Abstract
Cysteinyl-tRNA synthetase (CysRS) is highly specific for synthesis of cysteinyl adenylate, yet does not possess the amino acid editing activity characteristic of many other tRNA synthetases. To elucidate how CysRS is able to distinguish cysteine from non-cognate amino acids, crystal structures of the Escherichia coli enzyme were determined in apo and cysteine-bound states. The structures reveal that the substrate cysteine thiolate forms a single direct interaction with a zinc ion bound at the base of the active site cleft, in a trigonal bipyramidal geometry together with four highly conserved protein side chains. Cysteine binding induces movement of the zinc ion towards substrate, as well as flipping of the conserved Trp205 indole ring to pack on the thiol side chain. The imidazole groups of five conserved histidines lie adjacent to the zinc ion, forming a unique arrangement suggestive of functional significance. Thus, amino acid discrimination without editing arises most directly from the favorable zinc–thiolate interaction, which is not possible for non-cognate substrates. Additional selectivity may be generated during the induced-fit conformational changes that help assemble the active site.
Keywords: aminoacyl-tRNA synthetase/induced fit/substrate specificity/translation/X-ray crystallography
Introduction
Aminoacyl-tRNA synthetases are a ubiquitous family of enzymes that ensure the fidelity of protein synthesis (Ibba and Soll, 2000). Each of the 20 aminoacyl-tRNA synthetases transfers its specific amino acid to the corresponding cognate tRNA in a two-step reaction. The amino acid first reacts with ATP to form an aminoacyl adenylate intermediate with release of pyrophosphate, and the activated amino acid is subsequently transferred to the 3′-end of the cognate tRNA. Each aminoacyl-tRNA synthetase is highly specific for both its cognate amino acid and tRNA, with misincorporation rates in vivo of approximately one in 104 reactions (Cusack, 1997). The enzymes must therefore distinguish between very similar and often closely isosteric amino acid substrates. Some tRNA synthetases have overcome this problem by employing editing mechanisms, in which incorrectly synthesized aminoacyl adenylate or aminoacyl-tRNAs are hydrolyzed. Among the misactivation reactions known to occur are the synthesis of valyl adenylate by isoleucyl-tRNA synthetase (IleRS; Schmidt and Schimmel, 1994), threonyl adenylate by valyl-tRNA synthetase (ValRS; Lin and Schimmel, 1996), alanyl adenylate by prolyl-tRNA synthetase (ProRS; Beuning and Musier-Forsyth, 2000) and seryl adenylate by threonyl-tRNA synthetase (ThrRS; Dock-Bregeon et al., 2000). In each case, the misactivated adenylate (or misacylated tRNA) is hydrolyzed by an editing site distinct from the site of the synthesis reaction.
Escherichia coli cysteinyl-tRNA synthetase (CysRS), the smallest monomeric tRNA synthetase in that organism (Hou et al., 1991), does not possess an editing mechanism to help discriminate against non-cognate amino acids (Fersht and Dingwall, 1979). However, comparative kinetic analysis of aminoacyl adenylate synthesis shows that the enzyme is nonetheless highly selective, activating serine, alanine and α-aminobutyrate at rates 108-, 2 × 107- and 3 × 106-fold lower, respectively, than cysteine (Fersht and Dingwall, 1979). The ability of the enzyme to discriminate the structurally similar serine is particularly noteworthy, because the two side chains are capable of similar polar interactions, yet the slightly smaller non-cognate hydroxyl group cannot be excluded from the amino acid binding pocket on steric grounds.
In the absence of a crystal structure for CysRS, the underlying mechanism for the high level of amino acid discrimination has remained unresolved. Previously, three distinct alternatives had been proposed (Fersht and Dingwall, 1979). First, the hydrogen bonding ability of the thiol group has been invoked to account for the ability of CysRS to discriminate against alanine, which lacks the potential to form hydrogen bonds with its side chain. However, this mechanism appears insufficient to explain the strong discrimination against serine. A second proposed mechanism is based on disulfide exchange, in which the thiol group of cysteine reacts with a disulfide bond in the active site to form a covalent intermediate with the enzyme. Thirdly, a thiophilic metal such as zinc might specifically ligate the cysteine substrate, to accurately position the α-carboxylate for nucleophilic attack on ATP. The thiol group of cysteine possesses a pKa of ∼8.5, and can be easily ionized in the presence of zinc.
CysRS is a member of the class I aminoacyl-tRNA synthetases, each of which possesses the Rossmann dinucleotide binding fold together with two conserved active site sequence motifs: HIGH and KMSKS. The HIGH motif, located within the first half of the Rossmann fold, helps to correctly position the adenine base of ATP and also provides interactions with the phosphates. The second lysine of the KMSKS motif, located on a flexible loop directly following the final C-terminal β-strand of the fold, has been implicated in stabilizing the transition state for the first step of aminoacylation (Fersht et al., 1988; Perona et al., 1993). The 10 members of the class I synthetases are also partitioned into three subclasses on the basis of sequence and structural homology outside of the dinucleotide binding fold (Nagle and Doolittle, 1995). CysRS is a member of subclass Ia, which also includes arginyl-tRNA synthetase (ArgRS), IleRS, leucyl-tRNA synthetase (LeuRS), methionyl-tRNA synthetase (MetRS) and ValRS. Among these, sequence analyses have suggested that CysRS most closely resembles MetRS.
A sequence alignment of CysRS sequences from 64 different species has elucidated a number of highly conserved residues outside of the HIGH and KMSKS motifs (Figure 1). These include two highly conserved cysteines at positions Cys28 and Cys209 in the first and second halves of the Rossmann fold, respectively, which are plausibly positioned near the substrate binding cleft. Additionally, five strictly conserved histidines (His206, His224, His234, His235 and His256) are located within the second half of the fold. The presence of these conserved amino acids in the active-site domain is consistent with both the proposed disulfide exchange and metal-thiolate mechanisms for amino acid selectivity.
Fig. 1. Representative alignment of CysRS sequences. The HIGH and KMSKS sequences are shown in dark blue; strictly conserved histidines and zinc-ligating cysteines are shown in red. The divergence of two archaebacterial enzymes at position 209 is shown in yellow. Other highly conserved residues are shown in light blue. Secondary structure elements are color-coded according to Figure 2. Numbering is based on the sequence of E.coli CysRS. Alignments were done with CLUSTAL_X and the figure drawn with Alscript.
To address the mechanism of substrate discrimination by CysRS in detail, we have determined the X-ray structure of the enzyme from a new tetrahedral crystal form (Newberry et al., 1999). The structure has been solved for both the unliganded enzyme at 2.3 Å resolution, as well as a cysteine-bound form at 2.6 Å resolution. The structures reveal a direct zinc–thiolate interaction that can account for the high levels of amino acid discrimination against serine. Substrate-induced conformational changes, as well as other features of the active-site cleft arising from uniquely conserved amino acids, also appear likely to play important roles in amino acid selectivity and catalysis.
Results
Two structures of CysRS, each co-crystallized with both cysteine and ATP, are reported. Because the inclusion of ligands in the cryoprotectant solution caused loss of diffraction quality, an apo structure to 2.3 Å resolution was first determined by multiple isomorphous replacement from crystals stabilized in solutions lacking ATP and cysteine. A second structure from the same co-crystals, in which the ligands were retained in the cryoprotectant, was then determined to 2.6 Å resolution by difference Fourier analysis.
Overall structure of CysRS
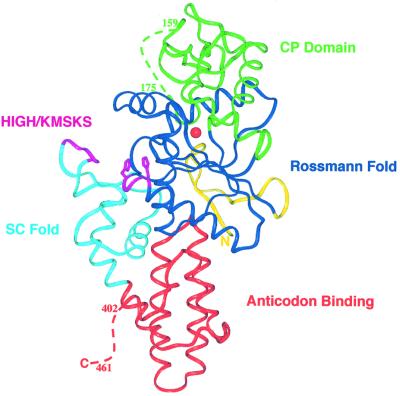
Escherichia coli CysRS, a monomeric enzyme of 461 amino acids, possesses an elongated structure consisting of four domains. As expected, the overall structural organization of CysRS is quite similar to those of the other class Ia tRNA synthetases (Figure 2) (Cavarelli et al., 1998; Nureki et al., 1998; Mechulam et al., 1999; Silvian et al., 1999; Cusack et al., 2000; Fukai et al., 2000; Sugiura et al., 2000). The four domains consist of: (i) the active site Rossmann fold; (ii) a connective polypeptide (CP) domain inserted between the two halves of the Rossmann fold; (iii) a C-terminal anticodon-binding domain; and (iv) a fourth domain connecting the Rossmann fold with the C-terminal domain (Figure 2), recently designated the stem-contact fold (SC-fold) (Sugiura et al., 2000). The model contains two monomers per asymmetric unit, which were built separately during refinement. Analysis of the structure refined without non-crystallographic symmetry constraints reveals no significant large-scale differences between the two polypeptide chains.
Fig. 2. Ribbon model of CysRS. The Rossmann fold is shown in dark blue, the connective polypeptide domain in green, the stem-contact fold in light blue and the anticodon binding domain in red. The positions of the conserved sequence motifs HIGH and KMSKS at the lip of the substrate binding cleft are shown in magenta. The solid red sphere represents the zinc ion at the base of the pocket.
The active site of CysRS is built from the canonical Rossmann fold domain seen in all other class I tRNA synthetase structures, and consists of a five-stranded parallel β-sheet surrounded by α-helices. As seen in the other structures, the Rossmann fold of CysRS is divided into two halves (residues 22–131 and 208–254) that are interrupted by the CP domain insertion (residues 132–207). The overall structure of CysRS is most similar to that of MetRS, as assessed by the similar size, topology and orientation of the C-terminal α-helical anticodon-binding domain. It was noted for MetRS that additional helices are inserted into the Rossmann fold at two positions: between the second strand and second helix within the first half of the fold, and between the first helix and first strand of the second half (these helices are designated α2 and α8, respectively, in the MetRS structure; Sugiura et al., 2000). CysRS contains the first of these inserted helices, but not the second.
All of the class I aminoacyl-tRNA synthases have a CP domain of variable size inserted between the two halves of the dinucleotide binding fold. The CP domain in CysRS is folded into a four-stranded antiparallel β-sheet motif common to all class Ia tRNA synthetase structures. This motif is also present in the class Ib tRNA synthetases, which comprise GluRS and GlnRS (Nagle and Doolittle, 1995). In CysRS, an insertion of 34 residues is found between the second and third strands of the β-sheet (residues 147–180), of which approximately half (residues 158–174) are disordered in this structure. An insertion at this position is also found in MetRS, and forms a zinc-binding subdomain in that enzyme.
The C-terminal anticodon binding domain in CysRS consists of a bundle of antiparallel α-helices, as seen in the other class Ia structures (Nureki et al., 1998; Mechulam et al., 1999; Cusack et al., 2000; Fukai et al., 2000). The overall orientation of the α-helices in the C-terminal domain with respect to the Rossmann fold is very similar in all the class Ia structures, with only the lengths of the helices and turns varying among the enzymes. CysRS contains the smallest C-terminal domain possessing only four antiparallel helices in the bundle as compared with five in IleRS, LeuRS, MetRS, ValRS and ArgRS. The 59 disordered amino acids at the C-terminus of CysRS (residues 403–461) may fold into a second domain that becomes ordered only upon tRNA binding. Such an ordering has indeed been observed for the homologous portion of IleRS, based on comparison of the unliganded and tRNA-bound structures of that enzyme (Silvian et al., 1999).
The SC-fold domain of CysRS consists of a β–α–α–β–α motif containing the KMSKS sequence, which is conserved in all class Ia and Ib tRNA synthetase structures. This motif binds at the inside corner of the L-shaped tRNA molecule, and is responsible for global positioning of the tRNA on the enzyme’s surface (Rould et al., 1989; Perona et al., 1991; Silvian et al., 1999; Delagoutte et al., 2000; Fukai et al., 2000). The loop region containing the KMSKS motif appears flexible in this structure. Although the polypeptide backbone can be clearly traced, the B-factors are high (40–50 Å2), and the lysine side chains of the motif are disordered (see Discussion).
tRNA binding
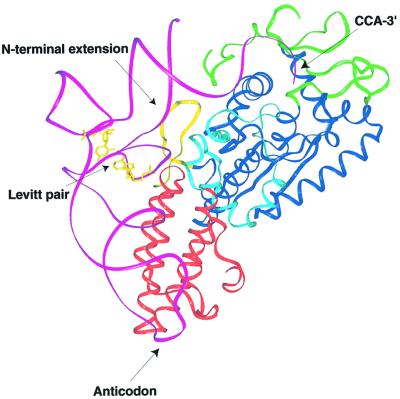
The availability of other class I synthetase-tRNA co-crystal structures allows straightforward modeling of how tRNA might bind to CysRS. Based on a superposition of the homologous Rossmann folds of CysRS and ArgRS (Delagoutte et al., 2000), it is clear that tRNACys will probably bind in a similar manner (Figure 3). It appears likely that the 3′-end of tRNACys adopts a hairpinned conformation, as observed in the GlnRS–tRNAGln and ArgRS–tRNAArg complexes (Rould et al., 1989; Delagoutte et al., 2000). Interestingly, the disordered residues of the CysRS CP domain (158–174) correspond to the region where the most important nucleotide for aminoacylation, the discriminator base U73 (Hamann and Hou, 1995; Hou et al., 2000), is predicted to interact. This suggests an induced-fit component to acceptor-end binding.
Fig. 3. Model of tRNA binding to CysRS. A superposition of equivalent backbone atoms of the Rossmann fold domains of CysRS and the ArgRS–tRNAArg complex was performed to position tRNA on the CysRS surface. Domains are colored as in Figure 2. tRNAArg is shown in magenta with the Levitt G15–C48 pair shown in yellow. The unique N-terminal β-hairpin of CysRS proposed to interact with the tRNACys tertiary core is also shown in yellow.
Also of interest is an N-terminal extension in CysRS (residues 1–21) that forms a β-hairpin structure. The hairpin is positioned adjacent to the tertiary core of the modeled tRNA (Figure 3). This is significant because E.coli tRNACys contains an unusual G15–G48 non-Watson–Crick base pair (‘Levitt pair’; Levitt, 1969) within the core region. This rare Levitt pair has been shown to be important to aminoacylation by CysRS (Hou et al., 1993; Hamann and Hou, 1997). Several basic residues from the N-terminus (K3, K10, K12) are positioned such that interactions with the core-region phosphates appear feasible. It is possible that CysRS recognizes the G15–G48 pair by virtue of its effect on the detailed conformation of the core region. There is precedence for this type of interaction with specific tRNA core region nucleotides: the recent co-crystal structure of yeast ArgRS bound to tRNAArg reveals a contact between an ArgRS-specific N-terminal domain and nucleotide 20 within the D-loop of the tRNA (Delagoutte et al., 2000).
CysRS contains one zinc ion
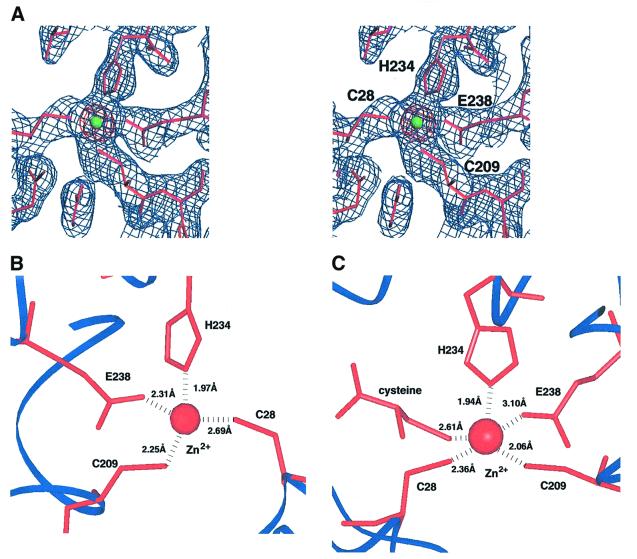
Atomic absorption analysis of CysRS samples indicates that one molecule of zinc is bound per monomer (see Materials and methods). To better localize the zinc ion within the structure, diffraction amplitudes were measured at the anomalous scattering K-edge for zinc (λ = 1.28 Å), and an anomalous difference Fourier map was calculated using these amplitudes together with solvent-corrected multiple isomorphous replacement phases. This map shows a very strong zinc peak at 41 σ above the mean (Figure 4A). The refined structure reveals that the zinc ion is co-ordinated to the side chains of Cys28, Cys209, His234 and Glu238, and is located at the very base of the substrate binding cleft (Figures 4B and 5). Although there are four ligands to the zinc, the inner-sphere ligation is not strictly tetrahedral. Instead, the bond angles are intermediate between tetrahedral and trigonal bipyramidal states. Sequence alignments of 64 canonical CysRS enzymes (Figure 1) show nearly complete conservation of the four zinc-ligating residues. The only exceptions are two sequences from the archaebacterial domain, in which Cys209 is replaced with aspartate. The zinc site in CysRS differs from those found in the other class I synthetases, in which the tetrahedral co-ordination consists of either four cysteines (IleRS, LeuRS, E.coli MetRS and ValRS), or a combination of histidine and cysteine ligands (Thermus thermophilus MetRS). More importantly, in all other structures the zinc ions are bound within the inserted CP domains and are far from the active sites.
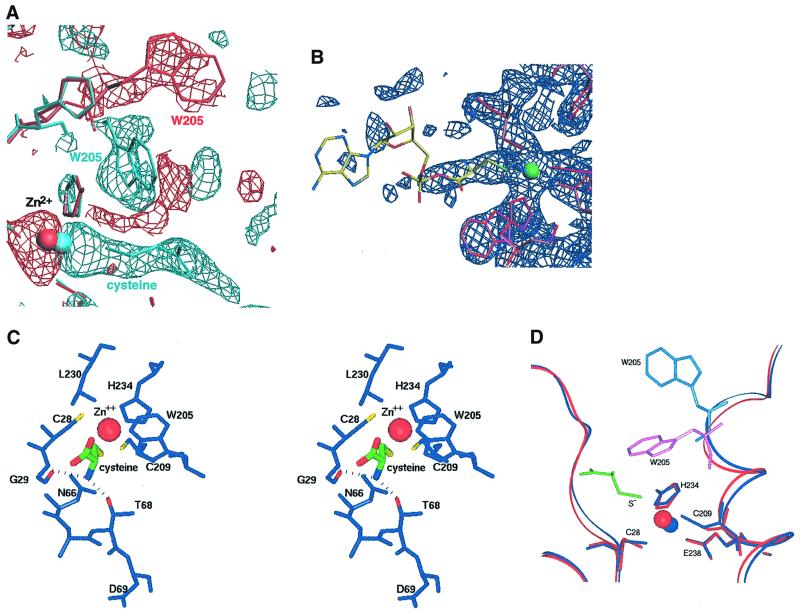
Fig. 4. Zinc binding sites in CysRS. (A) Stereo view of electron density around the zinc site in the apo structure. Shown in blue is a 2Fo – Fc map after completion of refinement, contoured at 1.3 σ. The anomalous difference map calculated from data collected at the zinc-edge, and contoured at 25 σ, is shown in red. The green sphere represents the zinc ion. (B) Detailed co-ordination sphere of the zinc ion bound in the apo CysRS structure. (C) Detailed co-ordination sphere of the zinc ion in the CysRS structure bound to the cysteine substrate. In the trigonal bipyramidal geometry, Cys28, Cys209 and His234 occupy the equatorial positions, while Glu238 and substrate cysteine occupy the apical positions.
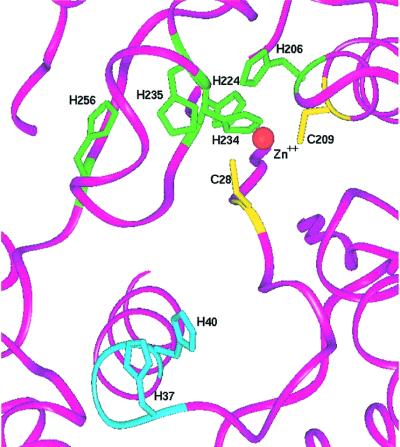
Fig. 5. Orientation of the five strictly conserved histidine residues (green) at the base of the amino acid binding cleft, with respect to the positions of the zinc ion (red sphere) and conserved cysteines (yellow). His37 and His40 from the active site HIGH motif are shown in light blue.
Five strictly conserved histidines (His206, His224, His234, His235 and His256) are positioned adjacent to each other at the base of the active site (Figure 5). Four of the imidazoles (His206, His234, His235 and His256) are arranged roughly in line with each other. The Nδ of His206 lies within hydrogen-bonding distance of His234 Nδ (3.12 Å). The Nε of His234, in turn, directly ligates the zinc. His235 and His256 are located at the opposite side of the zinc ion, but do not make direct hydrogen-bonding contacts with each other or with neighboring residues. However, the distances between the adjacent imidazoles are each 4.1 Å or less, so that small movements would suffice to permit hydrogen-bonding interactions. The fifth conserved histidine, His224, is positioned between the Cys28 and Glu238 zinc ligands, and is located 4.4 Å from the zinc ion. To our knowledge this remarkable arrangement of conserved imidazole groups is unique among known protein structures. The strict conservation suggests an important functional role, possibly in amino acid discrimination (see Discussion).
Cysteine binding site
Initial experiments aimed at optimizing the stabilization and cryoprotection solutions for the CysRS-cysteine-ATP co-crystals showed that inclusion of cysteine and ATP caused a loss of diffraction quality. For example, crystals soaked in stabilizing solution containing both ATP and cysteine for ∼12 h diffract to only 3.5 Å resolution. Because of this, we first determined the structure of the enzyme from crystals in which both ligands were omitted from stabilization/cryoprotectant solutions. To determine the co-crystal structure, experiments were conducted to progressively shorten the stabilization/cryoprotection time while retaining the ligands in the solutions. We found that crystals stabilized and cryoprotected over a 10-min time period in the presence of cysteine and ATP retained diffraction to 2.6 Å resolution, allowing determination of a ligand-bound complex.
Maps calculated with coefficients [Fo(lig) – Fo(unlig)] show a clear electron density peak at the base of the binding cleft, which can be well-fit with cysteine (Figure 6A). However, the density for the AMP moiety is poorly defined, even following refinement (Figure 6B). One possible explanation of this observation is that ATP does not bind in this crystal form, so that the electron density in fact represents free cysteine. This interpretation is supported by the finding that ATP is dispensable for crystal growth. Alternatively, it is conceivable that the electron density may correspond to the cysteinyl portion of the aminoacyl adenylate, with the AMP moiety disordered in this lattice environment. Although we have fit and refined free cysteine in this structure, the electron density at 2.6 Å resolution is not sufficiently well-defined to distinguish the α-carboxylate group from a partially disordered mixed anhydride linkage, which would occupy that position in the activated intermediate. The average B-factor for the cysteine is 47 Å2, somewhat higher than the overall Wilson B-factor of 35 Å2 for the ligand-bound structure.
Fig. 6. Structural changes upon cysteine binding. (A) [Fobs(cysteine-bound) – Fobs(apo)] electron density map contoured at –2.5 σ (red) and +2.5 σ (light blue), showing the movements of Trp205 and of zinc upon cysteine binding. (B) 2Fo – Fc electron density map contoured at 1.5 σ, after refinement against data from crystals soaked with cysteine and ATP. No density is observed for the AMP portion of cysteinyl adenylate, suggesting that the structure may represent free cysteine (see text). (C) Stereo view of the cysteine binding site. The cysteine substrate (center; thick bonds) is shown with atoms colored green (carbon), red (oxygen), blue (nitrogen) and yellow (sulfur). Dotted lines indicate hydrogen-bonding interactions made by the α-NH3+ group. (D) Superposition of the unliganded (dark blue) and cysteine-bound (red) structures in the active site region, showing the movements of Trp205 and zinc upon amino acid binding. The cysteine substrate is depicted in green.
Cysteine binds at the base of the active site cleft in a position similar to the amino acid binding sites of other class I tRNA synthetases, and makes interactions with amino acids in both the first and second halves of the Rossmann fold (Figures 4C and 6). The α-NH3+ group donates hydrogen bonds to both the main chain amide of Gly29 (adjacent to the Zn2+-binding Cys28 thiolate) and the side chain hydroxyl group of Thr68 (Figure 6C). Interestingly, the adjacent conserved Asp69 carboxylate group does not interact with substrate but instead points away from the binding site. While an equivalently positioned aspartate residue does bind the amino acid α-NH3+ group in many class I synthetases (Brick et al., 1989; Rath et al., 1998; Cusack et al., 2000; Serre et al., 2001), this contact is not made in all the enzymes. For example, the ArgRS/arginine and TrpRS/tryptophanyl adenylate structures each show that the equivalent aspartates do not directly bind amino acid (Doublie et al., 1995; Cavarelli et al., 1998).
Superposition analysis shows that binding of cysteine does not cause global rearrangements of the protein structure [root mean square deviation (r.m.s.d.) in position of equivalent Cα atoms = 0.29 Å]. However, inspection of the Flig – Fapo difference maps does reveal several peak/hole combinations that would be indicative of atomic movements. One of these differences corresponds to repositioning of the zinc ion toward the cysteine substrate by 0.8 Å. In the new position the zinc ion is co-ordinated by the thiol of the cysteine substrate, as well as by the side chains of the original ligands Cys28, Cys209, His234 and Glu238 (Figure 4C). The distance between the Glu238 carboxylate and the zinc ion increases from 2.27 to 3.09 Å, suggesting that this ligand may bind more weakly. However, the inner-sphere ligation of the zinc with cysteine bound corresponds to a nearly ideal trigonal bipyramidal geometry. Thus, the protein structural environment in this region appears optimally constructed for precise presentation of the zinc ion to the substrate thiolate group.
A second significant change in the active site upon substrate binding is the reorientation of the Trp205 indole side chain, which is oriented in the direction of the CP insertion domain in the unliganded structure. Upon cysteine binding, the indole flips down toward the active site, rotating nearly 180° around the Cα–Cβ bond from its original position (Figure 6A and D). This movement results in stacking of the substrate cysteine side chain between Leu230 and Trp205 (Figure 6C), and is thus necessary for complete formation of the amino acid binding site. Each of the amino acids in the immediate vicinity of the substrate is very highly conserved among the 64 known CysRS sequences (Figure 1).
Discussion
Zinc-ion mediated amino acid discrimination
The crystal structures of unliganded and cysteine-bound CysRS reveal a direct cysteine–zinc ion interaction, which appears to provide a major component of the selectivity against non-cognate amino acids. The zinc–thiolate interaction was indeed among the three possibilities originally suggested to provide discrimination (Fersht and Dingwall, 1979). Furthermore, a second suggested model invoking disulfide exchange appears unlikely in view of this structure, since the two active site Cys28 and Cys209 sulfhydryls interact directly with zinc rather than forming a disulfide bond with each other. Suggestive evidence against the disulfide mechanism also arises from sequence alignments of the canonical CysRS enzymes, as two archaebacterial enzymes exist in which Cys209 is replaced by aspartate (Figure 1). Since the Asp carboxylate can ligate zinc, while the disulfide mechanism requires two enzyme cysteines, only the direct zinc–thiolate mechanism can be universally conserved for all canonical CysRS enzymes.
Theoretical studies have shown markedly lower zinc binding affinity for the serine hydroxyl group (which is unlikely to ionize at physiological pH) versus the negatively charged cysteine thiolate (Rulisek and Havlas, 2000). These theoretical models also support what is seen in nature: to our knowledge there is currently only one example in the literature of a zinc enzyme (alkaline phosphatase) that uses serine as a zinc ligand (Kim and Wyckoff, 1991). It therefore seems reasonable that a significant part of the discrimination by CysRS against serine lies in the inherent much greater affinity of zinc for a cysteine thiolate compared with a serine hydroxyl.
ThrRS, a class II enzyme possessing a different active site domain, is the only other synthetase that also uses a zinc ion to help discriminate among amino acids (Sankaranarayanan et al., 2000). In both ThrRS and CysRS, the zinc is co-ordinated to four inner-sphere ligands in the unbound structure and to five when amino acid is bound. However, in ThrRS the threonine substrate is ligated to zinc via both its side chain hydroxyl and α-amino groups, and a water molecule present in the inner sphere is displaced upon threonine binding. The other three ThrRS zinc ligands comprise two histidines and a cysteine (Sankaranarayanan et al., 2000). Conversely, the zinc ion in CysRS ligates the cysteine substrate only through its side chain thiolate, while the enzyme contributes the same four ligands in both bound and unbound states (Figure 4). Interestingly, ThrRS is also a less discriminating enzyme than CysRS; it has been shown to misacylate serine and thus requires a second editing step to hydrolyze Ser-tRNAThr (Dock-Bregeon et al., 2000). Thus, even though ThrRS makes a specific zinc ion-mediated interaction with the threonine hydroxyl, it still cannot efficiently exclude serine, which is able to ligate zinc in a precisely analogous manner. In contrast, CysRS is able to utilize the discriminatory properties of zinc to directly reject serine without the need for editing, by virtue of the distinct strong interaction that can only be formed with the negatively charged thiolate (Rulisek and Havlas, 2000).
Induced-fit contributions to catalysis and amino acid selectivity
Analysis of the CysRS co-ordinates and comparison of the structures with those of other class I tRNA synthetases suggests that the enzyme is not poised for catalysis in the visualized conformation. This is suggested most clearly based on the position of the enzyme loop containing the KMSKS motif sequence. Structures of other class I tRNA synthetases have shown that binding of either ATP or aminoacyl adenylate analogs are required to position the loop in a ‘closed’ conformation in which the second lysine is positioned to stabilize the α-phosphate in the first transition state (Rould et al., 1989; Perona et al., 1993; Doublie et al., 1995; Cusack et al., 2000; Fukai et al., 2000). In contrast, unliganded synthetases or those bound to amino acid alone reveal the KMSKS loop in an ‘open’ conformation more distant from the ATP binding site (Brick et al., 1989; Nureki et al., 1995, 1998; Cavarelli et al., 1998; Mechulam et al., 1999; Cusack et al., 2000; Delagoutte et al., 2000; Sugiura et al., 2000). The loop conformation in both CysRS structures reported here is in the open state, with both lysine side chains of the KMSKS disordered, consistent with the binding of cysteine but not cysteinyl adenylate or ATP in this crystal form. Therefore, a further conformational change involving loop closure, and accompanying ATP binding, is expected prior to catalysis. Structures of LeuRS and TrpRS in the unliganded and ATP-bound states provide characterized examples in which structures of the loop before and after movement are known (Doublie et al., 1995; Cusack et al., 2000; Ilyin et al., 2000), and the loop motion has also been inferred from kinetic measurements on TyrRS (Fersht et al., 1988).
In addition to the likely requirement for further ordering of the KMSKS loop, we also emphasize that the structure of apo CysRS was determined by soaking substrates out of preformed crystals of the ligand-bound enzyme. Therefore, it is possible that some early conformational changes involved in cysteine binding have also been blocked by lattice forces in this crystalline environment. Clearly, additional structures of the enzyme in different lattices will be required for a full description of the catalytic pathway.
Despite these limitations, the two structures presented here do reveal two significant movements upon cysteine binding: translocation of the zinc ion by 0.8 Å towards substrate, and reorientation of the Trp205 indole ring. Each of these motions probably provides an indication of the larger-scale reorientations required to fully assemble the active site for cysteinyl-adenylate and cysteinyl-tRNA formation, and perhaps to discriminate against non-cognate amino acids. One possibility is suggested by the unusual arrangement of the strictly conserved His206, His224, His234, His235 and His256 residues around the zinc site at the base of the pocket. Given the observed move ment of zinc by 0.8 Å when cysteine binds, we speculate that alternative binding sites for the zinc ion may exist at different stages of the reaction, and possibly in the presence of non-cognate amino acid (or tRNA) binding. Modeling shows that movement of the zinc by ∼2.5 Å in the direction of His224 would allow formation of a second site with the ligands Cys28, His224, His234, His235 and Glu238. This movement is unencumbered by any intervening protein groups. A second zinc site would provide a direct role for three of the five conserved histidines, while His206 and His256 might be required for specific position ing of His234 and His235, respectively (Figure 5). One rationale for such a ‘zinc shuttle’ could be to help disrupt the strong zinc–thiolate interaction after the reaction, facilitating product release while retaining zinc within the enzyme for the next catalytic cycle. Alternatively, a second zinc site might help facilitate structural rearrangements leading to non-productive binding of non-cognate amino acids. This would provide a means for discrimination at the catalytic step in addition to the binding step of the activation reaction.
The Trp205 residue that rotates to clamp onto the cysteine side chain is strictly conserved in all 64 known CysRS sequences (Figures 1 and 6). In the cysteine-bound structure, the indole nitrogen is positioned 3.45 Å from the cysteine sulfur, suggesting a potential hydrogen-bonding interaction between the indole proton and cysteine thiolate at some stage during the catalytic cycle. In contrast to the negatively charged cysteine thiolate, a non-cognate serine hydroxyl group bound to zinc would presumably retain its proton, suggesting the potential for a steric clash or unfavorable electrostatic contact that might help to weaken the serine binding affinity. Thus, Trp205 may be involved in both discrimination against serine as well as in helping (along with zinc) to properly juxtapose and immobilize the amino acid α-carboxylate with respect to the α-phosphate of ATP.
Interestingly, Trp205 is conserved as an aromatic residue in all class I tRNA synthetases (Brick et al., 1989; Rould et al., 1989; Doublie et al., 1995; Nureki et al., 1995, 1998; Cavarelli et al., 1998; Mechulam et al., 1999; Cusack et al., 2000; Fukai et al., 2000; Sugiura et al., 2000). In each of the class I structures with amino acid or adenylate bound, the equivalent residue is oriented toward the R-group of the amino acid. Furthermore, the equivalent residues in both GlnRS (Tyr211) and ArgRS (Tyr347) are involved in a stacking interaction with the terminal A76 adenine base of the tRNA (Rould et al., 1989; Delagoutte et al., 2000). Thus, it appears that Trp205 may play an important role in facilitating required rearrangements of the active site for both steps of the aminoacylation reaction.
The highly differentiated amino acid specificity pocket in canonical CysRS enzymes raises the question of how the recently discovered non-canonical CysRS might recognize cysteine. The genomes of some archaebacteria lack a recognizable CysRS, and it appears that cysteinyl-tRNACys is derived instead from a dual-functioning enzyme capable of synthesizing both cysteinyl-tRNACys and prolyl-tRNAPro (Lipman et al., 2000; Stathopoulos et al., 2000, 2001). It is of interest to consider the sequences of these unusual enzymes in light of the E.coli CysRS structure reported here. The CysRS binding pocket is unique among class I tRNA synthetases, featuring a well co-ordinated zinc ion together with at least nine very highly conserved and potentially important amino acids (Cys28, Cys209, His206, His224, His234, His235, His256, Glu238 and Trp205) directly in or adjacent to the cysteine binding site. In contrast, the ProCys tRNA synthetases notably lack any conserved residues necessary for either the metal-binding or disulfide exchange specificity mechanisms, and their sequences closely align with those of canonical class II ProRS enzymes. The extreme conservation of the E.coli CysRS binding cleft implies functional importance, highlighting the disparity between the two types of CysRS enzymes and further underlining the possibility that the archael ProCys synthetases may reflect features of an ancient tRNA synthetase-like enzyme with broader specificity and a consequently much less well differentiated substrate binding cleft (Ribas de Pouplana et al., 2001).
Materials and methods
Protein expression, purification and crystallization
Escherichia coli CysRS was overexpressed and purified as described previously (Newberry et al., 1999). Intensity statistics from a previously published trigonal crystal form (Newberry et al., 1999) showed signs of merohedral twinning, as detected by the method of Yeates (1997). Analysis of the intensity distributions showed that the crystals are merohedrally twinned with a twinning factor (α) of 0.457. Data from crystals with a twinning factor at or near 0.5 cannot be successfully detwinned (Yeates, 1997); thus, a new crystal form was required. Crystallization experiments were performed at 17°C using the hanging-drop vapor diffusion method. New crystals were obtained by mixing 1 µl of protein solution [6 mg/ml CysRS, 10 mM HEPES pH 7.4, 50 mM NaCl, 1 mM dithiothreitol (DTT), 5 mM MgCl2, 5 mM ATP, 10 mM cysteine] with 1 µl of reservoir solution (15–17% PEG-8000, 100 mM sodium cacodylate pH 6.5, 200 mM magnesium acetate, 2% t-butanol). Tetragonal bipyramids appeared in 1 week and reached maximal dimensions (0.2 × 0.2 × 0.05 mm) in ∼3 weeks. Cysteine, but not ATP, is required for crystal growth. Heavy atom derivatives were prepared by soaking crystals in a stabilizing solution containing 25% PEG-8000, 100 mM sodium cacodylate pH 6.5, 200 mM magnesium acetate and 0.1–1 mM heavy atom compound (see Table I).
Table I. X-ray data collection and refinement statistics.
| Native | Zinc-edge | EMTSa | K2HgI4 | PIPa | Cysteinea | |
|---|---|---|---|---|---|---|
| Resolution (Å) | 2.3 | 2.3 | 2.7 | 2.6 | 3.0 | 2.6 |
| Wavelength (Å) | 1.03 | 1.28 | 1.08 | 1.08 | 1.08 | 0.978 |
| Rmergeb (%) | 6.9 | 8.0 | 8.9 | 9.6 | 9.5 | 10.0 |
| I/σ | 16.4 | 31.2 | 14.2 | 13.5 | 14.3 | 17.2 |
| Phasing power (FH/E) | 0.82 | 0.78 | 0.50 | |||
| Overall figure of merit (FOM) | 0.43 | |||||
| FOM after solvent flattening | 0.69 | |||||
| Refinement | ||||||
| Rcrystc (%) | 24.5 | 24.9 | ||||
| Rfreec (%) | 28.3 | 28.9 | ||||
| R.m.s.d. bonds (Å) | 0.010 | 0.009 | ||||
| R.m.s.d. angles (°) | 2.01 | 1.97 |
aEMTS, ethylmercurithiosalicylate; PIP, di-µ-iodobis(ethylenediamine) diplatinum (II) nitrate; cysteine is the dataset for which cysteine and ATP ligands were included in stabilizing and cryoprotecting solutions.
bRmerge = ∑hkl∑i(|Ihkl – <Ihkl>|)/∑hkl,i<Ihkl>, where Ihkl is the intensity of an individual measurement of the reflection with indices hkl and <Ihkl> is the mean intensity of that reflection.
cR-factor = ∑hkl|Fobs – Fcalc|/∑hklFobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rcryst and Rfree were calculated using the working and test set, respectively.
Determination of zinc content
Enzyme zinc content was determined by atomic absorption spectroscopy at the Marine Science Institute Laboratory (UC Santa Barbara). Enzyme samples were diluted to 10 mg/100 ml in 20 mM Tris–HCl pH 7.5, 0.2 mM DTT prior to analysis, and replicate readings were taken to obtain an average. The recombinant CysRS enzyme yielded a value of 0.89 ± 0.01 mol of zinc/mol of enzyme. The human carbonic anhydrase, which contains 1 mol of Zn/mol of enzyme, was used as a positive control and yielded a value of 0.98 ± 0.01 mol of zinc/mol of enzyme. The standard for zinc analysis was prepared from zinc oxide (Fisher certified atomic absorption standard, P/N SO-Z-13, 1000 p.p.m.) dissolved in diluted nitric acid.
X-ray data collection and structure determination
Experiments performed to optimize stabilization and cryoprotection conditions for the CysRS-cysteine-ATP co-crystals showed that inclusion of cysteine and/or ATP in these solutions resulted in loss of diffraction quality (see Results). Thus, crystals were first soaked in a stabilizing/cryoprotectant solution containing 25% PEG-8000, 100 mM sodium cacodylate pH 6.5, 200 mM magnesium acetate and 20% (v/v) glycerol, and flash-frozen at 100°K. These solutions omit both substrates. Diffraction amplitudes from these apo crystals were measured to 2.3 Å resolution at BNL beamline X12C, and the data were processed with MOSFLM (Leslie, 1999) and SCALA (CCP4, 1994).
X-ray diffraction amplitudes were measured at SSRL beamlines 9-2 and 7-1, and NSLS beamline X12C. These crystals belong to the tetragonal space group p41212 with unit cell dimensions a = b = 119.2 Å, c=144.3 Å, and contain two monomers per asymmetric unit (Table I). Heavy atom sites were found and initial MIR phases determined by SOLVE (Terwilliger and Berendzen, 1999) to 2.8 Å. Soaking conditions for the three derivatives (Table I) were as follows: EMTS, 1 mM, 54 h soak; K2HgI4, 0.1 mM, 9 h soak; PIP, 1 mM, 5 h soak.
Initial phases for the apo structure were improved by reciprocal-space solvent flattening using RESOLVE (Terwilliger, 1999). Two-fold NCS averaging was attempted using DM (CCP4, 1994), but did not significantly improve the resulting maps and so was not used. The model was built manually using the program O (Jones et al., 1991) and the structure was refined using CNS (Brünger et al., 1998). The refinement included several rounds of simulated annealing refinement, using a starting temperature of 5000°K, as well as individual B-factor refinement. A random 10% of the starting data was set aside for cross validation (Rfree calculation). After the majority of the model was built, the model was refined against native data to 2.3 Å and water molecules were picked automatically as implemented in CNS. A 4 σ cut-off for picking waters in the difference electron density maps was employed. The refinement converged to a final R-factor of 24.5% with an Rfree of 28.3% (20–2.3 Å resolution range). The refined model comprises two monomers with 386 residues per monomer and 102 water molecules. Electron density was not defined for 59 residues at the C-terminus or for 17 residues in the CP domain of each monomer. Electrospray mass spectroscopy analysis of CysRS used for crystallization showed that the last 59 residues of CysRS had not been proteolyzed. The quality of the refined structure was analyzed with PROCHECK (CCP4, 1994), which showed that 98.5% of the model lies within the core or allowed regions of the Ramachandran plot. The relatively high Rcryst is probably a consequence of the significant portion of the model which could not be built.
To determine the structure of the enzyme–substrate complex, experiments were carried out to determine the conditions in which cysteine and ATP could be tolerated in the stabilization/cryoprotectant solutions. In these experiments, crystals were first stabilized by soaking in a solution containing 25% PEG-8000, 100 mM sodium cacodylate pH 6.5, 200 mM magnesium acetate, 5 mM ATP, 10 mM cysteine, for 10 min, followed by transfer to a cryoprotectant consisting of the stabilizer solution with addition of 20% (v/v) glycerol. In contrast to initial experiments in which the stabilization was performed over a 12 h time period, we found that the 10-min stabilization period allowed retention of diffraction to 2.6 Å resolution. Data collected from crystals prepared in this manner were used to determine the ligand-bound structure by difference Fourier analysis. Difference electron density maps with coefficients (Flig – Fapo) and phased with the 2.3 Å model were calculated using CNS. The data were refined against the model of the apo structure, and the cysteine substrate was built into the model and refined using CNS (Brünger et al., 1998). The refinement for the final model with 386 residues, 73 water molecules and cysteine converged to a final R-factor of 24.9% with an Rfree of 28.9% (20–2.6 Å resolution range) (Table I). Structure superpositions were performed with the program LSQMAN (Kleywegt and Jones, 1994).
Data collected at zinc K-edge
In order to clearly localize the zinc ion within the asymmetric unit, a dataset was collected at the anomalous absorption K-edge for zinc (1.28 Å). Crystals were cryoprotected and frozen as described above for the apo crystal. Diffraction amplitudes were measured at APS beamline 17-ID and data were processed as previously described for native data. An anomalous difference map was computed with coefficients |F+| – |F–| using the MIR, solvent-flattened phase information. The anomalous difference map revealed two peaks at 41 σ corresponding to the zinc ion in each monomer.
Accession codes
Coordinates have been submitted to the Protein Data Bank and assigned codes 1LI5 (apoenzyme) and 1LI7 (cysteine bound).
Acknowledgments
Acknowledgements
We thank Nancy Horton for assistance in data collection and computations. This work was supported by NSF grant MCB-9600971 and NIH grant GM-63713 (to J.J.P.), and by NIH grant GM-56662 (to Y.M.H.). X-ray data collection was carried out in part at Brookhaven National Laboratories and the Advanced Photon Source at Argonne National Laboratory. Portions of this research were also performed at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the National Institute of General Medical Sciences.
References
- Beuning P.J. and Musier-Forsyth,K. (2000) Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 97, 8916–8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick P., Bhat,T.N. and Blow,D.M. (1989) Structure of tyrosyl-tRNA synthetase refined at 2.3 Å resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J. Mol. Biol., 208, 83–98. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Delagoutte,B., Eriani,G., Gangloff,J. and Moras,D. (1998) l-arginine recognition by yeast arginyl-tRNA synthetase. EMBO J., 17, 5438–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cusack S. (1997) Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol., 7, 881–889. [DOI] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J., 19, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delagoutte B., Moras,D. and Cavarelli,J. (2000) tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J., 19, 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock-Bregeon A., Sankaranarayanan,R., Romby,P., Caillet,J., Springer, M., Rees,B., Francklyn,C.S., Ehresmann,C. and Moras,D. (2000) Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell, 103, 877–884. [DOI] [PubMed] [Google Scholar]
- Doublie S., Bricogne,G., Gilmore,C. and Carter,C.W.,Jr (1995) Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure, 3, 17–31. [DOI] [PubMed] [Google Scholar]
- Fersht A.R. and Dingwall,C. (1979) Cysteinyl-tRNA synthetase from Escherichia coli does not need an editing mechanism to reject serine and alanine. High binding energy of small groups in specific molecular interactions. Biochemistry, 18, 1245–1249. [DOI] [PubMed] [Google Scholar]
- Fersht A.R., Knill-Jones,J.W., Bedouelle,H. and Winter,G. (1988) Reconstruction by site-directed mutagenesis of the transition state for the activation of tyrosine by the tyrosyl-tRNA synthetase: a mobile loop envelopes the transition state in an induced-fit mechanism. Biochemistry, 27, 1581–1587. [DOI] [PubMed] [Google Scholar]
- Fukai S., Nureki,O., Sekine,S., Shimada,A., Tao,J., Vassylyev,D.G. and Yokoyama,S. (2000) Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell, 103, 793–803. [DOI] [PubMed] [Google Scholar]
- Hamann C.S. and Hou,Y.M. (1995) Enzymatic aminoacylation of tRNA acceptor stem helices with cysteine is dependent on a single nucleotide. Biochemistry, 34, 6527–6532. [DOI] [PubMed] [Google Scholar]
- Hamann C.S. and Hou,Y.M. (1997) A strategy of tRNA recognition that includes determinants of RNA structure. Bioorg. Med. Chem., 5, 1011–1019. [DOI] [PubMed] [Google Scholar]
- Hou Y.M., Shiba,K., Mottes,C. and Schimmel,P. (1991) Sequence determination and modeling of structural motifs for the smallest monomeric aminoacyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 88, 976–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.M., Westhof,E. and Giege,R. (1993) An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl Acad. Sci. USA, 90, 6776–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.M., Sundaram,M., Zhang,X., Holland,J.A. and Davis,D.R. (2000) Recognition of functional groups in an RNA helix by a class I tRNA synthetase. RNA, 6, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M. and Soll,D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., 69, 617–650. [DOI] [PubMed] [Google Scholar]
- Ilyin V.A., Temple,B., Hu,M., Li,G., Yin,Y., Vachette,P. and Carter,C.W.,Jr (2000) 2.9 Å crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci., 9, 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim E.E. and Wyckoff,H.W. (1991) Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J. Mol. Biol., 218, 449–464. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) A super position. CCP4/ESF-EACBM Newsl. Protein Crystallogr., 31, 9–14. [Google Scholar]
- Leslie A.G. (1999) Integration of macromolecular diffraction data. Acta Crystallogr. D, 55, 1696–1702. [DOI] [PubMed] [Google Scholar]
- Levitt M. (1969) Detailed molecular model for transfer ribonucleic acid. Nature, 224, 759. [DOI] [PubMed] [Google Scholar]
- Lin L. and Schimmel,P. (1996) Mutational analysis suggests the same design for editing activities of two tRNA synthetases. Biochemistry, 35, 5596–5601. [DOI] [PubMed] [Google Scholar]
- Lipman R.S., Sowers,K.R. and Hou,Y.M. (2000) Synthesis of cysteinyl-tRNACys by a genome that lacks the normal cysteine-tRNA synthetase. Biochemistry, 39, 7792–7798. [DOI] [PubMed] [Google Scholar]
- Mechulam Y., Schmitt,E., Maveyraud,L., Zelwer,C., Nureki,O., Yokoyama,S., Konno,M. and Blanquet,S. (1999) Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J. Mol. Biol., 294, 1287–1297. [DOI] [PubMed] [Google Scholar]
- Nagle G.M. and Doolittle,R.F. (1995) Phylogenetic analysis of the aminoacyl-tRNA synthetases. J. Mol. Evol., 40, 487–498. [DOI] [PubMed] [Google Scholar]
- Newberry K., Kohn,J., Hou,Y.M. and Perona,J.J. (1999) Crystallization and preliminary diffraction analysis of E. coli cysteinyl-tRNA synthetase. Acta Crystallogr. D, 55, 1046–1047. [DOI] [PubMed] [Google Scholar]
- Nureki O., Vassylyev,D.G., Katayanagi,K., Shimizu,T., Sekine,S., Kigawa,T., Miyazawa,T., Yokoyama,S. and Morikawa,K. (1995) Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science, 267, 1958–1965. [DOI] [PubMed] [Google Scholar]
- Nureki O. et al. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science, 280, 578–582. [DOI] [PubMed] [Google Scholar]
- Perona J.J., Rould,M.A., Steitz,T.A., Risler,J.L., Zelwer,C. and Brunie,S. (1991) Structural similarities in glutaminyl- and methionyl-tRNA synthetases suggest a common overall orientation of tRNA binding. Proc. Natl Acad. Sci. USA, 88, 2903–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona J.J., Rould,M.A. and Steitz,T.A. (1993) Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry, 32, 8758–8771. [DOI] [PubMed] [Google Scholar]
- Rath V.L., Sylvian,L.F., Beijer,B., Sproat,B.S. and Steitz,T.A. (1998) How glutaminyl-tRNA synthetase selects glutamine. Structure, 6, 439–449. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana L., Brown,J.R. and Schimmel,P. (2001) Structure-based phylogeny of class IIa tRNA synthetases in relation to an unusual biochemistry. J. Mol. Evol., 53, 261–268. [DOI] [PubMed] [Google Scholar]
- Rould M.A., Perona,J.J., Soll,D. and Steitz,T.A. (1989) Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science, 246, 1135–1142. [DOI] [PubMed] [Google Scholar]
- Rulisek L. and Havlas,Z. (2000) Theoretical studies of metal ion selectivity. 1. DFT calculations of interaction energies of amino acid side chains with selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+). J. Am. Chem. Soc., 122, 10428–10439. [Google Scholar]
- Sankaranarayanan R., Dock-Bregeon,A.C., Rees,B., Bovee,M., Caillet,J., Romby,P., Francklyn,C.S. and Moras,D. (2000) Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase. Nature Struct. Biol., 7, 461–465. [DOI] [PubMed] [Google Scholar]
- Schmidt E. and Schimmel,P. (1994) Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science, 264, 265–267. [DOI] [PubMed] [Google Scholar]
- Serre L., Verdon,G., Choinowski,T., Hervouet,N., Risler,J.L. and Zelwer,C. (2001) How methionyl-tRNA synthetase creates its amino acid recognition pocket upon l-methionine binding. J. Mol. Biol., 306, 863–876. [DOI] [PubMed] [Google Scholar]
- Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- Stathopoulos C., Li,T., Longman,R., Vothknecht,U.C., Becker,H.D., Ibba,M. and Soll,D. (2000) One polypeptide with two aminoacyl-tRNA synthetase activities. Science, 287, 479–482. [DOI] [PubMed] [Google Scholar]
- Stathopoulos C., Jacquin-Becker,C., Becker,H.D., Li,T., Ambrogelly,A., Longman,R. and Soll,D. (2001) Methanococcus jannaschii prolyl-cysteinyl-tRNA synthetase possesses overlapping amino acid binding sites. Biochemistry, 40, 46–52. [DOI] [PubMed] [Google Scholar]
- Sugiura I. et al. (2000) The 2.0 Å crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules. Structure Fold. Des., 8, 197–208. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (1999) Reciprocal-space solvent flattening. Acta Crystallogr. D, 55, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates T.O. (1997) Detecting and overcoming crystal twinning. Methods Enzymol., 276, 344–358. [PubMed] [Google Scholar]