Abstract
In yeast, the differentiation process at the end of meiosis generates four daughter cells inside the boundaries of the mother cell. A meiosis-specific plaque (MP) at the spindle pole bodies (SPBs) serves as the starting site for the formation of the prospore membranes (PSMs) that are destined to encapsulate the post-meiotic nuclei. Here we report the identification of Ady3p and Ssp1p, which are functional components of the leading edge protein (LEP) coat, that covers the ring-shaped opening of the PSMs. Ssp1p is required for the assembly of the LEP coat, which consists of at least three proteins (Ssp1p, Ady3p and Don1p). The assembly of the LEP coat starts with the formation of cytosolic precursors, which then bind in an Ady3p-dependent manner to the SPBs. Subsequent processes at the SPBs leading to functional LEP coats require Ssp1p and the MP components. During growth of the PSMs, the LEP coat functions in formation of the cup-shaped membrane structure that is indispensable for the regulated cellularization of the cytoplasm around the post-meiotic nuclei.
Keywords: Ady3p/meiosis/membrane organization/spindle pole body/Ssp1p
Introduction
Meiosis is a unique type of cell division in which four haploid daughter cells are formed from a diploid mother cell. It begins with the duplication of the genome, followed by two subsequent rounds of chromosome segregation. The meiotic cell differentiation process is called gametogenesis and involves dramatic morphological changes that lead to the formation of specialized daughter cells, the gametes.
In the baker’s yeast Saccharomyces cerevisiae, the gametes are termed spores. They are formed at the end of meiosis II inside the boundaries of the mother cell. This meiotic differentiation process involves the formation of the so-called prospore membranes (PSMs), the plasma membrane equivalent encapsulating the spores. PSM formation is dependent on a developmentally controlled reprogramming of the secretory pathway and was shown to rely on secretory processes equivalent to the fusion of vesicles with the plasma membrane (Neiman, 1998; Neiman et al., 2000). The differentiation process starts with the formation of cytosolic precursors of the PSMs during meiosis I (Knop and Strasser, 2000). The formation of the PSMs is then initiated on the cytoplasmic side of the spindle pole bodies (SPBs) (Moens, 1971; Davidow et al., 1980; Knop and Strasser, 2000). SPBs are the centrosome equivalents of yeast and are always embedded in the nuclear envelope, which stays intact during all mitotic and meiotic division processes (Moens and Rapport, 1971; Byers and Goetsch, 1975). At the beginning of meiosis II, all precursors of the PSMs are found at the SPBs where they assemble into continuous PSMs, one per SPB. This step requires the assembly and function of an extended outer plaque of the SPB, the so-called meiotic plaque (MP) (Knop and Strasser, 2000). During meiosis II, the PSMs grow and form cup-shaped structures that engulf lobes of the nucleus. The leading edges of the PSMs are covered by a proteinaceous coat (Davidow et al., 1980; Knop and Strasser, 2000; web-companion of Chu et al., 1998; http://cmgm.stanford.edu/pbrown/sporulation/em/fig1.html) termed here the leading edge protein (LEP) coat. Upon completion of meiosis II, the nuclear lobes pinch off and the PSMs close to form two membranes each, stacked on top of one another (Lynn and Magee, 1970; Moens and Rapport, 1971; Zickler and Olson, 1975). The closed PSMs then mature to become the spore walls.
Although the meiotic differentiation pathway in yeast, leading to the formation of spores, has been described morphologically, the underlying molecular mechanisms that control the various structural processes are largely unknown. In this field, the most important questions address the targeting of membranes to the SPBs, the initiation of PSM formation, the subsequent growth and shaping of the membranes and finally the closure of the PSMs.
Recently, while investigating the meiosis-specific function of the SPB, we have identified two novel proteins, Mpc54p and Mpc70p, which form the MP of the SPB. During meiosis II, the MP replaces the (mitotic- and meiosis I-specific) cytoplasmic microtubule-organizing machinery of the SPB and is absolutely required for prospore membrane assembly (Knop and Strasser, 2000).
We began this study by asking whether other proteins are associated with the MP function in the assembly of the PSM. We purified a Mpc54p-protein A (ProA)-containing subcomplex from meiotic SPBs and found Ady3p as a co-purifying protein. We demonstrate that Ady3p transiently interacts during early steps of PSM assembly with the SPBs, and thereafter localizes to the LEP coats where it is required for the efficient formation of spores. The study was extended further to investigate the function of Ssp1p sporulation-specific protein 1; Nag et al., 1997), a two-hybrid interaction partner of Ady3p (Uetz et al., 2000; Ito et al., 2000). We show that Ssp1p is a key component in the pathway that organizes the shaping and sizing of the PSM. Ssp1p is required for the formation of Ady3p- and Don1p-containing protein coats at the leading edge of the PSMs. These investigations led us to propose a model for the assembly pathway of PSMs and for the role of Ady3p, Ssp1p and the MP components Mpc54p and Mpc70p during this process.
Results
Identification of Ady3p and Ssp1p
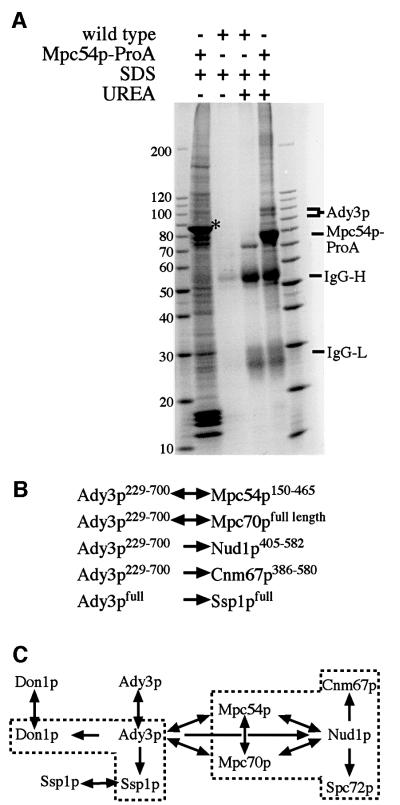
Searching for additional MP components, we purified Mpc54p from meiotic SPBs of cells expressing a fully functional Mpc54p–protein A fusion (Mpc54p–ProA; Knop and Strasser, 2000). We found one new protein co-purifying with Mpc54p–ProA (Figure 1A) which was identified as Ady3p by matrix-assisted laser desorption ionization (MALDI) mass spectrometry. Ady3p is necessary for efficient sporulation, as was shown recently in a genomics approach (Rabitsch et al., 2001). The binding of Ady3p to Mpc54p–ProA was stable in the presence of SDS and required addition of 6 M urea/dithiothreitol (DTT) to become fully resolved (Figure 1A), indicating a strong direct interaction between Ady3p and Mpc54p–ProA. We tested whether the Mpc54p–Ady3p interaction could be reproduced by the yeast two-hybrid system. Ady3p interacted specifically with Mpc54p and also with Mpc70p, the other MP component of the SPB (Knop and Strasser, 2000). In addition, Ady3p interacted with Nud1p and with Cnm67p (Figure 1B), two other components of the SPB. Taking the previously described interactions of Mpc54p and Mpc70p with Nud1p (Knop and Strasser, 2000), of Nud1p with Spc72p (Gruneberg et al., 2000) and of Nud1p with Cnm67p (Elliott et al., 1999) into account, these proteins form a network of interactions, as summarized in Figure 1C.
Fig. 1. Identification of Ady3p. (A) Sporulating cells from strains YMK363 (MPC54-ProA) and YMK333 (MPC54-GFP) were used. Mpc54p–ProA was purified from an SPB-enriched and high salt-treated fraction, using IgG affinity purification. Bound proteins were first eluted using 1% SDS (SDS) and then using urea and DTT-containing SDS buffer (UREA). The eluates were concentrated and analyzed by SDS–PAGE (5–20% gradient gel) and Coomassie Blue staining. The asterisk (*) labels the position of the band of the L-A-virus capsid protein, a common contaminant in protein isolations from sporulating cells. (B) Summary of the observed two-hybrid interactions of Ady3p with the indicated proteins. Superscript numbers indicate amino acids. Arrows (↔ and →) were used to indicate whether the two-hybrid interaction was independent of the orientation or not. Measurement of the two-hybrid interactions has been performed as described (Knop and Strasser, 2000). (C) Additional two-hybrid interactions. The boxed interactions have been published before (Elliott et al., 1999; Gruneberg et al., 2000; Knop and Strasser, 2000; Ito et al., 2000; Uetz et al., 2000). Similarly to Ady3p, Mpc54p and Mpc70p also interact with Cnm67p (not shown); however, these interactions might be mediated by Nud1p, which is naturally present in the yeast two-hybrid tester strains.
Inspection of the available information about Ady3p from genomics approaches revealed an interaction with Ssp1p (Nag et al., 1997) which was detected by two different two-hybrid approaches (Uetz et al., 2000; Ito et al., 2000; Figure 1C). Using our two-hybrid system (Schramm et al., 2000), we were able to reconfirm this interaction with full-length constructs for both proteins (Figure 1B). Deletion of both genes revealed no mitotic growth phenotypes, and no defects in progression through the meiotic divisions were seen. However, ssp1Δ cells failed to form spores (Nag et al., 1997; Rabitsch et al., 2001). For these reasons, we expanded our analysis of the function of Ady3p to include Ssp1p.
Ssp1p and Ady3p are meiosis-specific phosphoproteins
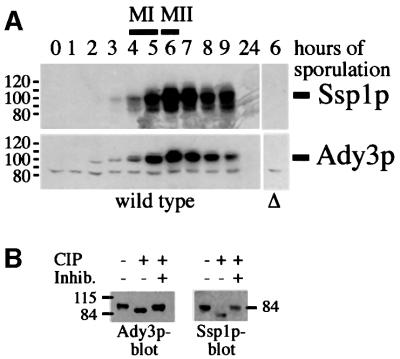
The ADY3 and SSP1 genes show similar expression profiles during sporulation, with maximal levels of the transcripts during mid to late phases of meiosis (Chu et al., 1998; Primig et al., 2000). Western blotting using specific antibodies for Ssp1p and Ady3p demonstrated that this was also true for the respective proteins (Figure 2A). Ady3p and Ssp1p were not detected in mitotic cells (Figure 2A, time point 0 h). Ady3p and Ssp1p appeared as doublets or sometimes also as diffuse bands on the western blots. This was due to phosphorylation, since treatment of the slower migrating bands of both proteins with alkaline phosphatase converted them into the faster migrating form (Figure 2B).
Fig. 2. Ady3p and Ssp1p are meiosis-specific proteins. (A) Ady3p and Ssp1p were detected in protein extracts prepared at different time points from cells of a meiotic time course (strain YKS32). Control extracts were prepared from either ady3Δ (YAM13-14) or ssp1Δ (YKS127) cells. MI and MII indicate the time points when most of the cells were in meiosis I and meiosis II, respectively, as revealed by DAPI staining. (B) Ady3p and Ssp1p are phospho-proteins. The slower migrating fractions of the proteins were isolated from crude meiotic extracts by SDS–polyacrylamide gel electrophoresis and subjected to treatment with alkaline phosphatase (CIP) with and without inhibitors as indicated. The samples were analyzed by western blotting using specific antibodies.
Localization of Ady3p and Ssp1p to the prospore membrane
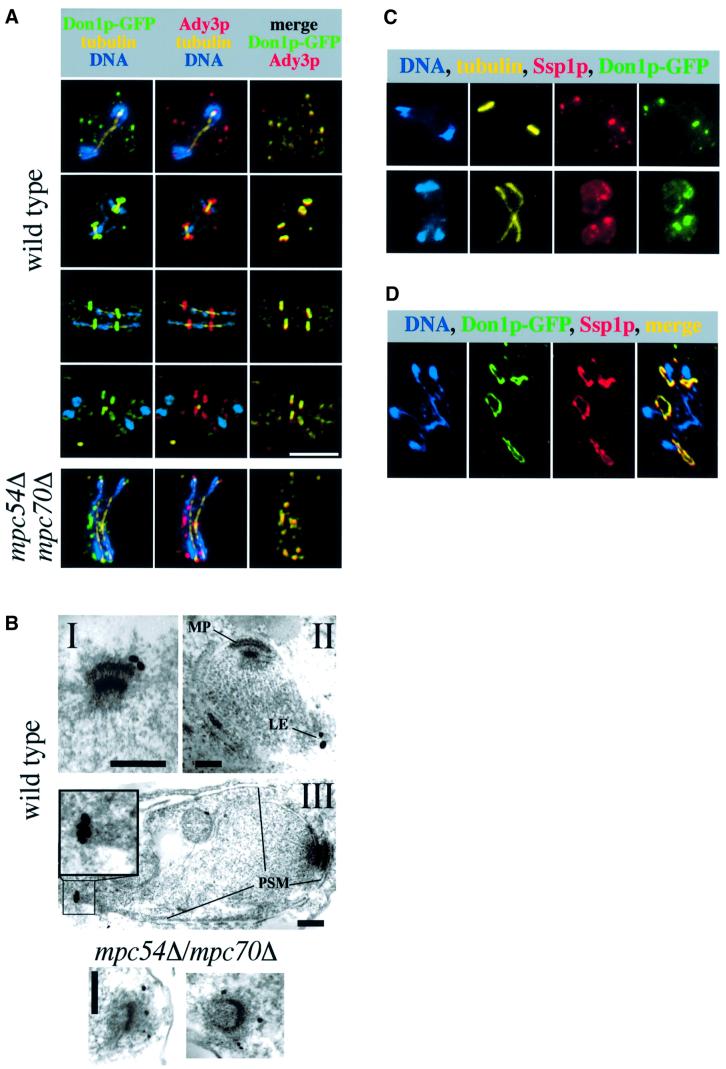
Cells from a culture midway through meiosis were prepared for immunofluorescence microscopy. Using specific antibodies, Ady3p was detected at precursors of the PSM in late phases of meiosis I and at the leading edge of the PSM during meiosis II (Figure 3A). Ady3p showed a perfect overlap with Don1p–green fluorescent protein (GFP) (Knop and Strasser, 2000), revealed by simultaneous co-detection of both proteins (Figure 3A). The localization of Ady3p to the leading edge of the PSM was confirmed using immunoelectron microscopy (Figure 3B). In a mpc54Δ mpc70Δ mutant (Knop and Strasser, 2000), Ady3p co-localized with Don1p–GFP to dots in the cytoplasm and at the SPBs (Figure 3A and B). No signal was detected in an ady3Δ strain (not shown). Taking into account that Ady3p interacts with SPB components (Figure 1), we speculated that only a fraction of Ady3p becomes localized to the leading edge of the PSM, with the rest remaining associated with the SPB during meiosis II. However, a functional N-terminally GFP-tagged Ady3p (GFP–Ady3p) no longer localized to the SPBs when the donut-like leading edge structures were visible (data not shown). Therefore, the interaction of Ady3p with the SPB is most probably of transient nature and must take place during the assembly of the MP (this issue is addressed in more detail in further experiments shown in Figures 7A and 8).
Fig. 3. Localization of Ady3p and Ssp1p. (A) Ady3p (red) and Don1p–GFP (green) were localized in cells of strains YKS53 (wild-type; DON1-GFP) and YKS65 (mpc54Δ mpc70Δ; DON1-GFP). Simultaneous detection of microtubules (yellow) and DNA (blue) allowed the estimation of the cell cycle stage of individual cells. These were (wild-type strain; from top to bottom): anaphase/telophase meiosis I; metaphase meiosis II; anaphase meiosis II; between telophase of meiosis II and post-meiotic cytokinesis. The mpc54Δ mpc70Δ cell is in anaphase of meiosis II. (B) Immunoelectron microscopic localization of Ady3p in the strains of (A). The black dots are silver-enhanced 1 nm gold particles coupled to the Fab′ fragments used to detect the anti-Ady3p antibody. MP, meiotic plaque; LE, leading edge; PSM, prospore membrane. Bar = 200 nm. (C) Localization of Ssp1p (red), Don1p–GFP (green), tubulin (yellow) and DNA (blue) in two cells of meiosis II (strain YKS53). (D) Localization of Ssp1p (red), Don1p–GFP (green) and DNA (blue) in a meiotic spread of a cell in anaphase/telophase of meiosis II. For this experiment, strain YKS53 was used.
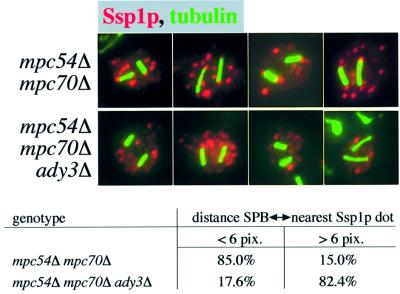
Fig. 7. Localization of Ady3p in the ssp1Δ and mpc54Δ mpc70Δ mutants. (A) Cells from sporulating cultures were processed for immunofluorescence microscopy, and Ady3p, microtubules and DNA were detected. Strains used were (from the left to the right): YKS65, YKS95 and YKS135. (B) Quantification of the Ady3p localization in cells of (A). Data obtained from ∼50 cells were evaluated for each strain. (C) Localization of Don1p–GFP depends on Ady3p. Sso1/2p, Don1p–GFP, tubulin and DNA were detected in cells of strain YKS53 (wild-type with Don1p–GFP) or YAM37-12 (ady3Δ with Don1p–GFP). One representative cell in meiosis II is shown for each strain. (D) Electron micrograph displaying a growing PSM from an ady3Δ cell (YAM37-12). A 2-fold magnification of the leading edge (LE) is shown in the inlay. For this experiment, the method of Byers and Goetsch (1975) was used with the described modifications (Knop and Strasser, 2000). (E) Localization of Ssp1p (red), tubulin (green) and DNA (blue) in an ady3Δ cell (strain YAM37-12).
Fig. 8. Ady3p mediates the interaction of Ssp1p-containing precursors with the SPB. Ssp1p was detected in sporulating cells of strains YKS65 (mpc54Δ mpc70Δ) and YAM96 (mpc54Δ mpc70Δ ady3Δ). From each strain, four individual cells are shown. Ssp1p (red) and microtubules (green) were visualized. Table: a statistical analysis of several cells per strain (n >50) was carried out using encrypted samples. Distances between the tips of the microtubules and the closest Ssp1p-containing dots were measured; 1 pixel corresponds to 0.1667 µm.
Localization of Ssp1p by immunofluorescence microscopy was performed using specific antibodies. The protein was detected at precursors of the PSM and at the PSM itself (Figure 3C). A fraction of Ssp1p co-localized with Don1p–GFP (Figure 3C) or GFP–Ady3p (not shown) at the leading edge. Detection of Ssp1p specifically at this site of the PSM seemed to be very fixation sensitive (data not shown). To obtain additional evidence for the localization of Ssp1p at the leading edge, we prepared meiotic spreads. This technique generates specimens that are devoid of soluble proteins, and also membranes become washed away. Only chromatin (and associated proteins) and large protein structures, such as microtubules, SPBs and apparently also Don1p–GFP-containing ring-like structures, most probably the LEP coats of the PSMs, remain on the slides (Figure 3D). Using this method, a clear co-localization of Ssp1p with Don1p–GFP (and with GFP–Ady3p, data not shown) was observed (Figure 3D). Taken together, Ady3p and Ssp1p both localize to precursors of the PSM and to the leading edge of the PSM. In addition, Ssp1p localizes all along the PSM.
Dyads formed in ady3Δ cells are not of ‘non-sister’ origin
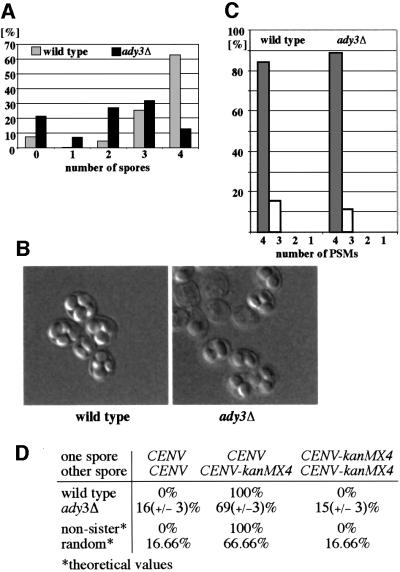
In order to investigate the function of Ady3p, we analyzed the diploid ady3Δ deletion strain for its ability to form spores. In the SK-1 genetic background, this led to the formation of asci containing less than wild-type levels of spores; mostly dyads and triads, with only few tetrads and monads observed (Rabitsch et al., 2001; Figure 4A and B). The spores were always haploid and showed wild-type viability (close to 100%, data not shown). This phenotype could be the result of an inefficient assembly of the PSMs at the SPBs. Alternatively, the closure and maturation of the PSMs during later stages of sporulation could be partially impaired. To distinguish between these possibilities, we investigated PSM formation in wild-type and ady3Δ cells using immunofluorescence microscopy and antibodies specific for the t-SNARES Sso1/2p (Neiman et al., 2000). We found a similar average number of PSMs formed per cell for both strains (Figure 4C). Therefore, Ady3p seems not to be required for PSM formation but for later steps during closure and/or maturation.
Fig. 4. ady3Δ mutants are defective in efficient maturation of the PSM. (A) Wild-type (YAM125) and ady3Δ cells (YAM127) were sporulated and the number of spores per cell was counted. (B) Normarski pictures of sporulated cells (36 h after induction of sporulation) from the above strains. (C) ady3Δ cells are not impaired in the formation of PSMs. Sporulating cells of the strains from (A) were prepared for immunofluorescence microscopy. Sso1/2p, microtubules and DNA were detected simultanously (as shown in Figure 7C). In cells of anaphase of meiosis II (cells with two elongated spindles; n = 100), the number of PSMs present was counted. (D) ady3Δ cells form dyads of random origin. Segregation of the heterozygous CENV-kanMX4 marker was investigated following ‘dyad’ dissection (see text) in the strains of (A). Each time, two independently constructed (but otherwise identical) strains were used (for ady3Δ cells, n = 2 × 100; for wild-type cells, n = 2 × 25).
Under non-optimal sporulation conditions, wild-type cells form asci with two haploid spores. Thereby, only one of the two haploid genomes per meiosis II spindle becomes encapsulated into a spore, resulting in the formation of ‘non-sister’ dyads (Davidow et al., 1980; Okamoto and Iino, 1981). Every mutation found so far that caused similar phenotypes showed defects associated with SPB function during PSM assembly (Okamoto and Iino, 1982; Bajgier et al., 2001; Deng and Saunders, 2001; Ishihara et al., 2001; Wesp et al., 2001). This prompted us to investigate whether such a preference for ‘non-sister’ dyads can be found in the ady3Δ strain, where a later step seems to be defective. We constructed wild-type and ady3Δ strains that contained centromere-linked heterozygous segregation markers (kanMX4; Wach et al., 1994) integrated between open reading frames (ORFs) YER001w and YER002w (between position 155 804 and 155 805 on chromosome V: CENV-kanMX4). After sporulation, we dissected the dyads (Davidow et al., 1980) and investigated the segregation of the marker. Statistical evaluation revealed a random segregation of CENV-kanMX4 for the ady3Δ mutant, while a ‘non-sister’ segregation was found in dyads from the wild-type strain (Figure 4D). This suggests that the preferential formation of ‘non-sister’ dyads can no longer take place when the PSMs have been formed, at least in the ady3Δ strain.
Ssp1p is required for PSM shaping
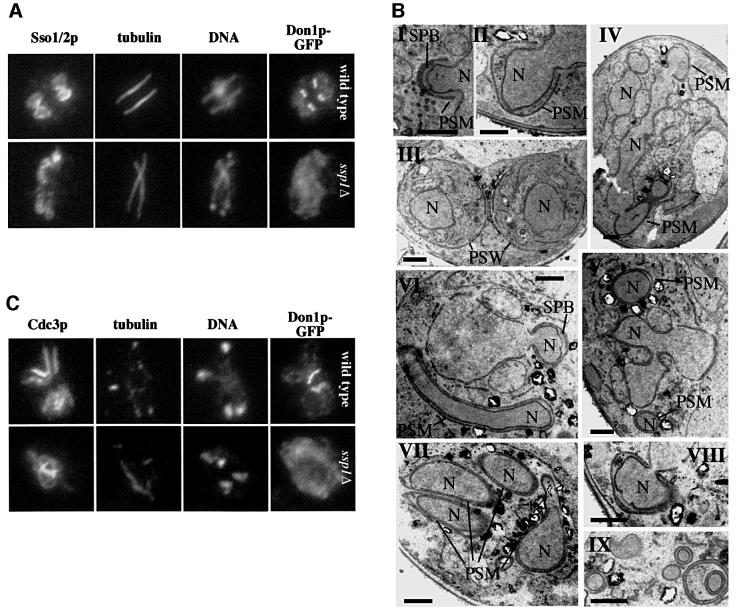
Ssp1p is essential for sporulation (Nag et al., 1997). We investigated PSM formation in the ssp1Δ mutant using immunofluorescence microscopy and the Sso1/2p antibodies. As shown in Figure 5A, ssp1Δ cells could still form PSM-like structures. Compared with PSMs in wild-type or ady3Δ cells, the membranes formed unnatural elongated ‘sausage-like’ structures. These always overlapped with the ends of the spindles, suggesting that the membranes originated from the SPBs (Figure 5A). Using electron microscopy (Figure 5B), this was confirmed [Figure 5B, I (wild-type), IV, VI and VIII (ssp1Δ)]. This investigation further revealed that in the ssp1Δ cells, the opening of the PSM at the leading edge was always very narrow. Also the PSMs stuck tightly to the surface of the nuclear envelope; no space was left between the two double membranes (Figure 5B, IV, VI and VIII). As a consequence, little or no cytoplasm was engulfed by the growing PSM in the ssp1Δ mutant [Figure 5B, compare III (wild-type) with VII (ssp1Δ)]. Tubular structures consisting of extended nuclear lobes surrounded by PSMs often could be seen (Figure 5B, IV and VI), explaining the Sso1/2p staining seen by immunofluorescence microscopy (Figure 5A). In cells with divided nuclei, small nuclear bodies surrounded by another membrane bilayer, presumably PSM-derived, were often visible (Figure 5B, V and IX). These may be caused by fragmentation of the observed tubular structures (Figure 5B, IV and VI).
Fig. 5. ssp1Δ cells show misshapen PSMs. (A) Localization of Sso1/2p, Don1p–GFP, tubulin and DNA in sporulating cultures of wild-type (YKS53) and ssp1Δ (YKS95) cells using immunofluorescence microscopy. (B) Defective PSM assembly in the ssp1Δ strain visualized by electron microscopy using KMnO4 fixation. Strains were: YKS32 (wild-type, I–III), YKS127 (ssp1Δ, IV–IX). Synchronously sporulating cultures were harvested at a time point where maximal levels of cells of meiosis II could be seen (as judged by DAPI staining and immunofluorescence microscopy, not shown) and processed for electron microscopy. Bar = 500 nm. PSW = immature spore walls. (C) Localization of Cdc3p, Don1p–GFP, tubulin and DNA in wild-type (YKS53) and ssp1Δ (YKS95) cells.
To characterize the ssp1Δ mutant further, we localized Cdc3p, a septin present in mitotic (Kim et al., 1991) and sporulating cells (Fares et al., 1996). In wild-type cells that have completed telophase of meiosis II but not yet closed the PSMs (indicated by fragmented bundles of microtubules and detectable Don1p–GFP signals at the leading edges), Cdc3p localized to parallel filaments (Figure 5C, wild-type). In the ssp1Δ cells, Cdc3p filaments were still detected but they appeared abnormally long and disorganized (Figure 5C, ssp1Δ).
Taken together, the analysis of the ssp1Δ phenotype suggests a specific role for Ssp1p in the proper shaping of domed PSMs and in the regulation of the size of the opening of the membrane.
Ssp1p is required for precursor membrane organization
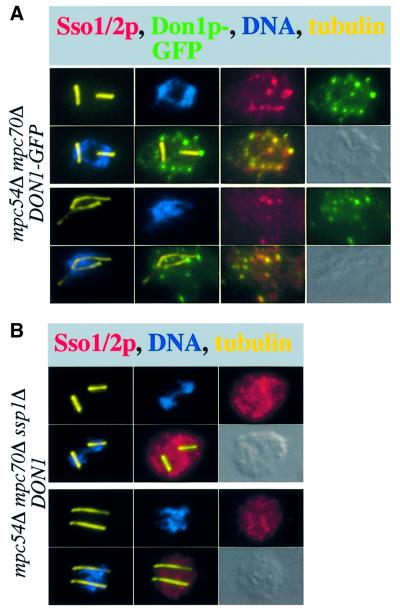
We investigated the localization of Ssp1p in the mpc54Δ mpc70Δ mutant by immunofluorescence microscopy and found Ssp1p co-localized with Don1p–GFP and GFP–Ady3p to precursors of the PSM (data not shown, and see Figure 8). In the mpc54Δ mpc70Δ cells, we could localize the t-SNARES Sso1/2p further to dots of differing brightness, either in the cytoplasm or in close proximity to the SPBs (Figure 6A). Several of the brightest Sso1/2p-containing structures co-localized with Don1p–GFP dots, indicating a co-localization of Don1p–GFP and membranes. Additional Don1p–GFP-containing dots were visible, which were not enriched for Sso1/2p. To determine if Ssp1p plays a role in the organization of these structures, we localized Sso1/2p in the mpc54Δ mpc70Δ ssp1Δ mutant. In this case, the Sso1/2p staining appeared much more disperse, and bright dots were not observed (Figure 6B). These observations suggest a further function for Ssp1p in clustering precursor membranes, presumably vesicles, into aggregates.
Fig. 6. Ssp1p is required for precursor membrane organization. (A) Localization of Sso1/2p, Don1p–GFP, tubulin and DNA in two cells in meiosis II of strain YKS65 using immunofluorescence microscopy. Various merges of the different single color pictures are shown to depict the co-localization of the different structures with each other and with the ends of the microtubules. (B) As (A) except that strain YKS135 was used. Since no GFP tag was present on Don1p in this strain, the corresponding pictures are not shown. The experiments shown in (A) and (B) were performed simultaneously under identical conditions.
Ady3p binding to the PSMs, but not to the SPBs, depends on Ssp1p
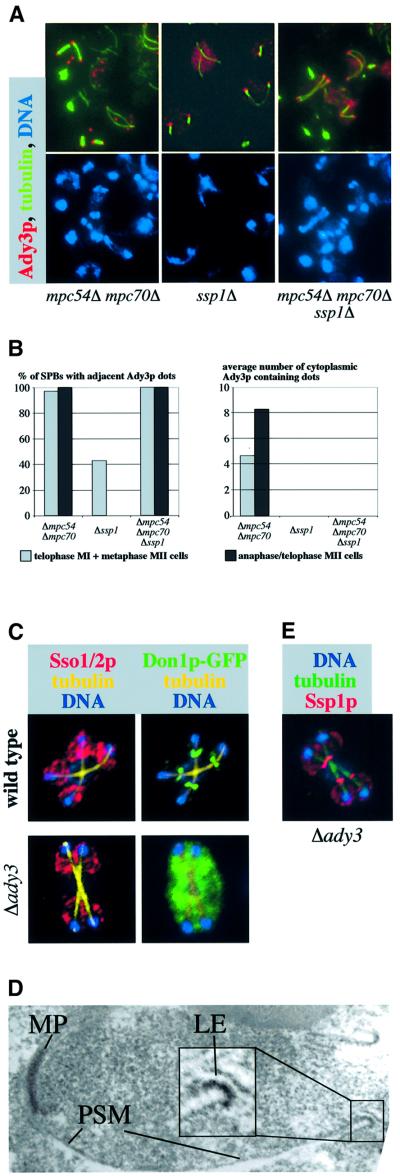
Deletion of SSP1 dramatically affected the structure of the PSMs (Figure 5A and B). To investigate whether this phenotype is associated with a defective LEP coat, we localized Don1p and Ady3p in ssp1Δ cells using immunofluorescence microscopy. As shown in Figure 5A, Don1p did not localize to the PSM like structures of ssp1Δ cells and showed a cytoplasmic staining. This was also the case for Ady3p (Figure 7A). In addition, we saw that Ady3p could be detected in these cells at 42% of the SPBs during telophase of meiosis I or metaphase of meiosis II (Figure 7A ssp1Δ; for statistical evaluation see Figure 7B). No staining at the SPBs was detected in cells from later stages of the meiotic cell cycle. A diffuse cytoplasmic staining of Ady3p was never observed in mpc54Δ mpc70Δ (Figure 7A) or wild-type cells (not shown, see also Figure 3A). This suggests an interaction of Ady3p with the SPBs during the progression from meiosis I to meiosis II. Thereafter, Ady3p becomes localized to the leading edge of the PSM in wild-type cells and to the cytoplasm in the ssp1Δ mutant. Concomitant with the disappearance of Ady3p from the SPBs, the MP assembles. Taking the observed interactions of Ady3p with Mpc54p and Mpc70p of the MP into account, this could mean that during the assembly of the MP components at the SPBs, they might interact with Ady3p. We also localized Ady3p in the mpc54Δ mpc70Δ ssp1Δ mutant. Ady3p showed persistent localization to the SPBs in this strain (Figure 7A) throughout later steps of meiosis II. Similar observations were also made when Don1p was localized in this strain.
Taken together, these data suggest that localization of Ady3p to the PSM precursors and the leading edge, but not to the SPBs, depends on Ssp1p. Furthermore, Ady3p might interact during the assembly of the MP with Mpc54p and Mpc70p.
Ady3p is required for the localization of Don1p, but not Ssp1p, to the PSM
Don1p and Ady3p always showed the same localization (Figure 3A). In their systematic analysis, Ito et al. (2000) found a two-hybrid interaction between Ady3p and Don1p. We failed to reproduce this interaction using our two-hybrid system, which relies on a plasmid system (Table I) different from that used by Ito et al. (2000). However, to address this independently, we localized Ady3p in don1Δ cells and Don1p in ady3Δ cells. In the first case, no difference in the localization of Ady3p was seen by immunofluorescence microscopy (data not shown). In the second case, however, Don1p was found exclusively in the cytoplasm (Figure 7C). This finding and the observation that Ady3p localization to the PSM depended on Ssp1p, but not vice versa (Figure 7A and E), explains the cytoplasmic localization of Don1p in the ssp1Δ cells (Figure 5A). Taken together, these findings provide evidence for the interaction of Don1p with Ady3p.
Table I. Yeast strains and plasmids.
| Name | Genotype/construction | Source or reference |
|---|---|---|
| Yeast strainsa | ||
| NKY289 | MATa ura3 lys2 ho::hisG | Alani et al. (1987) |
| NKY292 | MATα ura3 lys2 leu2::hisG ho::LYS2 | Alani et al. (1987) |
| YKS32 | diploid obtained by crossing NKY289 and NKY292 | Knop and Strasser (2000) |
| LH175 | MATa ho::hisG lys2 ura3 leu2 his3 trp1ΔFA | Linda S.Huang/Ira Herskowitz |
| LH176 | MATα ho::hisG lys2 ura3 leu2 his3 trp1ΔFA | Linda S.Huang/Ira Herskowitz |
| YMK333-2 | MATa/MATα his4-N/his4-G met4/MET4 MET13/met13 ura3/′′ leu2/′′trp1/′′lys2/′′ho::LYS2/′′ZIP1/Zip1-9Myc-klTRP1 MPC54/MPC54-eGFP-kanMX | Knop and Strasser (2000) |
| YMK363 | MATa/MATα his4-N/his4-G met4/MET4 MET13/met13 ura3/′′leu2/′′trp1/′′lys2/′′ho::LYS2/′MPC54-TEV-ProA-7HIS-kanMX4/′′ | Knop and Strasser (2000) |
| YKS53 | MATa/MATα ura3/′′LEU2/leu2::hisG lys2/′′ho::hisG/′′DON1-eGFP-kanMX4/′′ | Knop and Strasser (2000) |
| YKS65-1 | MATa/MATα LEU2/′′ho::LYS2/′′lys2/′′Δmpc70::kanMX4/′′Δmpc54::kanMX4/′′DON1-eGFP-kanMX4/′′ | Knop and Strasser (2000) |
| YKS95 | MATa/MATα DON1-eGFP-kanMX4/′′Δssp1::his3MX4/′′his3/′′LEU2/leu2 LYS2/lys2 TRP1/” ura3/′′ho::?/ho::? | this study |
| YKS127 | MATa/MATα LEU2/leu2 lys2/lys2 trp1/TRP1 Δssp1::his3MX4/′′ | this study |
| YKS135 | MATa/MATα HIS3/′′leu2/′′lys2/LYS2 trp1/′′Δssp1::his3MX4/′′Δmpc54::kanMX4/′′Δmpc70::kanMX4/′′ho::?/ho::? | this study |
| YAM13-14 | MATa/MATα lys2/′′ura3/′′LEU2/leu2::hisG Δady3::kanMX4/′′ho::LYS2/ho::hisG | this study |
| YAM37-12 | MATa/MATα lys2/′′ura3/′′LEU2/leu2::hisG Δady3::kanMX4/′′DON1-eGFP-kanMX4/′′ho::LYS2/ho::hisG | this study |
| YAM96 | MATa/MATα lys2/′′ura3/′′LEU/′′Δady3::hphMX/′′Δmpc70::kanMX4/′′Δmpc54::kanMX4/′′ΔDON1-eGFP-kanMX4/′′ho::LYS2/′′ | this study |
| YAM117 | LH176 with ady3::hphMX4 | this study |
| YAM118 | LH175 with ady3::hphMX4 | this study |
| YAM119 | YAM117 with CENV-kanMX4 | this study |
| YAM121 | LH176 with CENV-kanMX4 | this study |
| YAM125 | diploid obtained by crossing LH175 and YAM121 | this study |
| YAM127 | diploid obtained by crossing YAM118 and YAM119 | this study |
| ESM356-1 | MATa ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1GAL+ | Knop and Schiebel (1998) |
| Plasmids | ||
| pET28c(+) | E.coli expression vector | Novagen |
| pGEX-5X-1 | E.coli expression vector containing GST under control of the lacZ promotor | Pharmacia |
| pRS406 | URA3-based yeast integration vector | Sikorski and Hieter (1989) |
| pRS414 | CEN6-, TRP1- and ARS-containing yeast–E.coli shuttle vector | Sikorski and Hieter (1989) |
| pRS415 | CEN6-, LEU2- and ARS-containing yeast–E.coli shuttle vector | Sikorski and Hieter (1989) |
| pRS416 | CEN4-, URA3- and ARS-containing yeast–E.coli shuttle vector | Sikorski and Hieter (1989) |
| pRS425 | 2µm, LEU2-based yeast–E.coli shuttle vector | Sikorski and Hieter (1989) |
| pGAD424 | 2µm, LEU2-based vector carrying the GAL4 activation domain | Clontech |
| pEG202 | 2µm, HIS3-based vector carrying the lexA DNA-binding domain | Gyuris et al. (1993) |
| pAM6 | pRS416 containing ADY3 | this study |
| pAM9 | pET28c(+) containing ADY3 (codons 1–263) | this study |
| pAM10 | pGEX5X-1 containing ADY3 (codons 1–263) | this study |
| pAM34 | pRS406 containing eGFP fused to the N-terminus of ADY3 | this study |
| pAM12 | pMM5 containing ADY3 (codons 1–254) | this study |
| pAM13 | pMM6 containing ADY3 (codons 1–254) | this study |
| pAM14 | pMM5 containing ADY3 (codons 1–700) | this study |
| pAM15 | pMM6 containing ADY3 (codons 1–700) | this study |
| pAM16 | pMM5 containing ADY3 (codons 229–700) | this study |
| pAM17 | pMM6 containing ADY3 (codons 229–700) | this study |
| pKS26 | pGAD424 containing DON1 (codons 1–366) | this study |
| pKS27 | pEG202 containing DON1 (codons 1–366) | this study |
| pKS53 | pGEX5X-1 containing full-length SSP1 | this study |
| pKS54 | pET28c(+) containing full-length SSP1 | this study |
| pKS55 | pMM5 containing SSP1 | this study |
| pKS56 | pMM6 containing SSP1 | this study |
| pKS65 | pMM5 containing DON1 (codons 1–366) | this study |
| pKS66 | pMM6 containing DON1 (codons 1–366) | this study |
| pMF39 | pMM5 containing ADY3 (codons 700–791) | this study |
| pMF40 | pMM5 containing ADY3 (codons 730–791) | this study |
| pMF41 | pMM6 containing ADY3 (codons 700–791) | this study |
| pMF42 | pMM6 containing ADY3 (codons 730–791) | this study |
| pMM5 | p423-Gal1 carrying the the lexA DNA-binding domain and a Myc tag | Schramm et al. (2000) |
| pMM6 | p425-Gal1 carrying the the Gal4 activation domain and an HA tag | Schramm et al. (2000) |
aAll yeast strains listed are descendants of strain SK-1, with the exception of ESM356-1, which is a descendant of strain S288C.
To address further the consequences of the ady3Δ and ssp1Δ deletions, we attempted to compare the structure of the LEP coats from these mutants with LEP coats from wild-type cells. However, due to the difficulties associated with the reproducible visualization of the LEP coat by the available electron microscopic techniques, a reliable comparison between the strains was not possible. Nevertheless, we occasionally could visualize some sort of LEP coat in ady3Δ cells (Figure 7D), while this was not possible in the ssp1Δ cells (not shown). This is consistent with a continuous localization of Ssp1p (Figure 7E) and probably other proteins to the leading edge of the PSM in the ady3Δ mutant.
Ady3p is required for localization of Ssp1p-containing precursors of the PSM to the SPBs in cells lacking the meiotic plaque
To obtain further experimental evidence for the Ssp1p–Ady3p–SPB interaction, we analyzed the requirement for Ady3p for the localization of Ssp1p in the mpc54Δ mpc70Δ mutant that lacks the MP. In these cells, Ssp1p localized to accumulated PSM precursors in the cytoplasm and at the SPBs in meiosis II. When we additionally deleted ADY3, we found no Ssp1p-containing PSM precursors at the SPBs (Figure 8), while the cytoplasmic PSM precursors looked unchanged. Distance measurement between the tips of the spindles (which corresponds to the localization of the SPBs) and the next Ssp1p-stained dot confirmed the visual observation (table in Figure 8). This demonstrates that Ady3p can function as a physical link between Ssp1p-containing PSM precursors and the SPBs.
Discussion
It has long been recognized that cell differentiation during meiosis/sporulation in budding yeast involves a pathway of daughter cell formation completely different from that during mitotic growth (Lynn and Magee, 1970; Moens and Rapport, 1971). During the mitotic cell division, the nucleus is moved actively into the ready-made bud. This process depends on the cytoplasmic microtubule-organizing function of the SPB (for a review see Schiebel, 2000) and the regulated interactions of the microtubules with the cell cortex (Stearns, 1997; Adames and Cooper, 2000). During meiotic cell formation, nuclear movement does not rely on cytoplasmic microtubules, as suggested by the complete absence of cytoplasmic microtubules and microtubule-organizing machinery from the SPBs. Instead, the SPBs directly acquire the capability to fabricate new plasma membranes, the PSMs, discontinuous with and inside of the plasma membrane of the mother cell (Peterson et al., 1972; Davidow et al., 1980; Knop and Strasser, 2000). Subsequently, these PSMs grow at some distance around the nuclei, in order to cellularize the cytoplasm of the mother cell.
Here we describe the identification of two new proteins of the LEP coat of the PSM. We propose a model that describes how important aspects of PSM biogenesis could work.
Interaction of precursors of the PSM with the SPB
Purification of Mpc54p–ProA from meiotic SPBs identified one co-purifying protein, Ady3p. Further analysis using the yeast two-hybrid system pointed to additional interactions of Ady3p with the MP component Mpc70p and with Nud1p, a protein of mitotic and meiotic SPBs (Wigge et al., 1998; Knop and Strasser, 2000). To our surprise, Ady3p turned out to be not a primary component of the meiotic SPBs, but rather of the PSM precursors and the LEP coats (Figures 3A and B, and 9). Thus, further evidence was required to prove the interaction of Ady3p with the different SPB components. This analysis was possible since we identified Ssp1p, a two-hybrid interactor of Ady3p (Figure 1B and C), as the protein necessary for the localization of Ady3p to the PSM (Figure 7A). We were able to show that in the ssp1Δ mutant, Ady3p still bound to the SPB. This binding appeared to be transient (Figure 7A) during stages where the SPB must be in the process of acquiring the MP and where PSM assembly initiates (late stages of meiosis I, early stages of meiosis II; Knop and Strasser, 2000). In the absence of Mpc54p and Mpc70p, the interaction of Ady3p with the SPBs was stabilized and more Ady3p protein was bound to the SPBs (Figure 7A). Independent proof for an interaction of Ady3p with the SPB came from the observation that the localization of Ssp1p-containing precursors to the SPBs in the mpc54Δ mpc70Δ mutant was dependent on ADY3 (Figure 8).
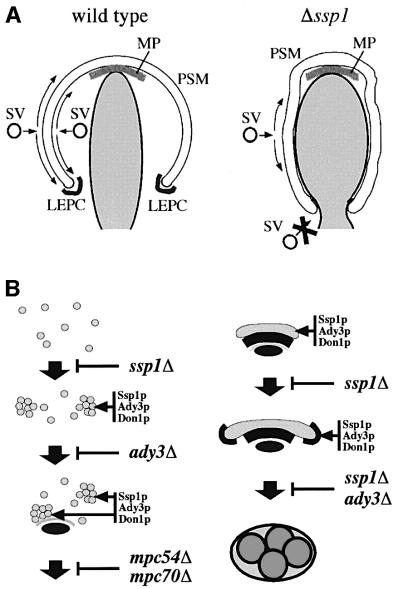
Fig. 9. Models for a possible function of the LEP coat, the formation pathway of the PSM and the function of Ssp1p, Ady3p, Mpc54p and Mpc70p within. (A) A failure in the assembly of a functional LEP coat seems to force the PSM to wrap around the nucleus. For explanation see Discussion. SV = secretory vesicle. (B) Ssp1p binds to secretory vesicles, which leads to the formation of vesicle clusters, the precursors of the PSM. Ssp1p binding causes Ady3p and Don1p to bind to the precursors. Ady3p can also bind to the SPBs, most probably via Nud1p. Subsequent assembly of the MP during meiosis II has at least three distinguishable consequences: (i) all precursor membranes become localized to the SPBs; (ii) a continuous PSM is formed; and (iii) Ady3p becomes displaced from the SPB and the LEP coat is formed at the leading edge of the membrane (further explanation in Discussion).
Taken together, these data may indicate two things. First, Ady3p seems to bind to Nud1p, which is a constituent of the outermost SPB layer (Adams and Kilmartin, 1999). Secondly, the binding of Mpc54p and Mpc70p to the SPB leads to the displacement of Ady3p from the SPB (Figure 7A). We observed only substoichiometric amounts of Ady3p bound to Mpc54p–ProA despite the high affinity of the interaction (Figure 1A) and the presence of enough Ady3p for stoichiometric binding to Mpc54p (data not shown). This again fits into a model where Ady3p interacts transiently with Mpc54p–ProA, during rearrangement processes at the SPB that lead to the assembly of the MP.
Dynamic interactions with the outer plaque of the SPB have also been described for other proteins. Spc72p, which is the anchor of the cytoplasmic microtubule-organizing machinery to the SPB (Knop and Schiebel, 1998), disappears from the outer plaque (together with all microtubules) at the end of meiosis I (Knop and Strasser, 2000). Also, during karyogamy, this interaction seems to be resolved (Pereira et al., 1999). Interestingly, Gruneberg et al. (2000) showed that Spc72p binds directly to Nud1p, which also binds to the central plaque of the SPB (Elliott et al., 1999) (Figure 1C). Nud1p is the only non-coiled-coil outer plaque SPB component and was also shown to function in the mitotic exit network (Adams and Kilmartin, 1999; Morgan, 1999; Bardin et al., 2000; Pereira et al., 2000). Based on these findings, it is tempting to speculate that Nud1p is the key adaptor molecule for the regulated binding of coiled-coil proteins to the cytoplasmic side of the SPBs, in concert with ongoing cell cycle events.
Despite the observed interactions of Ady3p with the SPBs, the ady3Δ phenotype is such that wild-type numbers of PSMs per cell are still formed (Figure 4C), indicating unaltered function of all SPBs. The defect appears later, during the maturation of the PSMs. As a consequence, reduced numbers of spores were formed (Figure 4A). To date, it is not clear whether closure of the PSMs or subsequent spore wall maturation is affected. Additionally, ineffective maturation of PSMs seems to be not surveyed by a mechanism that selects specific nuclei to become encapsulated (Figure 4D), as was proposed for mutations affecting meiotic SPB functions (Davidow et al., 1980; Bajgier et al., 2001; Ishihara et al., 2001; Wesp et al., 2001).
The function of Ssp1p and the LEP coat
Deletion of the SSP1 gene causes a dramatic disorganization of the PSM. Initial formation and curving of the membrane appeared normal. Then, the PSM seemed to grow in a manner that led to the formation of membranes with a very narrow opening and long tubular structures, which contrasted with the cup-shaped membranes with their wider opening in the wild-type (Figure 5B).
How can we explain this phenotype? The most straightforward explanation would be that in the ssp1Δ mutant, vesicle fusion with the inner membrane is abolished. In the beginning, this asymmetry could be ‘normal’ and caused by the presence of the MP at the inner side. With the lack of Ssp1p, nothing seems to counteract this asymmetric membrane flow later on, when the membrane starts to grow away from the SPBs. Consequently, the membrane would be forced to wrap around the nuclear lobes (see Figure 9A). As a further consequence, the opening of the PSM becomes very narrow, and is probably only prevented from complete closure by the nuclear envelope. This would argue for a function of Ssp1p in the assembly of a rigid proteinaceous ring that acts in determining the size of the opening of the membrane, right after initial PSM assembly at the MP. This is supported by the observations that Ssp1p is required for the localization of at least two other proteins, Don1p and Ady3p, to the leading edge of the PSM, both of which contain coiled-coil motifs which predict them to fulfil structural functions (Figures 5A, and 7A and C).
In meiosis, Ssp1p also binds to the precursors of the PSMs (Figures 3C and 8). We think that these precursors contain aggregates of unfused secretory vesicles, since deletion of SSP1 leads to a dispersed localization of a marker for such membranes, which is only possible if the structures consist of dispersible, e.g. vesicular, structures. Attempts to investigate this by immunoelectron microscopy did not give clear results, mainly because membranes are not preserved well during the procedures used/tested (method used for the experiment shown in Figure 3B, and Wright and Rine, 1989).
The assembly pathway of the PSM
The model presented in Figure 9B outlines a draft of the possible subsequent steps leading to the assembly of the PSMs at the SPBs. Binding of (most probably) secretory pathway (Golgi)-derived vesicles to the SPBs is followed by their homotypic fusion and subsequent enlargement of the nascent membrane system. With the focus on membrane fusion, the entire pathway seems to work independently of Ssp1p and Ady3p. The essential function of Ssp1p is to organize the membranes in such a way that they acquire a specific shape required for compartmentalization of the cytoplasm around the nuclei. Ssp1p seems to start working at the level of the precursor membranes, by promoting some clustering of the membranes. This can be seen best under artificial conditions where the membrane assembly is blocked by the deletion of the MP (Figure 6A and B). What function this clustering has under wild-type conditions is not clear. Ssp1p-dependent recruitment of Ady3p to the PSM also starts at the level of the precursors; the recruited Ady3p can then promote the binding of the membranes to the SPB. This function of Ady3p, however, is not very important (at least under the particular conditions used for sporulation), because wild-type levels of PSMs are formed in the ady3Δ mutant (Figure 4C). This suggests an additional mechanism by which precursors of the PSM can bind to the SPBs. One or both of the MP components could have such a function, since simultaneous deletion of both MPC genes was necessary to see the ADY3-dependent Ssp1p localization (Figure 8; data not shown). Ady3p might ensure that the initial steps of the membrane formation pathway, together with the Ssp1p-dependent membrane clustering, proceed efficiently. After the Mpc54p/Mpc70p-dependent assembly of the PSM, Ssp1p fulfills an essential function by way of organization of the LEP coat, while Ady3p functions at the leading edge to promote later steps. We speculate that Ady3p might be required for the structural integrity of the LEP coat, which could be important in promoting the faithful closure of the PSM. One could also speculate that Ady3p is required for the correct inheritance/localization of cellular components such as organelles or signaling molecules.
Materials and methods
Yeast strains, growth media and growth conditions
The genotypes of strains used in this work are listed in Table I. Basic yeast methods and growth media were as described (Guthrie and Fink, 1991). Chromosomal manipulations of yeast strains (gene deletions and C-terminal gene tagging) were performed using PCR-amplified cassettes as described (Wach et al., 1994; Knop et al., 1999). The hphMX4 cassette has been described (Goldstein and McCusker, 1999). PCR was used to verify the correct integration of the cassettes. Out-crossed strains were investigated by PCR for the presence of tags and/or deletions. Synchronous sporulation was performed using the pre-growth regimen from Alani et al. (1990). The sporulation medium used was 0.3% potassium acetate and 0.02% raffinose. 4′,6-Diamidino-2-phenylindole (DAPI) staining was performed routinely to monitor progression through meiosis.
Plasmid construction
Plasmids are listed in Table I. Construction was performed using standard procedures (Sambrook et al., 1989). The bacterial strains used were DH5α and SURE. SSP1 and ADY3 were cloned using chromosomal DNA of strain ESM356-1 with primers designed to amplify ∼350 bp of flanking sequences. Potential PCR errors were eliminated by the ‘gap repair’ technique. Two-hybrid plasmids were constructed using primers designed to amplify the desired part of an ORF.
Antibody production
Antibodies specific for the N-terminus of Ady3p were raised against a His6 fusion protein expressed from plasmid pAM9 (Table I) in strain BL21(DE3). The antiserum was affinity purified using GST–N-Ady3p (expressed from plasmid pAM10 in strain DH5α) that was coupled to BrCN–Sepharose (Pharmacia). Antibodies to Ssp1p were produced against His6-Ssp1p [expressed from plasmid pKS54-2 in strain BL21(DE3)] and purified using GST–Ssp1p (expressed from plasmid pKS53 in strain DH5α). The antibody specific for Sso1p and Sso2p (Sso1/2p) was a kind gift from Jussi Jäntti and was purified further using the blot purification method (Guthrie and Fink, 1991) and GST–Sso1p/GST–Sso2p (without transmembrane regions, both a gift of Jussi Jäntti).
Immunofluorescence, electron and immunoelectron microscopy, and mass spectrometry
Immunofluorescence microscopy was performed as described (Knop and Strasser, 2000). Primary antibodies were mouse monoclonal anti-β-tubulin Wa3, sheep anti-GFP and affinity-purified rabbit antibodies, as indicated in the figures. Secondary antibodies were donkey anti-mouse Cy5, donkey anti-sheep Cy2 and donkey anti-rabbit Cy3 (Jackson ImmunoResearch Laboratories). For image acquisition, sections throughout the cells (spacing of 0.4 µm) were collected using a Z-axis focus drive and a 100× Plan-Apo objective (Leica). Maximum projections of the image stacks are shown. For immunoelectron microscopy, the protocol of Adams and Kilmartin (1999) (2 h fixation) with the appropriate modifications (Knop and Strasser, 2000) was used. For the picture shown in Figure 5B, KMnO4 fixation electron microscopy was performed as described (Knop and Strasser, 2000). Meiotic spreads were performed using protocol 1 described by Loidl et al. (1998).
Coomassie Blue-stained protein bands were excised from one-dimensional polyacrylamide gels and in-gel digested with trypsin as described (Shevchenko et al., 1996a). Proteins were identified by MALDI peptide mass mapping on a modified REFLEX III mass spectrometer (Bruker Daltonics, Bremen, Germany) (Shevchenko et al., 1996b). Database searching was performed against a comprehensive non-redundant database using PeptideSearch v. 3.0 software developed at the EMBL Heidelberg, allowing for one miscleavage site. Eighteen peptides matched the Ady3p sequence (25% sequence coverage) with mass accuracy better than 60 p.p.m.
Two-hybrid assay
The two-hybrid tester plasmids used are listed in Table I. Cells harboring combinations of these plasmids, or different appropriate controls (empty plasmids and unrelated proteins), were assayed for two-hybrid interaction as described (Knop and Strasser, 2000). The two-hybrid plasmids for Nud1p, Spc42p, Cnm67p, Mpc54p and Mpc70p have been described (Elliott et al., 1999; Knop and Strasser, 2000; Schramm et al., 2000).
Cell lysis and immunoprecipitation
Extraction of proteins from yeast cells was done as described (Knop et al., 1999). For native protein purification, cells from sporulating cultures were incubated with phenylmethylsulfonyl fluoride (PMSF; 2 mM) for 5 min and harvested. Cell lysis, fractionation and high salt extraction prior to immunoprecipitation of Mpc54p–ProA were done essentially as described (Elliott et al., 1999). Extraction buffer was 0.5 M NaCl, 50 mM Tris–HCl pH 7.5, 10 mM EDTA, 2 mM EGTA, 1% Triton X-100. Immunoprecipitation was performed using non-specific rabbit IgGs (Dianova, Germany) covalently coupled to Dynabeads M-280 (Dynal, Norway). Bound proteins were eluted first with 1% SDS in water for 10 min at 65°C, followed by elution with 8 M urea, 5% SDS, 1 mM EDTA, 1.5% DTT, 100 mM NaxHYPO4 pH 6.8, for 10 min at 65°C.
For protein dephosphorylation, pieces from preparative SDS– polyacrylamide gels containing the higher molecular weight species of the proteins were excised. The gel cubes were then incubated first in acetonitrile (three times for 20 min) and then in dephosphorylation buffer (three times for 5 min) before homogenization. The supernatant obtained after a subsequent 300 000 g spin (30 min) was used for dephosphorylation as described (Pereira et al., 1998).
Acknowledgments
Acknowledgements
M.Matzner is acknowledged for her assistance with the electron microscope, and F.Barr for the sheep anti-GFP antibodies and critical reading of the manuscript. We are indebted to S.Jentsch for continuous support during the work, and for suggesting the abbreviation LEP coat. The Deutsche Forschungsgemeinschaft (KN498/1-1) supported part of this work. A.M.-B. is supported by the Studienstiftung des deutschen Volkes.
References
- Adames N.R. and Cooper,J.A. (2000) Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol., 149, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams I.R. and Kilmartin,J.V. (1999) Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol., 145, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Cao,L. and Kleckner,N. (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Padmore,R. and Kleckner,N. (1990) Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell, 61, 419–436. [DOI] [PubMed] [Google Scholar]
- Bajgier B.K., Malzone,M., Nickas,M. and Neiman,A.M. (2001) SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell, 12, 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J., Visintin,R. and Amon,A. (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell, 102, 21–31. [DOI] [PubMed] [Google Scholar]
- Byers B. and Goetsch,L. (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol., 124, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., Derisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- Davidow L.S., Goetsch,L. and Byers,B. (1980) Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics, 94, 581–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C. and Saunders,W. (2001) ADY1/PFS1, a novel gene required for prospore membrane formation at selected spindle poles in Saccharomyces cerevisiae. Mol. Biol. Cell, 12, 2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S., Knop,M., Schlenstedt,G. and Schiebel,E. (1999) Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl Acad. Sci. USA, 96, 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Goetsch,L. and Pringle,J.R. (1996) Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol., 132, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.L. and McCusker,J.H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast, 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Campbell,K., Simpson,C., Grindlay,J. and Schiebel,E. (2000) Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J., 19, 6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to yeast genetics and molecular biology. Methods Enzymol., 194. [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Ishihara S., Hirata,A., Minemura,M., Nogami,S. and Ohya,Y. (2001) A mutation in SPC42, which encodes a component of the spindle pole body, results in production of two-spored asci in Saccharomyces cerevisiae. Mol. Genet. Genomics, 265, 585–595. [DOI] [PubMed] [Google Scholar]
- Ito T., Tashiro,K., Muta,S., Ozawa,R., Chiba,T., Nishizawa,M., Yamamoto,K., Kuhara,S. and Sakaki,Y. (2000) Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl Acad. Sci. USA, 97, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.B., Haarer,B.K. and Pringle,J.R. (1991) Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol., 112, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. and Schiebel,S. (1998) Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J., 17, 3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. and Strasser,K. (2000) Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J., 19, 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Loidl J., Klein,F. and Engebrecht,J. (1998) Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol., 53, 257–285. [DOI] [PubMed] [Google Scholar]
- Lynn R.R. and Magee,P.T. (1970) Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol., 44, 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P.B. (1971) Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can. J. Microbiol., 17, 507–510. [DOI] [PubMed] [Google Scholar]
- Moens P.B. and Rapport,E. (1971) Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol., 50, 344–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. (1999) Regulation of the APC and the exit from mitosis. Nat. Cell Biol., 1, E47–E53. [DOI] [PubMed] [Google Scholar]
- Nag D.K., Koonce,M.P. and Axelrod,J. (1997) SSP1, a gene necessary for proper completion of meiotic divisions and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 7029–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A.M. (1998) Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol., 140, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A.M., Katz,L. and Brennwald,P.J. (2000) Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics, 155, 1643–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S. and Iino,T. (1981) Selective abortion of two nonsister nuclei in a developing ascus of the hfd-1 mutant in Saccharomyces cerevisiae. Genetics, 99, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S. and Iino,T. (1982) Genetic block of outer plaque morphogenesis at the second meiotic division in an hfd1-1 mutant of Saccharomyces cerevisiae. J. Gen. Microbiol., 128, 1309–1317. [DOI] [PubMed] [Google Scholar]
- Pereira G., Knop,M. and Schiebel,E. (1998) Spc98p directs the yeast γ-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol. Biol. Cell, 9, 775–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Grueneberg,U., Knop,M. and Schiebel,E. (1999) Interaction of the yeast γ-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J., 18, 4180–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Hofken,T., Grindlay,J., Manson,C. and Schiebel,E. (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell, 6, 1–10. [PubMed] [Google Scholar]
- Peterson J.B., Gray,R.H. and Ris,H. (1972) Meiotic spindle plaques in Saccharomyces cerevisiae. J. Cell Biol., 53, 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Williams,R.M., Winzeler,E.A., Tevzadze,G.G., Conway,A.R., Hwang,S.Y., Davis,R.W. and Esposito,R.E. (2000) The core meiotic transcriptome in budding yeasts. Nature Genet., 26, 415–423. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P. et al. (2001) A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol., 11, 1001–1009. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schiebel E. (2000) γ-Tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol., 12, 113–118. [DOI] [PubMed] [Google Scholar]
- Schramm C., Elliott,S., Shevchenko,A. and Schiebel,E. (2000) The Bbp1p–Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J., 19, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996a) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shevchenko A. et al. (1996b) Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl Acad. Sci. USA, 93, 14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. (1997) Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle. J. Cell Biol., 138, 957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wesp A., Prinz,S. and Fink,G.R. (2001) Conservative duplication of spindle poles during meiosis in Saccharomyces cerevisiae. J. Bacteriol., 183, 2372–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Jensen,O.N., Holmes,S., Souès,S., Mann,M. and Kilmartin,J.V. (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol., 141, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. and Rine,J. (1989) Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol., 31, 473–512. [DOI] [PubMed] [Google Scholar]
- Zickler D. and Olson,L.W. (1975) The synaptonemal complex and the spindle plaque during meiosis in yeast. Chromosoma, 50, 1–23. [DOI] [PubMed] [Google Scholar]