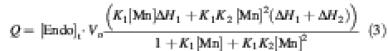Abstract
T7 endonuclease I is a nuclease that is selective for the structure of the four-way DNA junction. The active site is similar to those of a number of restriction enzymes. We have solved the crystal structure of endonuclease I with a wild-type active site. Diffusion of manganese ions into the crystal revealed two peaks of electron density per active site, defining two metal ion-binding sites. Site 1 is fully occupied, and the manganese ion is coordinated by the carboxylate groups of Asp55 and Glu65, and the main chain carbonyl of Thr66. Site 2 is partially occupied, and the metal ion has a single protein ligand, the remaining carboxylate oxygen atom of Asp55. Isothermal titration calorimetry showed the sequential exothermic binding of two manganese ions in solution, with dissociation constants of 0.58 ± 0.019 and 14 ± 1.5 mM. These results are consistent with a two metal ion mechanism for the cleavage reaction, in which the hydrolytic water molecule is contained in the first coordination sphere of the site 1-bound metal ion.
Keywords: calorimetry/crystal structure/DNA–protein interaction/Holliday junction resolvase/nuclease
Introduction
Homologous genetic recombination is important in the repair of double-strand breaks in DNA, in the rescue of stalled replication forks and in the creation of genetic diversity (reviewed in Lilley and White, 2001). The central intermediate in this process is the four-way (Holliday) DNA junction. This must ultimately be resolved by nucleases that are selective for the structure of the junction. The junction-resolving enzymes are widespread in nature, having been isolated from bacteriophage-infected Escherichia coli (Kemper and Garabett, 1981; de Massey et al., 1984), eubacteria (Connolly et al., 1991; Iwasaki et al., 1991), euryarchaea (Komori et al., 1999), crenarchaea (Kvaratskhelia and White, 2000), yeast (Symington and Kolodner, 1985; West et al., 1987; White and Lilley, 1997) and, finally, mammals (Constantinou et al., 2001) and their viruses (Garcia et al., 2000). In general, these are small, basic proteins that bind to four-way DNA junctions in dimeric form, with relatively high affinities. Resolution is achieved by the introduction of paired cleavages, generating nicked duplex species.
Most of the junction-resolving enzymes can be classified with one of two superfamilies of proteins (Lilley and White, 2000; Makarova et al., 2000). The resolving enzymes RuvC and Cce1 are placed in the integrase superfamily, with RNaseH, while T7 endonuclease I and the archaeal enzymes are classed along with λ-exonuclease and the restriction enzymes in the nuclease superfamily. All are probably evolved from an ancestral metal ion-binding domain. It is likely that the junction-resolving enzymes hydrolyse the phosphodiester bond using a metal ion-activated water molecule as a general mechanism. All the enzymes studied have essential acidic amino acids that could coordinate metal ions (Duckett et al., 1995; Saito et al., 1995; Giraud-Panis and Lilley, 1996, 1998; Bolt et al., 1999; Parkinson et al., 1999; Wardleworth et al., 2000) and, in each of the five known crystal structures, RuvC (Ariyoshi et al., 1994), T4 endonuclease VII (Raaijmakers et al., 1999), T7 endonuclease I (Hadden et al., 2001) and Hjc of Sulfolobus (Bond et al., 2001) and Pyrococcus (Nishino et al., 2001), the putative active sites contain clusters of acidic side chains that probably constitute metal ion-binding sites. A calcium ion was observed bound to the active site of T4 endonuclease VII (Raaijmakers et al., 1999).
We have recently presented the crystal structure of a catalytically defective mutant of T7 endonuclease I (E65K) (Hadden et al., 2001). The putative active site contains three acidic side chains (Glu20, Asp55 and Glu65) and a lysine (Lys67). These correspond closely to the active site residues of a number of type II restriction enzymes; for example the α and β carbon atoms of these four residues can be superimposed on the corresponding atoms of BglI to within an r.m.s.d. of 0.33 Å. The crystal structure of BglI was solved as a complex with a DNA oligonucleotide, and two calcium ions were observed bound in the active site (Newman et al., 1998). We have therefore proposed that two metal ions are bound in the active site of T7 endonuclease I (Déclais et al., 2001), and that both participate in the cleavage of the phosphodiester backbone.
In this study, we have sought direct evidence for the binding of metal ions to the active site of T7 endonuclease I. None was observed in our earlier crystal structure of the enzyme, but this may be a result of the E65K mutation that inverted the electrostatic charge of one of the active site residues. We have now solved the crystal structure of the wild-type protein, with and without bound manganese ions. Additionally, we have used isothermal titration calorimetry to provide strong evidence that two manganese ions bind to the enzyme in solution. These results are fully consistent with our proposed (Déclais et al., 2001) two metal ion mechanism for DNA junction cleavage.
Results
The activity of T7 endonuclease I in the presence of manganese ions
The striking similarity between the active sites of endonuclease I and type II restriction enzymes suggested that the resolving enzymes would be able to use manganese ions in place of magnesium ions for catalytic activity. We have sought to compare the site and rates of cleavage of a four-way junction by endonuclease I in the presence of either divalent metal ion.
Junction 3 (20 nM) radioactively 5′-32P-labelled on the x-strand was incubated with 40 nM endonuclease I in the presence of 10 mM either magnesium or manganese chloride. The products of cleavage were separated by sequencing gel electrophoresis, and the resulting autoradiograph (Figure 1A) shows that the enzyme has cleaved the junction at the same site in the presence of both metal ions.
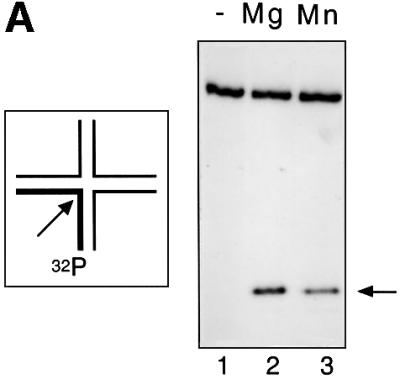
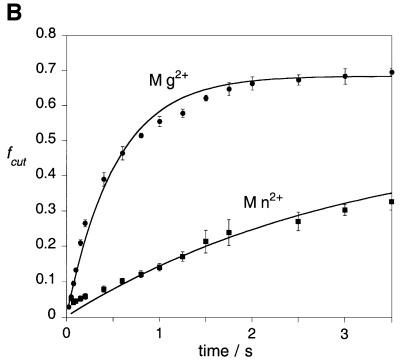
Fig. 1. Comparison of the catalytic activity of wild-type endonuclease I in the presence of magnesium or manganese ions. Junction 3 radioactively 5′-32P-labelled on the x-strand was cleaved with endonuclease I in the presence of magnesium or manganese ions at 20°C, using a quench-flow mixer. The products of the reaction were resolved by electrophoresis in denaturing polyacrylamide gels and analysed by phosphoimaging or autoradiography. (A) Autoradiograph showing the cleavage product after 2 s reaction. The reaction was initiated with no metal ions (track 1), magnesium ions (track 2) or manganese ions (track 3). (B) Time course of junction 3 cleavage by endonuclease I wt (ΔN11). The data are presented as fraction cut (fc) versus ageing time for reactions carried out in the presence of magnesium ions (circles) and manganese ions (squares). Experiments were performed in triplicate and the error bars represent standard deviations. The lines are best fits to single exponentials.
We have measured the rate of cleavage of the junction by endonuclease I in the presence of each metal ion. As we have shown earlier (Déclais et al., 2001), these rates are not accessible to manual sampling since the reaction is completed within 10 s. We thus used a quenched-flow mixer to start the reaction and quench it in a range of 50 ms to 5 s. The protein–DNA complex was loaded in the first syringe, the magnesium (or manganese) chloride solution in the second syringe and the EDTA solution in the third syringe. The reaction was started by mixing equal volumes (15 µl) from the first two syringes in the first mixer, allowed to proceed in the delay line and quenched in the second mixer with 15 µl from the third syringe. The ageing time was adjusted by adapting the flow rates. The reaction products were then resolved by polyacrylamide electrophoresis and quantified by phosphoimaging. The progress curves (Figure 1B) show that the enzyme is less active in manganese ions compared with magnesium ions. The two data sets were fitted by non-linear regression to single exponentials.
The rates of cleavage (kobs) at 20°C in the presence of either metal are of the same order, the enzyme being ∼6 times faster in magnesium (kobs = 1.90 ± 0.11/s) than in manganese ions (kobs = 0.32 ± 0.06/s). The absolute rates vary a little with the age of the enzyme preparation.
The crystal structure of wild-type sequence T7 endonuclease I
The structure of wild-type endonuclease I in the presence and absence of manganese ions was determined by molecular replacement using the E65K mutant variant structure (Hadden et al., 2001) with the side chain of Lys65 replaced by alanine as the model. The structure of the protein in the absence of metal ions was also determined by molecular replacement but using the wild-type metal-bound structure with the side chains Asp55, Glu65, Lys67 and Glu20 replaced by alanine as the model. The two dimers, 1 and 2, in the asymmetric unit consisting of chains A plus B and C plus D, respectively, were refined initially as rigid bodies. Final refinement and rebuilding were carried out without non-crystallographic symmetry restraints applied and, consequently, there are small structural variations between the chains in both the metal-bound and free structures. However, perhaps the most striking variation is that of the quality of the electron density for each chain. Dimer 1 has very well defined electron density, as does the majority of chain D in dimer 2. However, the majority of chain C (residues 44–145) and a small fragment of chain D (residues 17–44) in dimer 2 are less well defined. For these reasons, our description of the wild-type protein structure, and any comparisons between the wild-type protein and the E65K mutant, are confined to dimer 1 where the electron density is best defined. It should be noted that well-defined electron density was only present for residues 17–145 in all chains. In addition, a number of amino acid residues that lacked significant side chain electron density were identified. These were built and refined as alanine residues and are listed in Table I.
Table I. Statistics for data collection and refinement.
| T7 endonuclease I in the presence of 50 mM manganese chloride | T7 endonuclease I: no metal soak | |
|---|---|---|
| Maximum resolution | 1.9 Å | 2.55 Å |
| Observations | 419 157 | 168 267 |
| Unique observations | 81 257 | 34 014 |
| Completenessa | 99.9% (99.4%) | 99.8% (99.9%) |
| Average I/σIa | 7.6 (1.6) | 9.5 (2.4) |
| Multiplicity | 5.2 | 4.9 |
|
Rsymb |
0.085 |
0.063 |
| Refinement |
|
|
| Rcrystc | 21.0 | 22.2 |
| Rfreed | 23.4 | 27.3 |
| No. of amino acid residues | 516 | 516 |
| No. of water molecules |
473 |
53 |
| R.m.s. deviation from ideale |
|
|
| Bonds | 0.008 Å | 0.011 Å |
| Angles |
1.38° |
1.55° |
| Ramachandran plot |
|
|
| Most favouredf | 91.3% | 90.1% |
| Allowedf | 7.6% | 8.3% |
| Disallowedf | 0.9% | 0.9% |
Residues built as alanine in final model, metal-bound structure:
Chain A = Lys25, Gln26, Lys33, Lys103, Glu136, Lys143, Arg144, Lys145;
Chain B = Lys30, Lys143;
Chain C = Lys33, Glu71, Arg82, Glu87, Lys103, Lys115, Lys131, Lys135, Glu136, Val137, Pro138, Phe139, Asp140, Leu142, Lys143, Arg144, Lys145;
Chain D = Asp21, Lys25, Glu28, Lys30, Ile32, Lys33, Glu38, Trp39, Lys40, Arg98, Lys131, Lys145.
Residues built as alanine in final model, metal-free structure:
Chain A = Lys25, Lys33, Lys103, Glu136, Lys143, Lys145;
Chain B = Lys103, Glu128, Lys143;
Chain C = Lys33, Lys40, Asp55, Glu65, Lys67, Leu69, Glu71, Arg82, Glu87, Lys103, Ser105, Glu111, Lys115, Lys131, Lys135, Glu136, Val137, Pro138, Phe139, Asp140, Arg141, Leu142, Lys143, Arg144, Lys145;
Chain D = Glu20, Asp21, Lys25, Leu27, Glu28, Lys30, Ile32, Lys33, Glu38, Trp39, Lys40, Lys131, Glu136, Lys145.
aNumbers in parentheses refer to the outermost resolution shell.
bRsym (I) = ∑hkl∑i|Ihkl,i – <Ihkl>I|/∑hkl∑i|Ihkl,i|, where <Ihkl> is the mean intensity of the multiple Ihkl,i observations for symmetry-related reflections.
cRcryst = ∑hkl|Fobs – Fcalc|/∑hkl|Fobs|
dRfree = ∑hkl∑T|Fobs – Fcalc|/∑hkl∑T|Fobs|, where the test set T includes 5% of the data.
eReported by CNS (Brünger et al., 1998).
fDetermined using PROCHECK (Laskowski et al., 1993). Residue Lys123 was found to be in a disallowed region of the Ramachandran plot in all chains. However, the electron density supports this geometry.
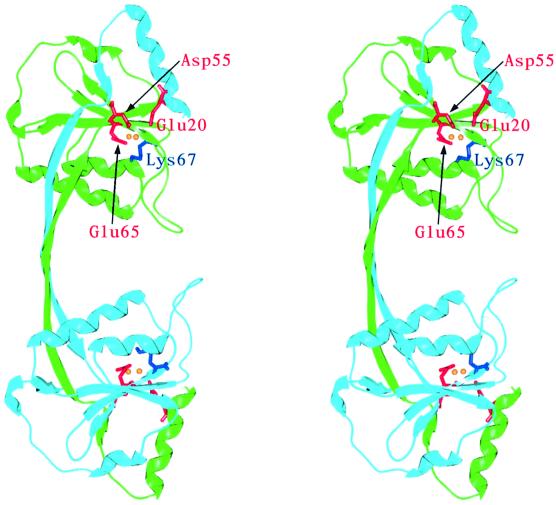
The overall structure of endonuclease I is, as expected, very similar to that of the E65K mutant. The r.m.s.d. over 128 Cα atoms is 0.53 Å (comparing chain A of the wild-type metal-bound structure with chain A of the E65K mutant). Briefly, endonuclease I forms a closely associated homodimer arranged in two domains (Figure 2). Each domain is composed of residues 17–44 from one chain and residues 49–145 from the other. The two domains are connected by a bridge that forms an extended two-stranded antiparallel β-sheet. Each domain is comprised of a central five-stranded mixed β-sheet, surrounded by five α-helices. One strand and one helix in each domain are contributed by the other subunit in the dimer. The four domains (comprising two dimers) in the asymmetric unit are named according to the polypeptide chains that contribute to them, e.g. domain AB is composed of residues A49–A145 and B17–B44, and domain BA of residues B49–B145 and A17–A44.
Fig. 2. Stereo view showing a ribbon representation of the structure of T7 endonuclease I with bound manganese ions. One subunit is drawn in light blue and the other in green. Active site residues are depicted in red (acidic amino acids) and blue (lysine). Manganese ions are represented as yellow spheres. All figures depicting the crystal structure of endonuclease I were generated using the program SPOCK (Christopher, 1998).
The active site of T7 endonuclease I
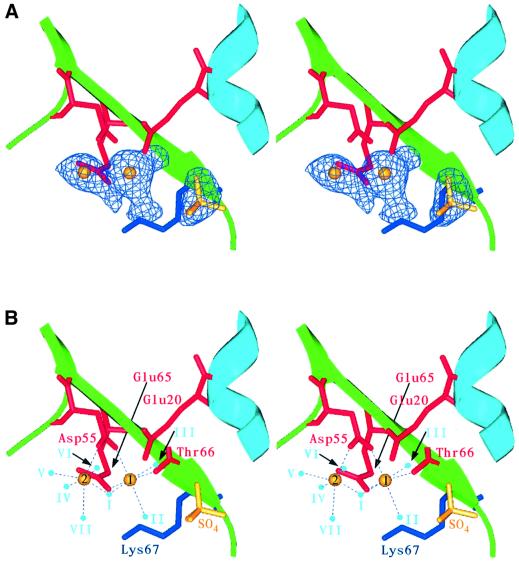
Based on site-directed mutagenesis studies and comparisons with a number of restriction endonucleases, we previously identified a number of potential active site residues in endonuclease I: Asp55, Glu65, Lys67 and Glu20 (Déclais et al., 2001) (Figure 2). These conformed well with the common catalytic sequence motif PD...(D/E)XK found in all the type II restriction endonucleases whose crystal structures have been solved (Pingoud and Jeltsch, 2001). Based on this similarity, we proposed that endonuclease I may cleave four-way DNA junctions using a mechanism involving two bound metal ions (Déclais et al., 2001). However, the E65K mutant variant is inactive and no metal ions could be located in the active site of the crystal structure. The crystal structure of the metal-bound wild-type protein reveals significant differences in the active site (Figure 3). In addition to clear density for Glu65 (replacing the lysine in the E65K mutant protein), two strong new peaks in the 2Fo – Fc electron density maps were observed. These are ∼3.5 Å apart and have been built and refined as manganese ions (Figure 3). All four active sites (two for each dimer) in the asymmetric unit showed equivalent peaks, resulting in eight metal sites per asymmetric unit. Interestingly, the heights of electron density peaks are different for the two metal-binding sites, and we have designated the higher peak in each domain metal site 1 and the lower one metal site 2. For example, in domain AB, sites 1 and 2 have heights of 18.9 and 9.9σ, respectively. A similar pattern is observed for all the domains in the asymmetric unit.
Fig. 3. Stereo images showing close-up views of the active site of T7 endonuclease I with bound manganese and sulfate ions. Endonuclease I subunits are shown in light blue and green. Active site residues are depicted in red (acidic residues and Thr66 CO) and blue (lysine). Manganese and sulfate ions are represented in yellow. (A) Fo – Fc electron density map in the vicinity of the active site contoured at 5.5σ with the manganese and sulfate ions and active site water molecules omitted from the model. Difference density can clearly be seen for the two manganese ions, the sulfate moiety and the three water molecules that are strongly associated with the metal bound at site 1. Reduction of the σ level to 3.0 would reveal electron density for the additional water molecules associated with metal site 2 (not shown). (B) The metal-ligating residues, manganese ions, sulfate ion and local solvent structure. Coordinating water molecules are shown in light blue. Potential interactions are highlighted with dashed lines.
When both sites 1 and 2 were refined with an occupancy of 1.0, it became apparent that the type 2 sites often had a large negative Fo – Fc electron density peak associated with them. This indicates that the type 2 sites may not be completely occupied under the conditions used to introduce metal ions into the crystal. We therefore refined the type 2 sites using an occupancy of 0.75. In domain AB, this leads to refined B-factors of 26.9 and 35.0 Å2 for sites 1 and 2, respectively. This value fits well with our solution measurements (see Discussion) and essentially removes any negative Fo – Fc electron density peaks.
When crystals were soaked in cryoprotectant that did not contain manganese chloride, no strong peaks corresponding to bound metals could be observed in the electron density maps. However, a small peak was present in a similar position to metal site 1. Due to the low intensity of this peak and the relatively long distances between it and any potential ligating residues, this probably represents a bound water molecule and has been built and refined as such. The orientations of the active site residues are similar in both the metal-bound and metal-free proteins, suggesting that the overall conformation of the active site is not greatly affected by the binding of manganese ions.
The metal ion-binding sites
The discussion that follows applies to the AB domain of dimer 1; where significant differences occur in the other domains in the asymmetric unit, these are noted. Metal site 1 has three protein ligands, Asp55 Oδ2, Glu65 Oε2 and Thr66 O, and three water ligands (I–III) (Table II). One of these water molecules (I) also interacts with the second metal site. Metal ions bound at type 2 sites appear to be more weakly associated with the protein, and have only one protein ligand Asp55 Oδ1. The number of water molecules associated with this site varies for the different subunits in the asymmetric unit. Two of the subunits (BA and DC) have four well-defined water molecules associated with metal site 2 (water positions I and IV–VI), whilst domain AB shows electron density for an additional water ligand (VII). In contrast, domain CD has only three water molecules associated with metal site 2 (I, IV and VII). It should be noted, however, that the electron density for the whole of domain CD is significantly less well defined than that of the other protein subunits in the asymmetric unit.
Table II. Active site manganese ion interactions in metal-bound endonuclease I.
| Metal site | Ligand | Distance (Å) | Domain | Water position |
|---|---|---|---|---|
| 1 | A55 Oδ2 | 2.12 | AB | – |
| A65 Oε2 | 2.07 | – | ||
| A66 O | 2.22 | – | ||
| Wat1 | 2.17 | I | ||
| Wat2 | 2.41 | II | ||
| Wat3 | 2.26 | III | ||
| 2 | A55 Oδ1 | 2.20 | – | |
| Wat4 | 2.24 | IV | ||
| Wat1 | 1.82 | I | ||
| Wat5 | 2.15 | V | ||
| Wat6 | 2.20 | VI | ||
| Wat7 | 2.46 | VII | ||
| 1 | B55 Oδ2 | 2.20 | BA | – |
| B65 Oε2 | 2.09 | – | ||
| B66 O | 2.26 | – | ||
| Wat8 | 2.08 | I | ||
| Wat9 | 2.32 | II | ||
| Wat10 | 2.17 | III | ||
| 2 | B55 Oδ1 | 2.16 | – | |
| Wat11 | 2.50 | IV | ||
| Wat8 | 1.92 | I | ||
| Wat12 | 2.53 | V | ||
| Wat13 | 2.29 | VI | ||
| No density | – | VII | ||
| 1 | C55 Oδ2 | 1.68 | CDa | – |
| C65 Oε2 | 2.21 | – | ||
| C66 O | 2.38 | – | ||
| Wat14 | 2.67 | I | ||
| Wat15 | 1.65 | II | ||
| Wat16 | 2.40 | III | ||
| 2 | C55 Oδ1 | 2.02 | – | |
| Wat17 | 2.32 | IV | ||
| Wat14 | 2.32 | I | ||
| No density | – | V | ||
| No density | – | VI | ||
| Wat18 | 2.38 | VII | ||
| 1 | D55 Oδ2 | 2.10 | DC | – |
| D65 Oε2 | 2.06 | – | ||
| D66 O | 2.29 | – | ||
| Wat19 | 2.23 | I | ||
| Wat20 | 2.28 | II | ||
| Wat21 | 2.23 | III | ||
| 2 | D55 Oδ1 | 2.09 | – | |
| Wat22 | 2.50 | IV | ||
| Wat19 | 1.96 | I | ||
| Wat23 | 2.57 | V | ||
| Wat24 | 2.31 | VI | ||
| No density | – | VII |
Manganese ion–ligand distances for the eight metal ions (comprising the four active sites) in the crystallographic asymmetric unit are listed. Water molecules located within each active site have been designated by Roman numerals (I–VII) and those located in equivalent positions in different active sites within the asymmetric unit are labelled identically.
aElectron density for domain CD is significantly less well defined than that for domains AB, BA and DC.
A sulfate ion bound in the active site
In addition to the peaks associated with metal or water sites, one additional strong feature could also be seen in the electron density map for each subunit of the metal-bound and free structures (this feature was not present in one of the metal-free subunits). Each of these was located in an equivalent position relative to each domain, in close proximity to the active site (4.6–4.9 Å from metal site 1 in the metal-bound structure). In the initial stages of refinement, these peaks were not assigned. However, as refinement proceeded, the peaks developed a tetrahedral shape, characteristic of sulfate ions. Given that crystals of endonuclease I were grown in the presence of 100 mM ammonium sulfate, and that sulfate ions have previously been seen to bind to the histone protein HMfA (Decanniere et al., 2000) in close proximity to the proposed DNA-binding site, these peaks were modelled as sulfate ions.
Some details of the sulfate-binding site, as with the metal-binding sites, are subunit dependent (Table III), but a number of interactions are conserved in all the sites. Two protein side chains are important in ligating the sulfate ion, Gly68 N and Ser17 Oγ, along with a number of water molecules. In particular, water molecules II and III form hydrogen bonds to the sulfate ion in each binding site in the metal-bound form of the protein. These form part of the hydration sphere of the type 1 metal site.
Table III. Active site sulfate ion interactions in metal-bound endonuclease I.
| Subunit | Atom | Ligand | Distance (Å) | Water position |
|---|---|---|---|---|
| AB | O1 | A68 N | 2.90 | – |
| O1 | Wat2 O | 2.53 | II | |
| O2 | B17 Oγ | 3.04 | – | |
| O2 | Wat3 O | 2.74 | III | |
| O3 | – | |||
| O4 | Wat284O | 2.65 | – | |
| BA | O1 | B68 N | 2.94 | |
| O1 | Wat9 | 2.63 | II | |
| O1 | Wat455 | 2.97 | ||
| O2 | A17 Oγ | 2.91 | ||
| O2 | Wat10 | 2.83 | III | |
| O3 | – | – | ||
| O4 | Wat176 | 2.85 | ||
| CDa | O1 | C68 N | 3.24 | |
| O1 | Wat15 | 2.73 | II | |
| O2 | D17 Oγ | 3.60 | ||
| O2 | Wat16 | 2.78 | III | |
| O4 | – | – | ||
| DC | O1 | D68 N | 2.96 | |
| O1 | Wat20 | 2.54 | II | |
| O1 | Wat351 | 2.97 | ||
| O2 | C17 Oγ | 2.69 | ||
| O2 | Wat21 | 2.76 | III | |
| O3 | Wat413 | 2.75 | ||
| O4 | Wat78 | 2.99 | ||
| O4 | Wat429 | 2.69 |
Sulfate ion–ligand distances for the four sulfate ions located in the crystallographic asymmetric unit are listed. Water positions are listed as in Table II.
aElectron density for domain CD is significantly less well defined than that for domains AB, BA and DC.
Binding of manganese ions to T7 endonuclease I in solution
We have sought confirmation of the binding of manganese ions to endonuclease I in solution using isothermal titration calorimetry (ITC) (Wiseman et al., 1989). This approach has been applied successfully to study the binding of metal ions to several endonucleases (Jose et al., 1999; Kvaratskhelia et al., 1999; Cowan et al., 2000). In these experiments, aliquots of a manganese chloride solution were injected into a solution of T7 endonuclease I contained in the calorimeter sample cell, and the heat energy changes accompanying the protein–Mn2+ association were detected. Absorption or evolution of heat energy upon injection of manganese was measured as the power (dq/dt; µcal/s) that was required to maintain zero temperature difference between the sample and the reference cell. Analysis of the titration data provides direct information on the energetics and stoichiometry of the interaction.
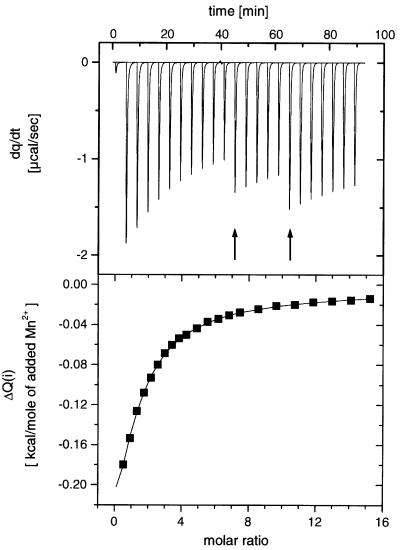
The titration experiments show that the binding of manganese ions to T7 endonuclease I is exothermic (Figure 4A). The raw data were corrected for dilution effects by subtracting heat energy changes per unit time measured in an identical series of titrations into buffer, and the corrected data were integrated to give the heat ΔQ(i) (cal/mol of injected manganese ions) removed to restore the thermal equilibrium between sample and reference cells after the ith injection of manganese ions to the protein. The series of ΔQ(i) values was plotted as a function of manganese ion concentration, representing the differential of the total heat Q [i.e. enthalpy (ΔH) at constant pressure] for each ion concentration (Figure 4B). These data were fitted to a variety of binding models. They were found to give an excellent fit (χ2 = 0.786) to a model of sequential binding of two manganese ions per active site, i.e.
Fig. 4. Isothermal titration calorimetry of the interaction of manganese ions with endonuclease I wt (ΔN11). Manganese chloride was titrated into a 343 µM protein solution, and the heat evolved was monitored by ITC. Upper panel: raw data for the sequential injection of various volumes (4, 6 or 10 µl) of a 50 mM manganese chloride solution into 1.4 ml of protein solution in 50 mM Tris–HCl pH 7.5, 400 mM NaCl at 25°C. The apparent discontinuities in the heat profile are due to changes in injection volumes (arrowed). Lower panel: integrated heat data (dilution corrected) with a fit according to a sequential two ion binding model (continuous line). The derived thermodynamic parameters are summarized in Table IV.
where K1 and K2 are the association constants for the first and second ion binding, respectively. The binding of two ions is clearly in good agreement with the crystal structure described above. The scheme corresponds to the equations:
where Q is the total heat content, V0 is the cell volume, [Mn] is the free manganese ion concentration and [Endo]t is the total endonuclease I concentration. The dissociation constants were found to be in the millimolar range, with KD1 = 0.58 ± 0.019 mM and KD2 = 14 ± 1.5 mM at 298 K. The thermodynamic data are summarized in Table IV.
Table IV. Thermodynamic parameters for manganese ion binding to wild-type endonuclease I, measured by calorimetry.
| Site 1 | Site 2 | |
|---|---|---|
| K (M–1) | 1716 ± 58.63 | 72.7 ± 9.183 |
| KD (mM) | 0.58 ± 0.019 | 14 ± 1.5 |
| ΔH° (kcal/mol) | –557.4 ± 10.85 | –786.2 ± 63.37 |
| ΔS° (cal/deg/mol) | 12.93 | 5.88 |
Discussion
Bacteriophage T7 endonuclease I is active in the presence of manganese ions, with a rate that is one-sixth of that measured in magnesium ions. In contrast, the enzyme is totally inactive in the presence of calcium ions. These properties are closely similar to those of many restriction enzymes (for an excellent current review see Pingoud and Jeltsch, 2001), suggesting an evolutionary relationship between these proteins (Lilley and White, 2000). We have previously noted a close similarity between the active site of endonuclease I and a number of restriction enzymes (Hadden et al., 2001). However, in our earlier work, we presented the structure of a severely catalytically impaired mutant of T7 endonuclease I. In the E65K mutant, the catalytically important Glu65 in the active site is replaced by a positively charged lysine residue, thus altering the charge characteristics of the active site in a very significant way. In particular, the altered active site would be unlikely to bind metal ions in the same way as the wild-type enzyme; this is significant because we expected that metal ions would play an important role in the catalytic mechanism. For this reason, we have solved the crystal structure of the wild-type enzyme. The overall structure of the wild-type enzyme is closely similar to that of the E65K mutant, with an r.m.s.d. of 0.53 Å over 128 Cα atoms (comparing chain A of the metal-bound wild-type structure with chain A of the E65K mutant).
The active site of wild-type endonuclease I comprises Glu20, Asp55, Glu65 and Lys67, and conforms to the catalytic sequence motif PD55...(D/E65)XK67 found in a number of restriction enzymes (Pingoud and Jeltsch, 2001). Similar motifs also exist in λ-exonuclease (Kovall and Matthews, 1998), MutH (Ban and Yang, 1998) and TnsA (Hickman et al., 2000), and have been found in the archaeal junction-resolving enzymes (Bond et al., 2001; Nishino et al., 2001). This suggests a common mechanism of DNA cleavage, requiring the coordination of divalent metal ions by carboxylate groups as we have proposed previously (Déclais et al., 2001). However, before any detailed mechanism can be considered, it is necessary to establish the number of bound metal ions in the active site. The active site of endonuclease I shares a particularly close similarity with that of BglI (Newman et al., 1998), with an r.m.s.d. of 0.48 Å (metal-bound endonuclease I) over 10 atoms in each protein comprising the α and β carbon atoms of the four critical side chains (E20, D55, E65 and K67) plus the conserved proline (P54). Two calcium ions were found coordinated to the active site of BglI complexed with DNA (Newman et al., 1998). Other restriction enzymes, including PvuII (Cheng et al., 1994) and BamHI (Newman et al., 1995), also appear to coordinate two divalent metal ions, but this is apparently not universal. Crystallographic studies of EcoRI (coordinates deposited by V.V.Horvath, J.Choi, Y.Kim, P.Wilkosz and J.M.Rosenberg, PDB 1QPS) and BglII (Lukacs et al., 2000) reveal the coordination of a single metal ion, while the situation with EcoRV is complex, and appears to depend upon conditions (Kostrewa and Winkler, 1995). Indeed, the results for any particular enzyme will depend on the relative binding affinities for different sites and the concentration of metal ions in the crystallization conditions. Given the close similarity between the active sites of endonuclease I and BglI, we previously felt it probable that the resolving enzyme would coordinate two metal ions.
Using calorimetry, we have now shown that wild-type endonuclease I binds two manganese ions in a sequential process, with significantly differing affinities. Moreover, diffusion of manganese chloride into crystals of wild-type endonuclease I resulted in the appearance of two well-defined peaks in the 2Fo – Fc electron density difference map, within the active site of the enzyme. It therefore seems probable that endonuclease I does indeed bind two divalent metal ions in its active site, at least as a free protein. The binding of manganese ions does not, however, appear to affect the conformation of the active site residues greatly.
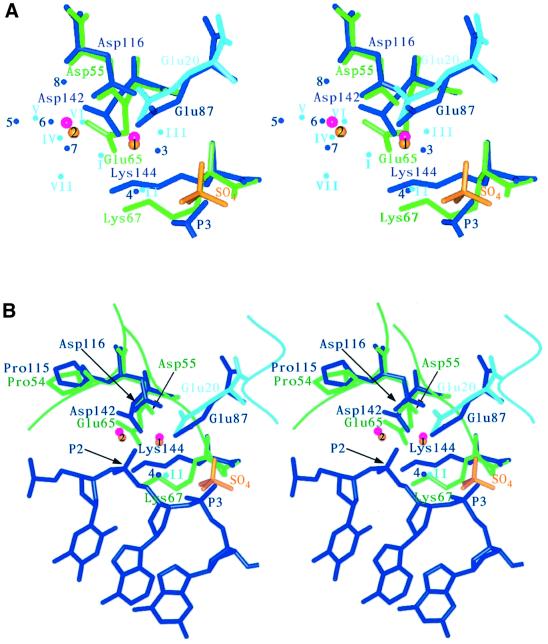
The positions of the bound metal ions, deduced from the new peaks observed in the difference map, are in close proximity to the calcium ions observed in the BglI–DNA complex, which are compared in Figure 5 and Table V. This suggests that the metal-binding sites may not be significantly perturbed by DNA binding. In addition, the sulfate group bound to endonuclease I is close to the site of a phosphate group in the BglI complex structure and is in a similar orientation. This is further evidence for the relevance of the structure of the free resolving enzyme. We term the two sites for metal ion binding as sites 1 and 2.
Fig. 5. Stereo views showing the superimposition of the active site residues in T7 endonuclease I (metal-bound protein) with those of the restriction enzyme BglI. Endonuclease I active site residues are shown in light blue and green, and manganese ions are represented as yellow spheres. Bound sulfate is shown in yellow, and water molecules are also shown in light blue. For BglI, active site residues, bound DNA and water molecules are depicted in dark blue. Calcium ions are shown in magenta. (A) A comparison of the metal-binding sites, showing the active site residues, metal ions, local solvent structure and sulfate moiety. One phosphate molecule (P3) from the BglI–DNA complex structure is also illustrated. (B) A comparison of the overall active site structural motif. The scissile phosphate in the BglI–DNA complex structure (P2) is highlighted with an arrow and the potential attacking water molecules are also shown in dark blue (4; BglI) and light blue (II; endonuclease I).
Table V. A comparison of the active site of metal-bound endonuclease I (chain A) and the restriction endonuclease BglI.
| Endo I (domain AB) | BglI | Displacement (Å) | Water position in Endo I |
|---|---|---|---|
| Mn1 | Ca1 | 0.34 | – |
| Mn2 | Ca2 | 0.89 | – |
| Wat2 | Wat4a | 0.91 | II |
| Wat3 | Wat3 | 1.01 | III |
| Wat4 | Wat6 | 1.01 | IV |
| Wat5 | Wat5 | 1.22 | V |
| Wat6 | Wat7 | 1.69 | VI |
The 10 Cα and Cβ atoms of Pro54, Asp55, Glu65, Lys67 and Glu20 in endonuclease I wt + Mn were superimposed on the corresponding atoms in BgII (Pro115, Asp116, Asp142, Lys144 and Glu87) giving an r.m.s.d. of 0.48 Å. The matrix obtained from this superimposition was then used to transform the coordinates of BgII, and the displacement of equivalent atoms in the two proteins was then compared. Water positions are listed as in Table II.
aAttacking water molecule in the BgII structure.
Site 1. The manganese ion is coordinated by three protein ligands, the carboxylate groups of Asp55 and Glu65, and the main chain carbonyl of Thr66. The latter interaction corresponds to residue X of the PD...(D/E)XK motif, and is observed in a number of restriction enzymes. In addition, the hexacoordination of the ion is completed by three water molecules. The electron density map shows high occupancy at this site, and thus this site corresponds to the higher affinity ion binding observed by calorimetry.
Site 2. This ion is only coordinated by a single protein ligand, the remaining carboxylate oxygen atom of Asp55. The inner coordination sphere is completed by a variable number of water molecules. This site is not fully occupied in the crystal, and therefore corresponds to the weaker binding site. Using our measured dissociation constant of 14 mM for the second metal ion, we calculate an expected 78% occupancy in 50 mM manganese chloride used to soak the crystals. Thus there is very good correspondence between the solution results and those in the crystal.
Neither metal ion is directly coordinated to the side chain of Glu20, contributed by the other polypeptide of the resolving enzyme dimer, although there is a water-mediated (site III) contact with the metal ion bound to site 1. Thus this carboxylate group may have a more general electrostatic role; mutants in this residue are significantly impaired, although to a lesser extent than those in Asp55 or Glu65 (Déclais et al., 2001). However, it is fully possible that when the enzyme is bound to a DNA junction, there is some rearrangement of the metal ions, whereupon Glu20 might participate directly in ion binding.
In the light of these results, we can refine our view of the probable mechanism of endonuclease I. At this time, we have insufficient evidence to propose a complete mechanism, but we can identify some probable components. It is highly likely that the hydrolytic water molecule is coordinated by the metal ion bound in site 1, which would need to be located correctly to attack in-line. We note that the water molecule in the BglI structure implicated as carrying out the nucleophilic attack on the scissile phosphate (Wat4) is equivalent to the water molecule at position II in the endonuclease I structure. A number of potential functions could be ascribed to the metal ion bound to site 2. This could carry out electrophilic catalysis, possibly in conjunction with Lys67, helping to stabilize the negatively charged transition state. It could polarize the P–O bond, and a coordinated water molecule could act as a general acid to protonate the leaving group. The candidate attacking water molecule is also hydrogen bonded to the sulfate ion, which probably occupies the site where the phosphate of the DNA substrate that is 3′ to the cleavage site would be bound. This suggests that there could be substrate assistance, whereby the 3′-phosphate increases the nucleophilicity of the hydrolytic water molecule, as has been observed in a number of restriction enzymes (Jeltsch et al., 1993).
Ultimately, the structure of an endonuclease I–DNA complex will probably be required before we can identify conclusively the full mechanism by which endonuclease I hydrolyses specific phosphodiester bonds within the four-way DNA junction, but the major functionalities are probably now identified.
Materials and methods
Expression and purification of T7 endonuclease I
Wild-type endonuclease I was expressed using pET19endo I (Hadden et al., 2001). However, as the overexpressed protein was toxic to the host cells, it was necessary to modify the expression conditions from those previously reported (Hadden et al., 2001), in order to optimize the yield of protein. Escherichia coli (BL-21 DE3 pLysS; Novagen) was freshly transformed with pET19endo I and grown on LB agar containing 50 µg/ml ampicillin and 33 µg/ml chloramphenicol in a 9 cm diameter Petri dish at 37°C overnight. The entire contents of the Petri dish were then cut into small pieces and used to inoculate 250 ml of medium, which was then grown at 30°C to an absorbance of A600 = 0.4 and used to inoculate eight 2 l flasks, each containing 500 ml of medium. The protein was then expressed at 30°C and purified as previously described (Hadden et al., 2001). In order to remove the N-terminal His10 affinity tag, the protein was subjected to a mild tryptic digestion and purified further using anion exchange chromatography. The resulting protein, lacking its first 11 N-terminal amino acids, has been termed endonuclease I wt (ΔN11).
Crystallization of T7 endonuclease I
To grow crystals of endonuclease I wt (ΔN11), the protein was buffer exchanged into 20 mM Tris–HCl pH 8.0, 50 mM NaCl, and concentrated to ∼4 mg/ml. Hanging drop vapour diffusion experiments were set up by mixing 1 µl of protein solution with 1 µl of precipitant solution, 17–19% PEG 4000, 100 mM ammonium sulfate, 100 mM Tris–HCl pH 7.2. Drops were equilibrated against 500 µl of precipitant solution. Once the drops had been incubated for ∼3 h, they were streak seeded using extremely small crystals of endonuclease I (ΔN11, E65K) (Hadden et al., 2001). Small crystals could be seen within 24 h, and crystals grew to ∼400 µm in the largest dimension within 7 days.
Data collection
Prior to collecting data, crystals of wild-type endonuclease I were soaked in cryoprotectant (well solution containing 20% glycerol) for 7 min. For the metal-bound structure, the cryoprotectant additionally contained 50 mM manganese chloride. Crystals were then frozen immediately prior to data collection. Data were collected on station ID14 EH2, European Synchrotron Radiation Facility (ESRF) (metal-bound structure) or station 14.2 Daresbury SRS (metal-free structure). Data collection and processing statistics are shown in Table I.
Crystals of wild-type endonuclease I belong to the orthorhombic space group P21212 with unit cell dimensions a = 123.81 Å, b = 133.96 Å, c = 61.46 Å (metal-bound structure) or a = 123.44 Å, b = 134.55 Å, c = 61.39 Å (metal-free structure), and contain two dimers per asymmetric unit.
Data processing, model building and structure refinement
All data were integrated using MOSFLM (A.W.G.Leslie, MRC-LMB, Cambridge) and scaled using SCALA (CCP4, 1994). Approximately 95% of the data were used for refinement, whilst the remaining 5% were used to calculate Rfree. The program CNS (Brünger et al., 1998) was used to perform rigid body refinement to adjust our previously reported structure of endonuclease I (ΔN11, E65K, 2.1 Å) (Hadden et al., 2001) to the metal-bound protein data. With bulk solvent correction applied, data between 25 and 1.9 Å (metal-bound protein) or 33.15 and 2.55 Å (metal-free protein) were used in the refinement. Refinement proceeded with alternating use of CNS and O (Jones et al., 1991), and solvent molecules were added at positions with at least 3σ peaks in Fo – Fc maps and 1σ peaks in 2Fo – Fc maps. Final refinement statistics are given in Table I. A number of residues within the protein lacked any significant side chain electron density, and were built and refined as alanine residues (Table I). No significant electron density could be located for residues 11–16 and 146–149.
Analysis of cleavage of DNA junctions by T7 endonuclease I
The kinetics of junction cleavage were carried out with a QFM400 quench-flow apparatus (Bio-Logic, Grenoble, France), fitted with a 4 µl delay line and thermostatted at 20°C. Endonuclease I wt (ΔN11) at 80 nM was pre-incubated with 40 nM junction 3 radioactively 5′-32P-labelled on the x-strand in binding buffer [50 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EDTA, 800 µg/ml bovine serum albumin (BSA)] for at least 15 min to allow the complex to form. The reaction was initiated by mixing 15 µl of complex with 15 µl of 21 mM MgCl2 (or MnCl2) in EDTA-free binding buffer, and the reaction terminated at various time intervals by adding 15 µl of quenching buffer (120 mM EDTA). The exit line was washed with a mixture of 15 µl of binding buffer and 7.5 µl of quenching buffer to improve sample recovery. Samples were mixed with one volume of formamide, heat denatured and electrophoresed in a 15% polyacrylamide gel containing 7 M urea at 80 W for 45 min. The gel was dried onto Whatmann 3MM paper, exposed to storage phosphor screens (BAS-IP MP 2040; Fuji) and quantified using a BAS-1500 phosphoimager and the MacBAS v2.5 software. Data were analysed as fraction of DNA cleaved (fc) versus ageing time (ta), and were fitted by non-linear regression analysis to the equation:
Isothermal titration calorimetry
ITC measurements were carried out at 25°C using a VP-ITC titration calorimeter (MicroCal, Northampton, MD). All solutions were degassed before titrations. The endonuclease I wt (ΔN11) sample was dialysed extensively against 50 mM Tris–HCl pH 7.5, 400 mM NaCl. The high background concentration of monovalent cations was used to reduce non-specific electrostatic interactions with the negatively charged amino acids on the surface of the protein and to reduce heat effects due to the dilution of manganese ions. The 50 mM manganese chloride solution was prepared by dissolving the salt in the dialysis buffer. The titration was carried out using a 370 µl syringe, with stirring at 300 r.p.m. Each titration consisted of a preliminary 1 µl injection, followed by 22 subsequent injections of various increasing volumes (10 × 4 µl, 5 × 6 µl, 7 × 10 µl) into a cell containing ∼1.4 ml of a 343 µM endonuclease I sample. Calorimetric data were analysed using MicroCal ORIGIN software. To correct for dilution and mixing effects, a series of control injections was carried out, in which manganese chloride was injected into buffer alone. The heat signal of this control was then subtracted from the raw data for endonuclease I.
Coordinates
Coordinates for wild-type endonuclease I have been deposited in the Protein Data Bank with accession codes 1M0D (metal-bound protein) and 1M0I (metal-free protein).
Acknowledgements
We would like to thank the staff at the ESRF and the Daresbury SRS for assistance with data collection. We are grateful to Cancer Research UK (Dundee) and the Wellcome Trust (Leeds) for financial support and for facilities provided by the BBSRC-funded North of England Structural Biology Center (NESBIC).
References
- Ariyoshi M., Vassylyev,D.G., Iwasaki,H., Nakamura,H., Shinagawa,H. and Morikawa,K. (1994) Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E.coli. Cell, 78, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Ban C. and Yang,W. (1998) Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J., 17, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt E.L., Sharples,G.J. and Lloyd,R.G. (1999) Identification of three aspartic acid residues essential for catalysis by the RusA Holliday junction resolvase. J. Mol. Biol., 286, 403–415. [DOI] [PubMed] [Google Scholar]
- Bond C.S., Kvaratskhelia,M., Richard,D., White,M.F. and Hunter,W.N. (2001) Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc. Natl Acad. Sci. USA, 98, 5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cheng X.D., Balendiran,K., Schildkraut,I. and Anderson,J.E. (1994) Structure of PvuII endonuclease with cognate DNA. EMBO J., 13, 3927–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher J.A. (1998) The Spock Homepage. http://quorum.tamu.edu/spock/
- Connolly B., Parsons,C.A., Benson,F.E., Dunderdale,H.J., Sharples,G.J., Lloyd,R.G. and West,S.C. (1991) Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc. Natl Acad. Sci. USA, 88, 6063–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A., Davies,A.A. and West,S.C. (2001) Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell, 104, 259–268. [DOI] [PubMed] [Google Scholar]
- Cowan J.A., Ohyama,T., Howard,K., Rausch,J.W., Cowan,S.M. and Le Grice,S.F. (2000) Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J. Biol. Inorg. Chem., 5, 67–74. [DOI] [PubMed] [Google Scholar]
- de Massey B., Studier,F.W., Dorgai,L., Appelbaum,F. and Weisberg,R.A. (1984) Enzymes and the sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage λ. Cold Spring Harb. Symp. Quant. Biol., 49, 715–726. [DOI] [PubMed] [Google Scholar]
- Decanniere K., Babu,A.M., Sandman,K., Reeve,J.N. and Heinemann,U. (2000) Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J. Mol. Biol., 303, 35–47. [DOI] [PubMed] [Google Scholar]
- Déclais A.-C., Hadden,J.M., Phillips,S.E.V. and Lilley,D.M.J. (2001) The active site of the junction-resolving enzyme T7 endonuclease I. J. Mol. Biol., 307, 1145–1158. [DOI] [PubMed] [Google Scholar]
- Duckett D.R., Giraud-Panis,M.-E. and Lilley,D.M.J. (1995) Binding of the junction-resolving enzyme bacteriophage T7 endonuclease I to DNA: separation of binding and catalysis by mutation. J. Mol. Biol., 246, 95–107. [DOI] [PubMed] [Google Scholar]
- Garcia A.D., Aravind,L., Koonin,E. and Moss,B. (2000) Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl Acad. Sci. USA, 97, 8926–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis M.-J.E. and Lilley,D.M.J. (1996) T4 endonuclease VII: importance of a histidine–aspartate cluster within the zinc-binding domain. J. Biol. Chem., 271, 33148–33155. [DOI] [PubMed] [Google Scholar]
- Giraud-Panis M.-J.E. and Lilley,D.M.J. (1998) Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J. Mol. Biol., 278, 117–133. [DOI] [PubMed] [Google Scholar]
- Hadden J.M., Convery,M.A., Déclais,A.-C., Lilley,D.M.J. and Phillips,S.E.V. (2001) Crystal structure of the Holliday junction-resolving enzyme T7 endonuclease I at 2.1 Å resolution. Nat. Struct. Biol., 8, 62–67. [DOI] [PubMed] [Google Scholar]
- Hickman A.B., Li,Y., Mathew,S.V., May,E.W., Craig,N.L. and Dyda,F. (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell, 5, 1025–1034. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi,M., Shiba,T., Nakata,A. and Shinagawa,H. (1991) Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J., 10, 4381–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A., Alves,J., Wolfes,H., Maass,G. and Pingoud,A. (1993) Substrate-assisted catalysis in the cleavage of DNA by the EcoRI and EcoRV restriction enzymes. Proc. Natl Acad. Sci. USA, 90, 8499–8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jose T.J., Conlan,L.H. and Dupureur,C.M. (1999) Quantitative evaluation of metal ion binding to PvuII restriction endonuclease. J. Biol. Inorg. Chem., 4, 814–23. [DOI] [PubMed] [Google Scholar]
- Kemper B. and Garabett,M. (1981) Studies on T4 head maturation. 1. Purification and characterisation of gene-49-controlled endonuclease. Eur. J. Biochem., 115, 123–131. [PubMed] [Google Scholar]
- Komori K., Sakae,S., Shinagawa,H., Morikawa,K. and Ishino,Y. (1999) A Holliday junction resolvase from Pyrococcus furiosus: functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in bacteria, eukarya and archaea. Proc. Natl Acad. Sci. USA, 96, 8873–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D. and Winkler,F.K. (1995) Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 Å resolution. Biochemistry, 34, 683–696. [DOI] [PubMed] [Google Scholar]
- Kovall R.A. and Matthews,B.W. (1998) Structural, functional and evolutionary relationships between λ-exonuclease and the type II restriction endonucleases. Proc. Natl Acad. Sci. USA, 95, 7893–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaratskhelia M. and White,M.F. (2000) An archaeal Holliday junction resolving enzyme from Sulfolobus solfataricus exhibits unique properties. J. Mol. Biol., 295, 193–202. [DOI] [PubMed] [Google Scholar]
- Kvaratskhelia M., George,S.J., Cooper,A. and White,M.F. (1999) Quantitation of metal ion and DNA junction binding to the Holliday junction endonuclease Cce1. Biochemistry, 38, 16613–16619. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.W. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lilley D.M.J. and White,M.F. (2000) Resolving the relationships of resolving enzymes. Proc. Natl Acad. Sci. USA, 97, 9351–9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D.M.J. and White,M.F. (2001) The junction-resolving enzymes. Nat. Rev. Mol. Cell. Biol., 2, 433–443. [DOI] [PubMed] [Google Scholar]
- Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A.K. (2000) Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 Å resolution. Nat. Struct. Biol., 7, 134–140. [DOI] [PubMed] [Google Scholar]
- Makarova K.S., Aravind,L. and Koonin,E.V. (2000) Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res., 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M., Strzelecka,T., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1995) Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science, 269, 656–663. [DOI] [PubMed] [Google Scholar]
- Newman M., Lunnen,K., Wilson,G., Greci,J., Schildkraut,I. and Phillips,S.E.V. (1998) Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J., 17, 5466–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Komori,K., Tsuchiya,D., Ishino,Y. and Morikawa,K. (2001) Crystal structure of the archaeal Holliday junction resolvase Hjc and implications for DNA recognition. Structure, 9, 197–204. [DOI] [PubMed] [Google Scholar]
- Parkinson M.J., Pöhler,J.R.G. and Lilley,D.M.J. (1999) Catalytic and binding mutants of the junction-resolving enzyme endonuclease I of bacteriophage T7: role of acidic residues. Nucleic Acids Res., 27, 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers H., Vix,O., Toro,I., Golz,S., Kemper,B. and Suck,D. (1999) X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J., 18, 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Iwasaki,H., Ariyoshi,M., Morikawa,K. and Shinagawa,H. (1995) Identification of four acidic amino acids that constitute the catalytic centre of the RuvC Holliday junction resolvase. Proc. Natl Acad. Sci. USA, 92, 7470–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. and Kolodner,R. (1985) Partial purification of an endonuclease from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc. Natl Acad. Sci. USA, 82, 7247–7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardleworth B.N., Kvaratskhelia,M. and White,M.F. (2000) Site-directed mutagenesis of the yeast resolving enzyme Cce1 reveals catalytic residues and relationship with the intron-splicing factor Mrs1. J. Biol. Chem., 275, 23725–23728. [DOI] [PubMed] [Google Scholar]
- West S.C., Parsons,C.A. and Picksley,S.M. (1987) Purification and properties of a nuclease from Saccharomyces cerevisiae that cleaves DNA at cruciform junctions. J. Biol. Chem., 262, 12752–12758. [PubMed] [Google Scholar]
- White M.F. and Lilley,D.M.J. (1997) Characterization of a Holliday junction resolving enzyme from Schizosaccharomyces pombe. Mol. Cell. Biol., 17, 6465–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman T., Williston,S., Brandts,J.F. and Lin,L.N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem., 179, 131–137. [DOI] [PubMed] [Google Scholar]