, even in the weakest variants. We found that both weak and strong Sup35PSI+ converted soluble NM into amyloid remarkably well. Indeed, after comparing the amount of Sup35 in our assays with a known amount of NM, we estimated that the molar ratio of Sup35 to NM was between 1:1000 and 1:5000, yet we easily detected a [PSI+]-dependent stimulation in NM conversion, for both strong and weak Sup35PSI+. The likely explanation for this discrepancy is that the inhibitory activity we observed in cruder preparations reduced the conversion efficiency in the earlier study.
We also compared the conversion efficiency of NM amyloid fibers formed spontaneously in vitro with that of strong Sup35PSI+ from yeast cells. When comparable amounts of each were used, the rates of conversion were nearly identical, suggesting that the physical states that NM adopts when converted by strong Sup35PSI+ from yeast cells and by spontaneous nucleation in vitro are similar.
By comparison, conversion by the mammalian PrP is generally much less efficient than conversion by Sup35PSI+. It is usually limited to sub-stoichiometric yields in vitro (Caughey, 2000; Wong et al., 2001), although the yield can be substantially improved with repeated cycles of sonication (Saborio et al., 2001). This difference appears to be biologically relevant since yeast can mitotically divide in as little as 90 min, whereas neuronal cells are not mitotic. Thus, propagation of prions in yeast seems to necessitate much more efficient conversion to the prion state. When conversion of Sup35 is less efficient, as is the case with weak [PSI+] variants, more Sup35 is soluble and a greater proportion of progeny fail to inherit the prion.
Epigenetic differences in how well PrP converts the normal folded conformation into the prion conformation also occur with the mammalian PrP. Indeed, different efficiencies of conversion have been reported for two mink prion strains passaged in hamster, termed Drowsy (DY) and Hyper (HY). The PrP from the HY strain converts more efficiently in vitro and has greater infectivity in vivo than that from the DY strain (Bessen and Marsh, 1992b; Bessen et al., 1995). Intriguingly, the incubation period of DY in hamsters is much longer than that of HY (Bessen and Marsh, 1992b). Thus, conversion efficiency in vitro and in vivo for the mammalian prion as well as for [PSI+] correlates with their properties in vivo.
What leads to the differences in conversion efficiencies associated with [PSI+] variants? We suggest three possibilities that are not mutually exclusive. First, the Sup35 protein may itself adopt different conformations that have intrinsically different abilities to convert the normal form into the prion form. Using EM, Congo Red binding and protease digestion patterns, one study has shown that NM can form amyloid fibers with different structural states in vitro (Glover et al., 1997) and another has shown that a chimeric NM can form fibers with different biochemical properties (Chien and Weissman, 2001). However, the relevance to [PSI+] variants is unclear. By these and other techniques, we have not yet found any direct evidence for conformational differences between NM fibers that were nucleated by weak or strong Sup35PSI+ preparations, nor were we able to passage the characteristic conversion efficiencies of weak Sup35PSI+ to a second NM preparation in vitro. Instead, when such fibers were passaged, they converted as efficiently as fibers formed in the presence of strong Sup35PSI+ preparations. Perhaps conversion in vitro is not faithful enough to reproduce conformational differences throughout the >1000-fold amplification that we have employed. If NM occasionally switches from an efficient form to an inefficient form, this would have a negligible effect on the reaction. However, with occasional switching of inefficient to efficient forms, the latter would soon take over the reaction.
A second explanation for the different conversion efficiencies of weak and strong Sup35PSI+ is predicated upon the finding that fiber ends nucleate NM amyloid formation (Serio et al., 2000). If conversion of Sup35 ensues via a similar mechanism in vivo, the prion protein of a strong [PSI+] variant might contain more surfaces to promote conversion than weaker variants. This model does not require that the Sup35PSI+ aggregates vary in secondary or tertiary structure; rather, differences in quaternary structures could be sufficient. For example, aggregates of stronger variants could be smaller and more numerous than those of weaker variants, thus, they would presumably convert soluble Sup35 more effectively and be propagated more faithfully. Such differences in nucleation surfaces are not likely to be retained by NM fibers formed under the influence of Sup35PSI+ in vitro.
A third explanation is that interactions between other cellular factors and Sup35, which might be stochastic in origin but self-perpetuating in nature, may modulate the efficiency of conversion to the prion state in yeast [PSI+] variants. If so, these factors would be present in our initial preparations and would influence the first, but not subsequent rounds of conversions. Two likely candidates are molecular chaperones, which aid protein folding, and glutamine- and asparagine-rich proteins (QNrps). Changing the balance of molecular chaperones has pleiotropic effects on different [PSI+] variants (Chernoff et al., 1995, 1999; Newnam et al., 1999; Jung et al., 2000; Kushnirov et al., 2000b). Moreover, two chaperones, Sis1 and Ssa1, interact stably with another yeast prion protein, Rnq1, only when it is in the prion conformation (Sondheimer et al., 2001). At least 12 QNrps, including Rnq1, can influence the frequency at which [psi–] cells convert to [PSI+] (Derkatch et al., 2001; Osherovich and Weissman, 2001), but they are not required to maintain [PSI+]. Although there were no apparent differences in the distribution of several molecular chaperones and Rnq1 in our Sup35PSI+ preparations, this does not preclude variant-specific differences in the nature of these or other such protein associations. Phenomena of this type may be common. In mammalian cells, worms and yeast, diverse QNrps interact with each other and with molecular chaperones, and may thereby modulate the toxicity of protein aggregates in several neurodegenerative diseases (Cha, 2000; Chernoff, 2001; Nucifora et al., 2001; Perutz and Windle, 2001).
Our work establishes that the key feature underlying [PSI+] variants is a difference in conversion efficiency. Since our system preserves this feature in the initial rounds of conversion, it provides an assay for discovering the molecular mechanism(s) that might maintain and propagate such differences. Moreover, it will be interesting to see whether similar differences in conversion efficiencies of another yeast prion protein, Ure2, underlie the recently reported [URE3] prion variants (Schlumpberger et al., 2001). Ultimately, experiments of this type may contribute to our understanding of the molecular mechanism that underlies strains of mammalian prions, other neurodegenerative disease processes and diverse epigenetic phenomena that still defy explanation.
Materials and methods
Yeast strains
We used [PSI+] variants in 74-D694 [MATa ade1-14 (UGA) trp1-289 his3-Δ200 ura3-52 leu2-3,112] (Chernoff et al., 1995) and SL1010-1A [MATα ade1-14 (UGA) met8-1 (UAG) leu2-1 his5-2 trp1-1 ura3-52] (Zhou et al., 1999). The latter strain is related to 74-D694 by one out-crossing (Zhou et al., 1999). The 74-D694 strong [PSI+], [psi–] and the ΔNMSup35 strains were provided by I.Derkatch and Y.Chernoff. The other [PSI+] variants were provided by P.Zhou, I.Derkatch and S.Liebman: 74-D694 derivatives weak [PSI+] 13 (Derkatch et al., 1997, 1998; Zhou et al., 2001) and weak [PSI+] 21 (Derkatch et al., 1998; Zhou et al., 2001) and SL1010-1A derivatives strong [PSI+], [ETA+] and [psi–] (Zhou et al., 1999).
Lysate preparation
Cells were grown in YPD at 25 or 30°C to an optical density of 0.4–0.6 at 600 nm (OD600), treated with 50 µg/ml zymolyase 100 T (Seikagaku America, Inc.) and incubated in YPD containing 1.2 M sorbitol and 1 mM NaN3 for 15 min. Spheroplasts were suspended in lysis buffer [25 mM Tris–HCl pH 7.5, 100 mM NaCl, 5 mM MgCl2, 5% (v/v) glycerol, 4 mM AEBSF, 5 µg/ml aprotonin and 5 µg/ml leupeptin]. Lysis commenced upon sequential addition of sodium deoxycholate to 0.5% (w/v) and 5 min later Brij-58 to 0.5% (w/v). Lysates were partially clarified by centrifugation at 10 000 g for 20 min at 4°C. The protein concentration of the supernatants was normalized and they were processed in one of three ways.
In preparation 1, supernatants were sedimented at 100 000 g for 30 min at 4°C (TLA-100.2 rotor; Beckman). Pellets were suspended in the original volume of lysis buffer without AEBSF. After sedimentation was repeated, the pellets (P1) were suspended in lysis buffer without AEBSF, which was omitted since it retarded fiber formation (data not shown).
In preparation 2, LiCl was added to P1 pellets to 1 M. Suspensions were incubated for 30 min with gentle rotation at 4°C and layered upon 30% (w/v) sucrose prepared in lysis buffer without AEBSF. Samples were sedimented as described (Zhou et al., 1999). Sucrose pellets (P2) were suspended in lysis buffer without AEBSF.
In preparation 3, supernatants were layered upon 30% (w/v) sucrose prepared in lysis buffer without protease inhibitors and sedimented at 200 000 g for 103 min at 4°C (SW28 rotor; Beckman). The pellet fraction (P3) was suspended in lysis buffer without protease inhibitors. We estimate that ∼30 µg of Sup35 were present per 1 mg of total protein in each P3 Sup35PSI+ preparation (Figure 3E).
NM purification
Untagged NM was purified to homogeneity from Escherichia coli as described (Serio et al., 1999) with minor changes. Proteins were eluted from the Q-Sepharose fast flow column (Pharmacia) using a shallow linear gradient of 25 column vols from 40 to 240 mM NaCl in 8 M urea and 10 mM Tris–HCl pH 7.2. Fractions containing NM were pooled and loaded onto the Macro prep Ceramic hydroxyapatite Type 1 column (HA; Bio-Rad). Sodium phosphate replaced potassium phosphate in all HA column solutions. HA wash buffer II contained 40 mM sodium phosphate pH 6.8 in 8 M urea. Protein was eluted using 125 mM sodium phosphate pH 6.8 in 8 M urea. Purified NM was dialyzed against 8 M urea, 20 mM Tris–HCl pH 7.5, precipitated with 4 vols of 100% anhydrous methanol and stored at –80°C.
NM conversion and detection
Recombinant NM (10 µM) was incubated in 1× PBS pH 7.4, 5 µg/ml aprotonin, 5 µg/ml leupeptin with or without yeast protein fractions according to the figure legends. Conversion was performed at 7°C for all experiments except for those shown in Figures 3F and 4C, which were performed at 25°C. Conversion was monitored by fluorescence emission of 20 µM ThT (Sigma) and in the presence of 0.2 µM NM using a Jasco FP-750 (bandpass 5 nm ex., 10 nm em.) or a Jobin Yvon Horiba Fluormax-3 (bandpass 3 nm ex. and em.) fluorescence spectrophotometer. The excitation and emission wavelengths were 450 nm and between 481 and 486 nm, respectively. Sigmoid curves [SigmaPlot 2000 for Windows (SPSS Inc.)] in Figures 2A, B, 3A, D and F indicate only trends and are not intended as correlated fits. Error bars represent the standard error of between 3 and 5 measurements of one sample at each time point. The activity of any contaminating proteases during fiber assembly was determined throughout each time course by Coomassie Blue R-250 staining of proteins separated by SDS–PAGE.
For AFM, 50 µl of 1.0 µM fibers were applied to a freshly cleaved mica surface for 10 min. The mica was rinsed once with 150 µl of 1× PBS, rinsed five times with 150 µl of distilled water and allowed to air dry overnight. Samples were imaged in tapping mode with a Multimode scanning probe microscope (Digital Instruments, Santa Barbara, CA) using a type NCH etched silicon probe (Nanosensors, Wetzlar-Blankenfeld, Germany).
For FTIR spectroscopy, NM fibrils were pelleted by centrifugation at 11 000 g for 10 min at 4°C. Aliquots (2 µl) of slurried pellets were applied to a Golden Gate single reflection diamond attenuated total reflectance unit (Graseby-Specac) purged with dry air and covered to prevent sample evaporation. Data collection was performed on a System 2000 FT-IR Instrument (Perkin Elmer). Test conditions: 20°C, 2 cm–1 resolution, 5 cm/s OPD velocity, 1000 scans, 2500–1000 cm–1 scan range, 0.5 cm–1 data interval. The detector was an nb1 MCT cooled by liquid nitrogen. Primary spectra were obtained by individually subtracting spectra of corresponding buffer and water vapor. Second-derivative spectra were calculated from smoothed primary spectra using five data points and normalized to adjust for differences (of less than a factor of two) in NM concentration. The software used for spectral analyses was Spectrum v2.00 (Perkin Elmer).
Dot-blot analysis
The amount of soluble Sup35 in [PSI+] variants was determined as described (Zhou et al., 1999), except that crude lysates were partially clarified by sedimentation at 12 000 g rather than 4000 g. All strains were grown at 25°C to an OD600 of ∼1.5 and a cell density of 4–5 × 107 cells/ml.
Quantification of translational read through
[PSI+] variants expressing a translational fusion of phosphoglycerate kinase to β-gal either with or without an intervening UGA, UAG or UAA stop codon (Stansfield et al., 1995) were grown at 25°C to an OD600 between 0.4 and 0.6. β-gal activity was measured as described (Chernoff et al., 2001) using a GalactoStar kit (Tropix). The average values from two independent experiments (three replicates each) for each plasmid are plotted as percentage β-gal specific activity of strains with the plasmid lacking a stop codon. Error is reported as percentage population standard deviation (Taylor, 1982).
Acknowledgments
Acknowledgements
We thank Yury Chernoff, Ping Zhou, Irina Derkatch and Sue Liebman for yeast strains, Anthony Kowal for performing EM, Thomas Scheibel for the NM expression plasmid, John Lis and members of the Lindquist laboratory for their comments on this manuscript. This work was supported by the Training Program in Cancer Biology and a postdoctoral fellowship from the American Cancer Society awarded to S.M.U. and funding from the NIH (GM57840) and HHMI awarded to S.L.L.
References
- Bailleul P.A., Newnam,G.P., Steenbergen,J.N. and Chernoff,Y.O. (1999) Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics, 153, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen R.A. and Marsh,R.F. (1992a) Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol., 66, 2096–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen R.A. and Marsh,R.F. (1992b) Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J. Gen. Virol., 73, 329–334. [DOI] [PubMed] [Google Scholar]
- Bessen R.A., Kocisko,D.A., Raymond,G.J., Nandan,S., Lansbury,P.T. and Caughey,B. (1995) Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature, 375, 698–700. [DOI] [PubMed] [Google Scholar]
- Bruce M.E. (1993) Scrapie strain variation and mutation. Br. Med. Bull., 49, 822–838. [DOI] [PubMed] [Google Scholar]
- Bruce M.E. and Dickinson,A.G. (1987) Biological evidence that scrapie agent has an independent genome. J. Gen. Virol., 68, 79–89. [DOI] [PubMed] [Google Scholar]
- Bruce M.E. and Fraser,H. (1991) Scrapie strain variation and its implications. Curr. Top. Microbiol. Immunol., 172, 125–138. [DOI] [PubMed] [Google Scholar]
- Caughey B. (2000) Transmissible spongiform encephalopathies, amyloid oses and yeast prions: common threads? Nature Med., 6, 751–754. [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond,G.J. and Bessen,R.A. (1998) Strain-dependent differences in β-sheet conformations of abnormal prion protein. J. Biol. Chem., 273, 32230–32235. [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond,G.J., Callahan,M.A., Wong,C., Baron,G.S. and Xiong,L.W. (2001) Interactions and conversions of prion protein isoforms. Adv. Protein Chem., 57, 139–169. [DOI] [PubMed] [Google Scholar]
- Cha J.H. (2000) Transcriptional dysregulation in Huntington’s disease. Trends Neurosci., 23, 387–392. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O. (2001) Mutation processes at the protein level: is Lamarck back? Mutat. Res., 488, 39–64. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Derkatch,I.L. and Inge-Vechtomov,S.G. (1993) Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet., 24, 268–270. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist,S.L., Ono,B., Inge-Vechtomov,S.G. and Liebman,S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+]. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Newnam,G.P., Kumar,J., Allen,K. and Zink,A.D. (1999) Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol., 19, 8103–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y.O., Uptain,S.M. and Lindquist,S. (2001) Analysis of prion factors in yeast. Methods Enzymol., in press. [DOI] [PubMed] [Google Scholar]
- Chien P. and Weissman,J.S. (2001) Conformational diversity in a yeast prion dictates its seeding specificity. Nature, 410, 223–227. [DOI] [PubMed] [Google Scholar]
- Collinge J. (2001) Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci., 24, 519–550. [DOI] [PubMed] [Google Scholar]
- Cox B. (1965) [PSI+], a cytoplasmic suppressor of super-suppression in yeast. Heredity, 20, 505–521. [Google Scholar]
- Cox B.S., Tuite,M.F. and Mundy,C.J. (1980) Reversion from suppression to nonsuppression in SUQ5 [psi+] strains of yeast: the classification of mutations. Genetics, 95, 589–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePace A.H., Santoso,A., Hillner,P. and Weissman,J.S. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell, 93, 1241–1252. [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Chernoff,Y.O., Kushnirov,V.V., Inge-Vechtomov,S.G. and Liebman,S.W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics, 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E., Zhou,P., Chernoff,Y.O. and Liebman,S.W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics, 147, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E. and Liebman,S.W. (1998) Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc. Natl Acad. Sci. USA, 95, 2400–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E., Hong,J.Y. and Liebman,S.W. (2001) Prions affect the appearance of other prions: the story of [PIN+]. Cell, 106, 171–182. [DOI] [PubMed] [Google Scholar]
- Dickinson A.G. and Outram,G.W. (1979) The scrapie replication-site hypothesis and its implications for pathogenesis. In Prusiner,S.B. (ed.), Slow Transmissible Diseases of the Nervous System. Academic Press, New York, NY, pp. 13–31.
- Doel S.M., McCready,S.J., Nierras,C.R. and Cox,B.S. (1994) The dominant PNM2– mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics, 137, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S.S., Cox,B.S. and Tuite,M.F. (1999) Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J., 18, 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Kowal,A.S., Schirmer,E.C., Patino,M.M., Liu,J.J. and Lindquist,S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S.cerevisiae. Cell, 89, 811–819. [DOI] [PubMed] [Google Scholar]
- Honey S., Schneider,B.L., Schieltz,D.M., Yates,J.R. and Futcher,B. (2001) A novel multiple affinity purification tag and its use in identification of proteins associated with a cyclin–CDK complex. Nucleic Acids Res., 29, E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Jones,G., Wegrzyn,R.D. and Masison,D.C. (2000) A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics, 156, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N.V., Chechenova,M.B., Valouev,I.A., Kushnirov, V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (2001) [PSI+] prion generation in yeast: characterization of the ‘strain’ difference. Yeast, 18, 489–497. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. and Ter-Avanesyan,M.D. (1998) Structure and replication of yeast prions. Cell, 94, 13–16. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V., Kochneva-Pervukhova,N.V., Chechenova,M.B., Frolova,N.S. and Ter-Avanesyan,M.D. (2000a) Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J., 19, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V.V., Kryndushkin,D.S., Boguta,M., Smirnov,V.N. and Ter-Avanesyan,M.D. (2000b) Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol., 10, 1443–1446. [DOI] [PubMed] [Google Scholar]
- Liebman S.W. and Derkatch,I.L. (1999) The yeast [PSI+] prion: making sense of nonsense. J. Biol. Chem., 274, 1181–1184. [DOI] [PubMed] [Google Scholar]
- Liu J.J. and Lindquist,S. (1999) Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature, 400, 573–576. [DOI] [PubMed] [Google Scholar]
- Lund P.M. and Cox,B.S. (1981) Reversion analysis of [psi–] mutations in Saccharomyces cerevisiae. Genet. Res., 37, 173–182. [DOI] [PubMed] [Google Scholar]
- Newnam G.P., Wegrzyn,R.D., Lindquist,S.L. and Chernoff,Y.O. (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol., 19, 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora F.C. Jr et al. (2001) Interference by Huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science, 291, 2423–2428. [DOI] [PubMed] [Google Scholar]
- Osherovich L.Z. and Weissman,J.S. (2001) Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell, 106, 183–194. [DOI] [PubMed] [Google Scholar]
- Patino M.M., Liu,J.J., Glover,J.R. and Lindquist,S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1997) In vitro propagation of the prion-like state of yeast Sup35 protein. Science, 277, 381–383. [DOI] [PubMed] [Google Scholar]
- Perutz M.F. and Windle,A.H. (2001) Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature, 412, 143–144. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1998) Prions. Proc. Natl Acad. Sci. USA, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborio G.P., Permanne,B. and Soto,C. (2001) Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature, 411, 810–813. [DOI] [PubMed] [Google Scholar]
- Schirmer E.C. and Lindquist,S. (1997) Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc. Natl Acad. Sci. USA, 94, 13932–13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumpberger M., Prusiner,S.B. and Herskowitz,I. (2001) Induction of distinct [Ure3] yeast prion strains. Mol. Cell. Biol., 21, 7035–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M., DeArmond,S.J., Prusiner,S.B., Ridley,R.M. and Baker,H.F. (1999) Transgenic investigations of the species barrier and prion strains. In Prusiner,S.B. (ed.), Prion Biology and Diseases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 307–347.
- Serio T.R. and Lindquist,S.L. (1999) [PSI+]: an epigenetic modulator of translation termination efficiency. Annu. Rev. Cell Dev. Biol., 15, 661–703. [DOI] [PubMed] [Google Scholar]
- Serio T.R., Cashikar,A.G., Moslehi,J.J., Kowal,A.S. and Lindquist,S.L. (1999) Yeast prion [PSI+] and its determinant, Sup35p. Methods Enzymol., 309, 649–673. [DOI] [PubMed] [Google Scholar]
- Serio T.R., Cashikar,A.G., Kowal,A.S., Sawicki,G.J., Moslehi,J.J., Serpell,L., Arnsdorf,M.F. and Lindquist,S.L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science, 289, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Sondheimer N., Lopez,N., Craig,E.A. and Lindquist,S. (2001) The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J., 20, 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I., Akhmaloka and Tuite,M.F. (1995) A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet., 27, 417–426. [DOI] [PubMed] [Google Scholar]
- Taylor J.R. (1982) An Introduction to Error Analysis: The Study of Uncertainties in Physical Measurements. University Science Books, Mill Valley, CA.
- Tuite M.F., Mundy,C.R. and Cox,B.S. (1981) Agents that cause a high frequency of genetic change from [PSI+] to [psi–] in Saccharomyces cerevisiae. Genetics, 98, 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Czaplinski,K., Rao,Y. and Peltz,S.W. (2001) The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J., 20, 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner R.B. and Chernoff,Y.O. (1999) Prions of fungi: [URE3], [PSI], and [Het-s] discovered as heritable traits. In Prusiner,S.B. (ed.), Prion Biology and Diseases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 229–272.
- Wong C., Xiong,L.W., Horiuchi,M., Raymond,L., Wehrly,K., Chesebro, B. and Caughey,B. (2001) Sulfated glycans and elevated temperature stimulate PrPSc-dependent cell-free formation of protease-resistant prion protein. EMBO J., 20, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Derkatch,I.L., Uptain,S.M., Patino,M.M., Lindquist,S. and Liebman,S.W. (1999) The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J., 18, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Derkatch,I.L. and Liebman,S.W. (2001) The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+]. Mol. Microbiol., 39, 37–46. [DOI] [PubMed] [Google Scholar]