Abstract
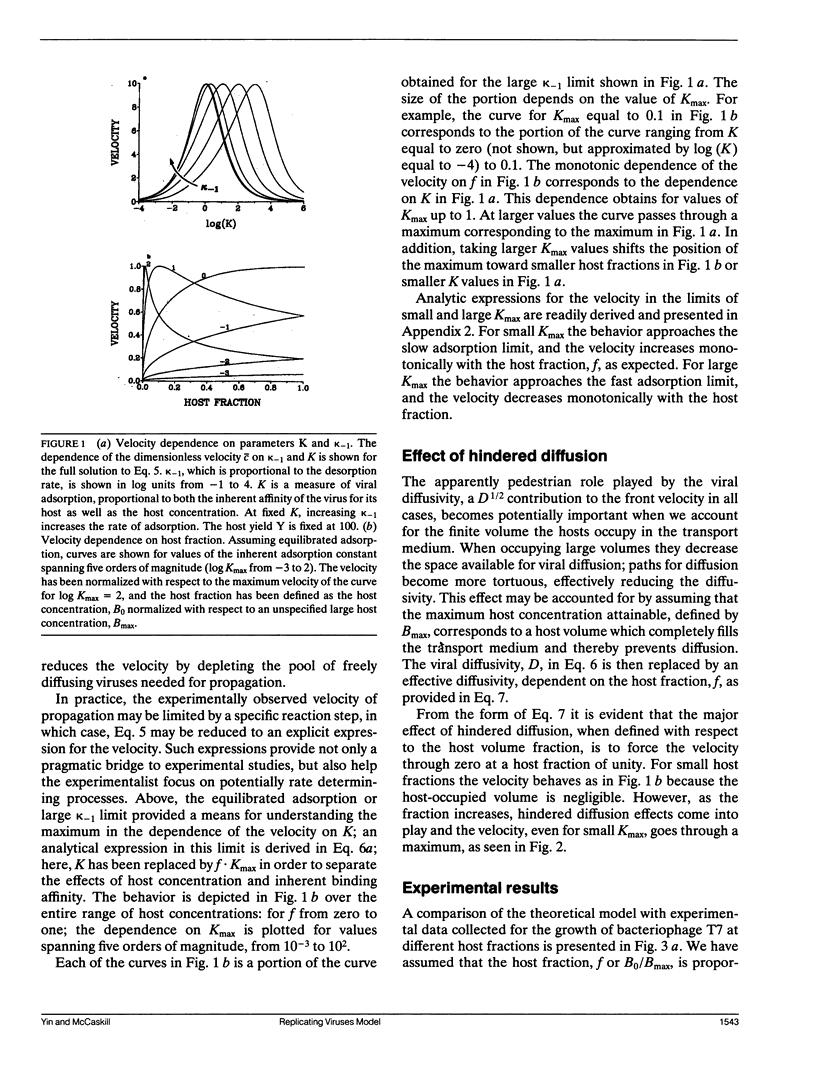
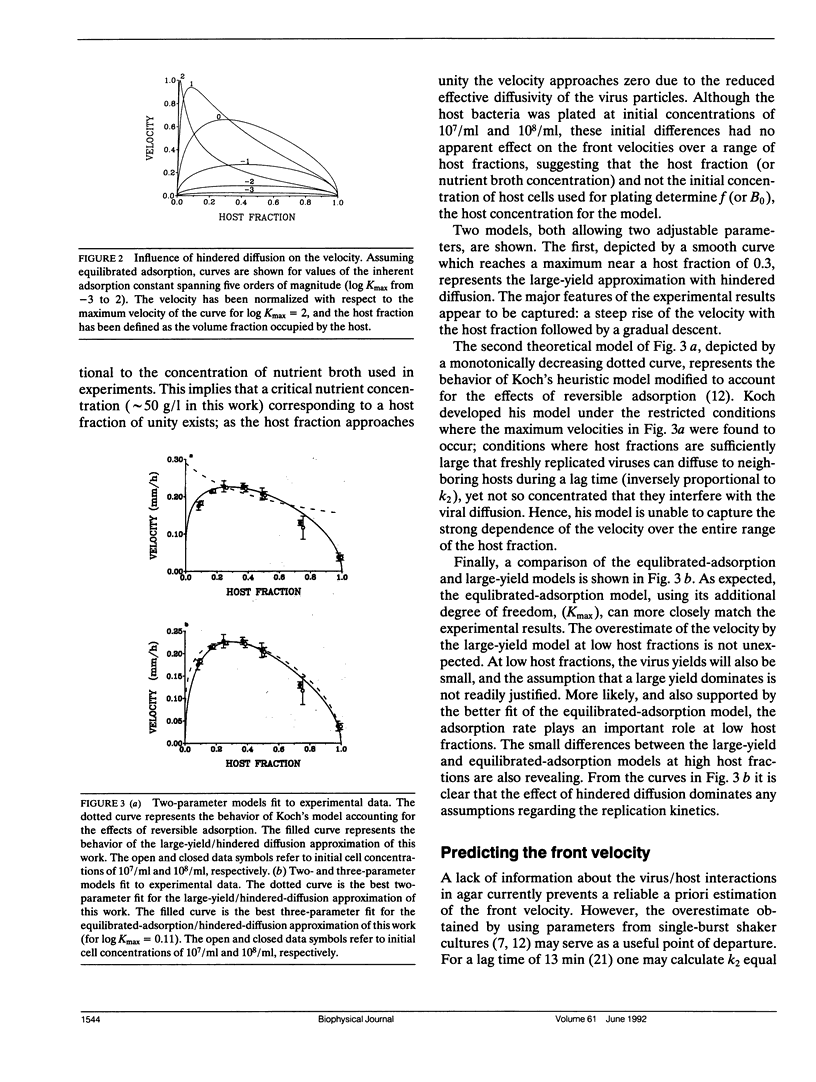
An understanding of the viral replication process commonly referred to as "plaque growth" is developed in the context of a reaction-diffusion model. The interactions among three components: the virus, the healthy host, and the infected host are represented using rates of viral adsorption and desorption to the cell surface, replication and release by host lysis, and diffusion. The solution to the full model reveals a maximum in the dependence of the velocity of viral propagation on its equilibrium adsorption constant, suggesting that conditions can be chosen where viruses which adsorb poorly to their hosts will replicate faster in plaques than those which adsorb well. Analytic expressions for the propagation velocity as a function of the kinetic and diffusion parameters are presented for the limiting cases of equilibrated adsorption, slow adsorption, fast adsorption, and large virus yields. Hindered diffusion at high host concentrations must be included for quantitative agreement with experimental data.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann H. W. La classification des phages caudés des entérobactéries. Pathol Biol (Paris) 1976 May;24(5):359–359. [PubMed] [Google Scholar]
- Bauer G. J., McCaskill J. S., Otten H. Traveling waves of in vitro evolving RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7937–7941. doi: 10.1073/pnas.86.20.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COOPER P. D. The plaque assay of animal viruses. Adv Virus Res. 1961;8:319–378. doi: 10.1016/s0065-3527(08)60689-2. [DOI] [PubMed] [Google Scholar]
- DIAMOND L., CRAWFORD L. V. SOME CHARACTERISTICS OF LARGE-PLAQUE AND SMALL-PLAQUE LINES OF POLYOMA VIRUS. Virology. 1964 Feb;22:235–244. doi: 10.1016/0042-6822(64)90008-x. [DOI] [PubMed] [Google Scholar]
- Domingo E., Sabo D., Taniguchi T., Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978 Apr;13(4):735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Freund R., Garcea R. L., Sahli R., Benjamin T. L. A single-amino-acid substitution in polyomavirus VP1 correlates with plaque size and hemagglutination behavior. J Virol. 1991 Jan;65(1):350–355. doi: 10.1128/jvi.65.1.350-355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAREN A. Thermodynamic and kinetic studies on the attachment of T1 bacteriophage to bacteria. Biochim Biophys Acta. 1954 Jun;14(2):163–172. doi: 10.1016/0006-3002(54)90155-9. [DOI] [PubMed] [Google Scholar]
- Husimi Y., Nishigaki K., Kinoshita Y., Tanaka T. Cellstat-a continuous culture system of a bacteriophage for the study of the mutation rate and the selection process of the DNA level. Rev Sci Instrum. 1982 Apr;53(4):517–522. doi: 10.1063/1.1137002. [DOI] [PubMed] [Google Scholar]
- Kennedy C. R., Aris R. Traveling waves in a simple population model involving growth and death. Bull Math Biol. 1980;42(3):397–429. doi: 10.1007/BF02460793. [DOI] [PubMed] [Google Scholar]
- Koch A. L. The growth of viral plaques during the enlargement phase. J Theor Biol. 1964 May;6(3):413–431. doi: 10.1016/0022-5193(64)90056-6. [DOI] [PubMed] [Google Scholar]
- Krüger D. H., Mann W., Hansen S., Bläsing G., Bläsing M., Schroeder C. A simple method of distinguishing the bacterial viruses T3 and T7, and a critical reevaluation of their heterologous and homologous exclusion. J Basic Microbiol. 1988;28(1-2):45–53. doi: 10.1002/jobm.3620280107. [DOI] [PubMed] [Google Scholar]
- MAYR-HARTING A. Die Entwicklung von Phagenlöchern und der Mechanismus der Phagenwirkung in festen Nährböden. Zentralbl Bakteriol Orig. 1958 Jun;171(6-7):380–392. [PubMed] [Google Scholar]
- Parvin J. D., Moscona A., Pan W. T., Leider J. M., Palese P. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol. 1986 Aug;59(2):377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SYMONDS N. The properties of a star mutant of phage T2. J Gen Microbiol. 1958 Apr;18(2):330–345. doi: 10.1099/00221287-18-2-330. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Genetic analysis of non-essential bacteriophage T7 genes. J Mol Biol. 1973 Sep 15;79(2):227–236. doi: 10.1016/0022-2836(73)90002-8. [DOI] [PubMed] [Google Scholar]
- Studier F. W. The genetics and physiology of bacteriophage T7. Virology. 1969 Nov;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Yin J. A quantifiable phenotype of viral propagation. Biochem Biophys Res Commun. 1991 Jan 31;174(2):1009–1014. doi: 10.1016/0006-291x(91)91519-i. [DOI] [PubMed] [Google Scholar]
- de la Torre J. C., Holland J. J. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J Virol. 1990 Dec;64(12):6278–6281. doi: 10.1128/jvi.64.12.6278-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


