Abstract
Replication through (6–4)TT and G-AAF lesions was compared in Saccharomyces cerevisiae strains proficient and deficient for the RAD30-encoded DNA polymerase η (Pol η). In the RAD30 strain, the (6–4)TT lesion is replicated both inaccurately and accurately 60 and 40% of the time, respectively. Surprisingly, in a rad30Δ strain, the level of mutagenic bypass is essentially suppressed, while error-free bypass remains unchanged. Therefore, Pol η is responsible for mutagenic replication through the (6–4)TT photoproduct, while another polymerase mediates its error-free bypass. Deletion of the RAD30 gene also reduces the levels of both accurate and inaccurate bypass of AAF lesions within two different sequence contexts up to 8-fold. These data show that, in contrast to the accurate bypass by Pol η of TT cyclobutane dimers, it is responsible for the mutagenic bypass of other lesions. In conclusion, this paper shows that, in yeast, translesion synthesis involves the combined action of several polymerases.
Keywords: AAF adducts/Pol η/semi-targeted frameshifts/translesion synthesis/(6–4)TT photoproduct
Introduction
Mutations in the human RAD30A gene encoding DNA polymerase η (hPol η) were found to be responsible for an autosomal recessive disease, namely the variant form of xeroderma pigmentosum (XP-V), characterized by a high incidence of sunlight-induced skin cancer (Johnson et al., 1999a; Masutani et al., 1999). Pol η belongs to the recently identified Y-family of DNA polymerases (Ohmori et al., 2001) characterized by their low fidelity and their ability to read across lesions in DNA templates. At first sight, it may appear paradoxical that the inactivation of a low fidelity DNA polymerase, such as Pol η in XP-V cells, renders cells hypermutable to UV light. This paradox is best understood on the basis of the capacity of Pol η to read efficiently through the major UV lesion, a TT cyclobutane dimer, by inserting two adenines across from the dimer, thus restoring the correct sequence (Johnson et al., 1999b; Masutani et al., 1999). In its absence, another as yet unidentified DNA polymerase copies past UV lesions, introducing an increased number of point mutations in XP-V cells. On this basis, Pol η has been regarded as being a DNA polymerase capable of bypassing DNA lesions without introducing mutations. Although this appears to be the case for the cis–syn TT dimer, it is of course unlikely that Pol η can perform such a function for any type of lesion. Numerous studies investigating the capacity of purified yeast or human Pol η to elongate in vitro oligonucleotide primers annealed to templates containing various DNA lesions have been published and have indeed shown that, for some lesions, Pol η is reasonably accurate, while for others it is clearly mutagenic (Haracska et al., 2000; Masutani et al., 2000; Yuan et al., 2000; Zhang et al., 2000; Johnson et al., 2001; Kuraoka et al., 2001; Minko et al., 2001). The physiological significance of such in vitro studies is also questionable as multiple DNA polymerases and various accessory proteins are present simultaneously in vivo, making the process of translesion synthesis (TLS) far more complex than a simple primer elongation reaction. Analysis of TLS in vivo is made possible by the use of double-stranded plasmids containing a single adduct located within a short heteroduplex sequence (Figure 1; Koffel-Schwartz et al., 1996; Baynton et al., 1998, 1999). In this paper, we have implemented this strategy to study the role of Pol η in the bypass of a pyrimidine (6–4) pyrimidone photoproduct, namely (6–4)TT, and of the N-2-acetylaminofluorene (AAF) adduct to the C8 position of guanine (G-AAF). The present results show that Pol η participates in the replication of both lesions in combination with at least one additional translesion polymerase, namely Pol ζ. In addition, mutagenic bypass of the (6–4)TT photoproduct is unequivocally attributed to Pol η.
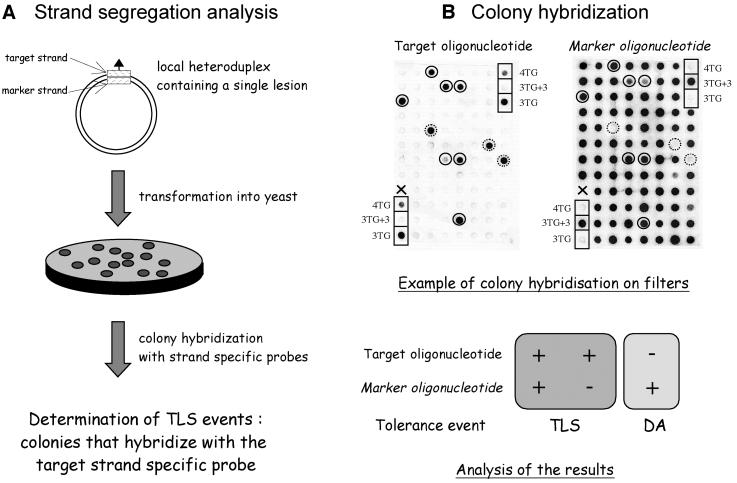
Fig. 1. Determination of TLS in vivo. Double-stranded plasmids carrying a single adduct (triangle), located within a short heteroduplex region (hatched rectangles), are constructed as described in Materials and methods, and used for strand segregation analysis as shown in (A). The sequence context of the heteroduplex region is shown in Figure 2 for the different lesion bypass assays implemented in the present paper. In (B), an example of a filter containing colonies obtained following transformation of plasmid pKB-3TG in wild-type yeast cells is shown. The left and right images are autoradiographs obtained upon hybridization with the target (3TG probe) and marker (3TG+3 probe) oligonucleotides, respectively. Control colonies transformed with plasmids pKB-4TG, pKB-3TG+3 and pKB-3TG are shown in boxes. Plasmid pKB-4TG is derived from plasmid pKB-3TG containing an additional T residue within sequence 5′-TTTG, thus mimicking the +1 mutation induced by AAF adducts in this sequence context (see Figure 2). As expected, the colonies containing control plasmid pKB-3TG+3 light up with probe 3TG+3 only. Conversely, colonies containing control plasmids pKB-3TG and pKB-4TG light up strongly and weakly with the 3TG probe only, respectively. Among the colonies analyzed, seven colonies (circles with solid outline) hybridize with both target and marker strand probes, suggesting that TLS occurred during replication. Three colonies (circles with broken outline) hybridize with target strand probe only, suggesting that TLS occurred during a gap-filling event. These latter TLS events most likely occur during gap filling of excision tracks generated during mismatch repair and have been shown to exhibit the same genetic requirements as TLS events that occur during replication (Baynton et al., 1998; data not shown). All other colonies responding to probe 3TG+3 only are scored as damage-avoidance events. A single colony marked with an X failed to grow (no signal with either probe).
Results and discussion
Aim and strategy
The aim of this paper is to investigate the involvement of Pol η in the bypass of (6–4)TT photoproducts and G-AAF adducts in vivo. The (6–4)TT photoproduct is a UV-induced lesion known for its capacity to induce base substitution mutations (Gibbs et al., 1995), while G-AAF adducts mainly induce frameshift mutations (Fuchs et al., 1981; Koffel-Schwartz et al., 1984; Roy and Fuchs, 1994). Our assay allows for the determination of both error-free and mutagenic bypass events (Koffel-Schwartz et al., 1996; Baynton et al., 1998, 1999). Quantification of TLS is made possible by the use of double-stranded plasmids that contain a single adduct located within a short heteroduplex sequence. The strand containing the lesion and the complementary strand are called the target and marker strands, respectively (Figures 1A and 2). Following transformation of the heteroduplex plasmid and replication in yeast cells, the presence, in individual colonies, of plasmids derived from the target and/or marker strands is detected by means of oligonucleotide hybridization with strand-specific probes (Figure 1B). Colonies that hybridize with the target strand probe are scored as colonies in which TLS has occurred. Colonies that hybridize only with the marker strand-specific probe (Figure 1B) result from damage-avoidance pathways (Baynton and Fuchs, 2000). Mutagenic TLS events are detected phenotypically on the basis of their Lac+ phenotype and confirmed by sequencing as described in Materials and methods.
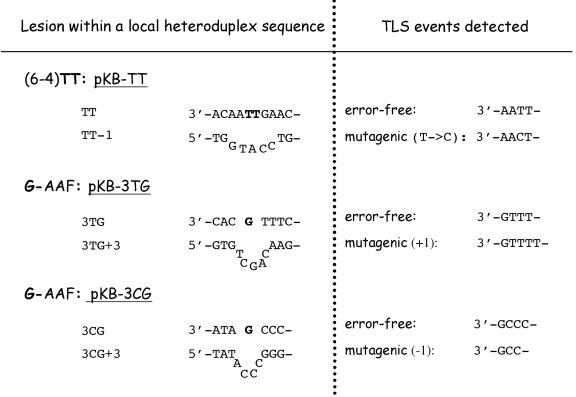
Fig. 2. Specific lesion bypass assays. The location of the adduct within the target strand and the sequence of the marker strand are shown for all three lesion bypass assays that are implemented in the present work. The damaged nucleotides are shown in bold. The sequences of the error-free and mutagenic TLS events that are detected for each assay are shown.
Bypass of a single (6–4)TT lesion in vivo
A plasmid containing a single pyrimidine (6–4) pyrimidone photoproduct [(6–4)TT] (Figure 2) was introduced into yeast cells by electroporation in order to analyse its replication in vivo. The proportion of transformed yeast colonies resulting from error-free and mutagenic bypass of the photoproduct was determined by colony hybridization followed by sequencing as described in Materials and methods. In the RAD30 strain, colonies in which TLS occurred represent ∼4% of the transformants, the majority (96%) of the transformed colonies containing only plasmids with the marker strand sequence. These latter colonies arise from damage-avoidance mechanisms and will not be discussed further here. Among the 4% of colonies resulting from TLS, 1.6% are error-free replication products, while the remaining 2.4% are mutagenic bypass events all corresponding to a single 3′-T→C transition (Figure 3). Among the TLS events, mutagenic bypass represents 60%, a proportion higher than the 30% previously determined in yeast for a (6–4)TT lesion located in a different sequence context (Gibbs et al., 1995; Nelson et al., 2000).
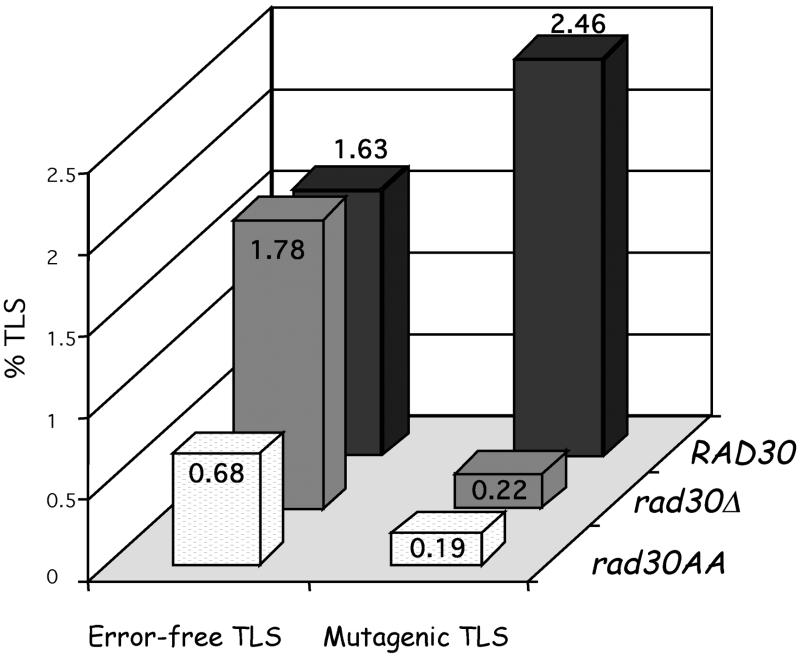
Fig. 3. Bypass of a (6–4)TT photoproduct in S.cerevisiae wild-type and rad30 mutant strains. Error-free and mutagenic TLS events are determined as described in Materials and methods. Mutagenic TLS (T→C transitions) is strongly reduced either when Pol η is absent (rad30Δ) or catalytically defective (rad30AA). Error-free TLS is unaffected in the Pol η deletion strain, but is significantly reduced in the catalytically defective Pol η strain as if the mutant protein is able to compete with the DNA polymerase that mediates error-free bypass. Over 1000 colonies were analyzed by hybridization in each strain.
Pol η mediates mutagenic (3′-T→C) bypass of the (6–4)TT photoproduct. Surprisingly, in the rad30Δ strain, the level of error-free TLS remains constant, while mutagenic bypass products decrease ∼10-fold when compared with the corresponding wild-type background (Figure 3). A similar decrease in mutagenic TLS (Figure 3) is observed in a strain containing a two-amino-acid substitution in the catalytic domain of Pol η (strain rad30AA), which inactivates its DNA polymerase activity (Johnson et al., 1999c). These data strongly suggest that, in vivo, Pol η ‘misinserts’ G opposite the 3′-T of the (6–4)TT photoproduct. Based on the severe reduction of the UV-induced reversion frequency in a rad30Δ strain at the arg4-17 locus, Zhang and Siede (2002) recently suggested that Pol η might either be responsible for an unprecedented error-prone processing of a TT cyclobutane dimer in that sequence context or for mutagenic replication of a (6–4)TT photoproduct. Whereas their photoreactivation experiments hinted at the former, our present data clearly validate the latter hypothesis.
These in vivo data are in good agreement with both structural and biochemical data. Indeed, (6–4)TT photoproducts are known to induce large structural distortions in DNA, the 3′-T in the (6–4) lesion being oriented perpendicular to the 5′-T (Kim and Choi, 1995; Kim et al., 1995). As a consequence, a G residue can form hydrogen bonds with the 3′-T of a (6–4)TT photoproduct (Lee et al., 1999). In vitro data, involving primer template elongation experiments with a single (6–4)TT lesion, have shown that both yeast and human Pol η insert G opposite the 3′-T 8-fold more efficiently than A (Zhang et al., 2000; Johnson et al., 2001).
Error-free bypass of the (6–4)TT photoproduct is mediated by another polymerase. As error-free bypass products are detected to a similar level in both the wild-type and rad30Δ strains, another DNA polymerase is able to insert the ‘correct’ A nucleotide across the 3′-T, yielding an error-free bypass product (Figure 3). Curiously, error-free TLS is significantly decreased in the rad30AA strain (Figure 3), suggesting that the catalytically inactive form of Pol η may in fact interfere with the error-free pathway. The nature of the DNA polymerase that inserts A across the 3′-T remains to be determined.
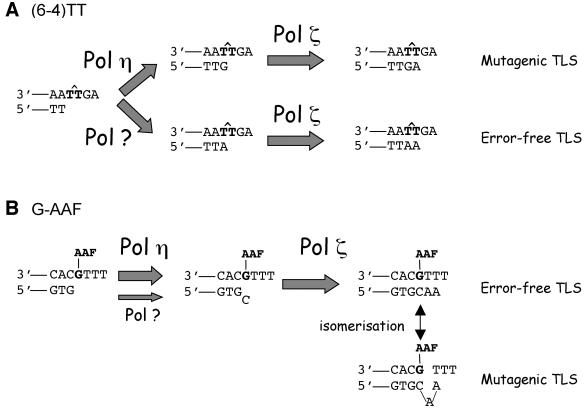
Our data show that two distinct DNA polymerases (Pol η and another DNA Pol) can gain access to the primer template and compete at the sole insertion step across the 3′-T of the (6–4)TT photoproduct. Similar observations have been made in Escherichia coli for several lesion pathways (Becherel and Fuchs, 2001; Wagner et al., 2002). In addition, the post-insertion step(s) during (6–4)TT photoproduct bypass is most likely mediated by Pol ζ, encoded by the REV3 and REV7 genes, as supported by in vivo data showing that the level of bypass of this lesion is <1% in a rev3Δ strain (Nelson et al., 2000). Pol ζ has indeed been shown to extend (6–4)TT replication intermediates containing a nucleotide across the 3′-T efficiently and accurately (Johnson et al., 2001). A model involving Pol η and another DNA polymerase at the insertion step and Pol ζ at the extension step(s) is presented in Figure 4A.
Fig. 4. Model for the bypass of (6–4)TT and G-AAF lesions in S.cerevisiae. (A) (6–4)TT. Data presented here suggest that, in vivo, Pol η inserts G ‘incorrectly’, while another polymerase inserts A ‘correctly’ opposite the 3′-T of the (6–4)TT lesion. In a wild-type background, both events occur at similar frequencies: 60% of misinsertion versus 40% of error-free insertion. Following this insertion step, both replication intermediates are likely to be extended by Pol ζ, yielding mutagenic and error-free bypass products, respectively (see text for discussion). (B) G-AAF. Within the 3TG sequence context, G-AAF adducts are mostly bypassed in an error-free manner. However, the adduct may also trigger a slippage event in the primer strand during replication of three Ts located 5′ to the adduct, thus yielding a +T frameshift mutation (semi-targeted frameshift event). As inactivation of Pol η suppresses 75% of all bypass events, we suggest that Pol η is involved mainly at the insertion step, and Pol ζ at the extension steps, yielding both error-free and frameshift bypass events. In the absence of Pol η, yet another DNA polymerase appears to perform limited insertion, as shown by the thin arrow (see text for further details).
Bypass of a single G-AAF lesion in vivo
AAF is a chemical carcinogen that predominantly forms adducts at the C8 position of guanine residues (Kriek et al., 1967). This adduct introduces a major local perturbation in the DNA structure by triggering an anti→syn conformational change of the adducted G-AAF nucleoside (Fuchs and Daune, 1972). Such adducts cause strong replication delays or blocks. However, when bypass occurs, C is almost always ‘accurately’ incorporated across from the G-AAF adduct by many different DNA polymerases (Belguise-Valladier et al., 1994; Lindsley and Fuchs, 1994; Masutani et al., 2000; Yuan et al., 2000). The apparent fidelity of DNA polymerases when replicating through a G-AAF adduct can be best understood by the fact that AAF binds the C8 position of guanine, while not altering the positions that base pair with cytosine. Sequence-dependent frameshift mutation hot spots have been identified in short runs of guanine-containing mono- or di-nucleotides (Fuchs et al., 1981; Koffel-Schwartz et al., 1984; Roy and Fuchs, 1994). For example, when a G-AAF adduct is located within a short run of Gs, the lesion terminus can form a slipped intermediate in which the terminal C in the primer strand base pairs with an undamaged G located 5′ to the adduct, yielding a frameshift intermediate with a looped-out adduct (Lambert et al., 1992; Veaute and Fuchs, 1993; Napolitano et al., 1997; Becherel and Fuchs, 2001). In addition to these targeted frameshift mutation hot spots, G-AAF lesions were also found to trigger frameshifts in short runs located immediately 5′ to the adducted G. These events have been referred to as semi-targeted frameshift events (Lambert et al., 1992). Using a forward mutation assay involving the inactivation of the URA3 gene in the yeast Saccharomyces cerevisiae, we have shown previously that randomly distributed G-AAF adducts efficiently trigger +1 frameshift mutations in short monotonous runs of AT base pairs, provided a G residue is located 3′ to the run (Roy and Fuchs, 1994). We hypothesized that the putative G-AAF adduct sufficiently perturbs replication to trigger a +1 slippage event in the short run of AT base pairs located downstream from the adduct. In order to prove that an AAF adduct can trigger semi-targeted frameshift mutations in nearby sequences, we constructed a double-stranded plasmid containing a single G-AAF adduct in a 5′-TTTG- sequence context (Figure 2). Introduction of a sequence heterology in the vicinity of the adduct site allows the extent of TLS to be determined, as discussed above for the UV photoproduct. While error-free bypass is determined by colony hybridization (Figure 1B), mutagenic (+1) events are detected by a phenotypic assay that involves lacZ expression in the yeast cells. Their molecular nature (+T) is confirmed by sequencing (see Materials and methods).
In the RAD30 strain, colonies in which the lesion in the 5′-TTTG- context was actually bypassed represent ∼7% of the transformants (Table I). As for the UV photoproduct, the majority of transformed colonies only contained plasmids with the marker strand sequence, thus representing damage-avoidance events. Among the transformed colonies analysed, 6.7 and 0.16% were error-free and +1 frameshift bypass products, respectively (Table I). In other words, among the colonies resulting from TLS of the G-AAF adduct, 2% [i.e. 0.16/(6.7 + 0.16) = 2%] carry a +T mutation within the run of 3 Ts that is located 5′ to the G-AAF adduct, while the remaining 98% correspond to error-free replication (Table I). In the rad30Δ strain, both error-free and mutagenic TLS events decrease by 75%, implying that Pol η plays a major role in the bypass of the G-AAF adduct in vivo (Table I). At the same time, these data point to the fact that, in the absence of Pol η, yet another DNA polymerase can mediate residual bypass (Figure 4B). A similar decrease in the extent of bypass has also been observed in the rad30AA strain (data not shown). We have previously shown that bypass of a G-AAF adduct, within several different sequence contexts, absolutely requires the REV3 and REV7 gene products (Baynton et al., 1998, 1999). In the present sequence context, the level of induced +1 frameshift mutations was also reduced >100-fold in a rev3Δ strain compared with the corresponding REV3 wild-type strain (data not shown). Taken together, these data indicate that G-AAF bypass requires the combined action of at least two translesion polymerases (see Conclusion).
Table I. Effect of Pol η on the bypass of AAF lesions in S.cerevisiae.
| Sequence context (mutation event) | Error-free TLS (%) |
Mutagenic TLS (%) |
||
|---|---|---|---|---|
| RAD30 | rad30Δ | RAD30 | rad30Δ | |
| 5′-TTTG- (+1T) | 6.7 (77/1151) | 1.6 (18/1143) | 0.16 (98/60 320) | 0.04 (23/51 307) |
| 5′-CCCG- (–1C) | 7.0 (79/1129) | 0.8 (9/1134) | 0.028 (9/31 448) | 0.004 (1/26 952) |
The base shown in bold indicates the location of the adduct. The numbers in parentheses represent the number of positive colonies over the total number of colonies. Error-free TLS fractions are measured by colony hybridization. Mutagenic TLS is determined phenotypically by a white (lacZ–) to blue (lacZ+) change in colony colour and the molecular nature of the mutation +T or –C in 5′-TTTG- and 5′-CCCG-, respectively, is confirmed by sequencing.
In the 5′-CCCG- context, error-free bypass is determined by colony hybridization (Figure 1B), while mutagenic (–1) events are detected phenotypically, as described above for the 5′-TTTG- context. Their molecular nature (–C) is confirmed by sequencing. Similarly to the model proposed above for semi-targeted +T mutations, bypass through the G-AAF adduct sufficiently perturbs replication to trigger a –1 slippage event in the short run of C nucleotides located immediately 5′ to the adduct (5′-CCCG-) (Figure 2). In the RAD30 strain, colonies resulting from error-free and mutagenic bypass represent 7 and 0.03% of the total transformants, respectively (Table I). In the rad30Δ strain, both error-free and mutagenic TLS events decrease 7- to 9-fold (Table I), implying that, as in the previous sequence context, Pol η plays a major role in the bypass of the G-AAF adduct in vivo, while another polymerase is still able to perform limited TLS in its absence.
Conclusion
‘Correct’ versus ‘incorrect’ bypass of lesions by Pol η
Within the context of this conclusion, we define a DNA lesion as a chemically altered base, although DNA lesions also include damage to the phosphodiester backbone. In most cases, a chemically damaged base loses its original coding properties, the most extreme example being the frequently formed abasic sites. How should we qualify the product resulting from the replication of a lesion by a DNA polymerase? The common usage qualifies ‘correct’ or ‘error free’ as when the nucleotide inserted across from the damaged base is that expected if the base were undamaged. Conversely, the replication product is referred to as ‘incorrect’ or ‘mutagenic’ when any other base is inserted across the lesion. Although this terminology may be convenient, it is obviously misleading as it tends to imply that polymerases know what is ‘correct’ or ‘incorrect’. We would like to suggest that DNA polymerases insert whatever base, among the four common bases, will structurally best fit across the lesion when positioned within the polymerase active site irrespective of Watson–Crick base pairing rules. Although Pol η may have been selected by evolution to insert As across the most abundant UV-induced lesion, the TT cyclobutane dimer, and thus prevent mutations caused by this slowly repaired lesion, it is not able to replicate ‘correctly’ any type of DNA lesion. Indeed, as shown here, the ‘incorrect’ bypass of the (6–4)TT lesions, yielding 3′-T→C transitions, depends upon Pol η, while another DNA polymerase appears to be able to insert the ‘correct’ A across the 3′-T (Figure 4A). In terms of avoiding UV mutagenesis, it is much more beneficial for a cell to replicate the abundant TT dimer accurately than the less frequent (6–4)TT photoproduct.
TLS often requires the combined action of several polymerases
Base substitution pathway. The bypass in vivo of (6–4)TT lesions yields both error-free and mutagenic (3′-T→C transition) replication products (Figure 4A). The insertion step is either ‘correct’ (insertion of A opposite T) or ‘incorrect’ (insertion of G opposite T), whereas the extension step of both intermediates appears to be accurate as A is inserted opposite the 5′-T in all cases. Pol ζ was shown to be required for the bypass of the (6–4)TT lesion in vivo (Nelson et al., 2000). We suggest that full bypass of the (6–4)TT lesion requires the combined action of two distinct DNA polymerases at the insertion step, Pol η for the mutagenic pathway and another polymerase for the error-free pathway, and Pol ζ at the extension steps (Figure 4A). This observation nicely illustrates the concept that several DNA polymerases can simultaneously gain access and compete for elongation of a given replication intermediate, as previously shown in E.coli (Becherel and Fuchs, 2001).
Frameshift pathways. Frameshift pathways induced by lesions such as G-AAF adducts entail an error-free insertion step of C opposite the adduct followed by a mis-elongation event of a slipped replication intermediate (Figure 4B; Lambert et al., 1992; Veaute and Fuchs, 1993; Napolitano et al., 1997; Becherel and Fuchs, 2001). The data presented here and in previous studies (Baynton et al., 1998, 1999) indicate that Pol η and Pol ζ play an important role in both error-free and mutagenic bypass of AAF adducts in vivo. In vitro, primer elongation experiments have shown that Pol η most efficiently inserts C across a single G-AAF adduct (Masutani et al., 2000; Yuan et al., 2000). On the other hand, the role of Pol ζ as a DNA polymerase specialized in extending distorted primer templates has been documented in vitro (Johnson et al., 2000; Haracska et al., 2001). Thus, in good agreement with the present genetic requirements for G-AAF bypass, the biochemical data suggest the combined action of Pol η and Pol ζ at the insertion and extension steps, respectively (Figure 4B).
The present study strongly suggests that the bypass of lesions such as (6–4)TT photoproducts and G-AAF adducts requires the combined action of several DNA polymerases in S.cerevisiae. Similarly, the combined role of yeast Pol δ and ζ and of Rev1 during the bypass of abasic sites in yeast has been suggested (Haracska et al., 2001). In E.coli, various examples of mutation pathways with requirements for a specific combination of DNA polymerases and, in contrast, other examples where two DNA polymerases exhibit functional redundancy within the same pathway have been reported (Napolitano et al., 2000; Becherel and Fuchs, 2001; Fuchs et al., 2001; Wagner et al., 2002).
Materials and methods
Yeast strains, bacterial strains and media
The yeast strains EMY74-7 (MATa his3Δ100, leu2-3,112, trp1Δ, ura3-52) carrying a RAD30, rad30Δ or rad30AA allele were kindly provided by L.Prakash (University of Texas, Galveston, TX). The two last strains are rad30 deleted and carry either a CEN LEU2 vector with no insert or the same plasmid containing the rad30D155A–E156A mutations (Johnson et al., 1999c). Bacterial strains used for mutational and sequencing analysis and conditions for transformation of both organisms are as described elsewhere (Baynton et al., 1998, 1999).
Plasmids with single lesions
Plasmids containing single lesions, either a (6–4)TT photoproduct or a G-AAF adduct, were constructed as described previously (Koehl et al., 1989; Baynton et al., 1998). Briefly, an oligonucleotide containing the single lesion is ligated into a gapped duplex structure to form a covalently closed circular plasmid that is subsequently isolated by centrifugation on CsCl/ethidium bromide gradients. Plasmid pKB-(6–4)TT is obtained by inserting a 13mer oligonucleotide containing a (6–4)TT photoproduct (Becherel and Fuchs, 1999) into a gapped duplex structure forming a local heteroduplex, as shown in Figure 2. Similarly, an AAF adduct was introduced in two distinct sequence contexts, pKB-3TG-AAF and pKB-3CG-AAF, using oligonucleotides with a single G-AAF (G*) residue: 5′-ATCACTTTG*CACACT-3′ (15mer) and 5′-TCATACCC G*ATACATCA-3′ (17mer), respectively. Previously, they were purified by reverse-phase HPLC chromatography (Koehl et al., 1989). The local heteroduplex structure formed by the lesion-containing (target strand) and marker strands is shown in Figure 2.
Determination of TLS in vivo
Following transformation and replication in yeast cells of the plasmids containing the single lesion, the presence of the target and marker strands is detected in individual colonies using the strand segregation analysis (SSA) assay as described elsewhere (Baynton et al., 1998, 1999). The SSA assay involves colony hybridization with oligonucleotides specific either for the target or marker strands (Figures 1A and 2). (6–4)TT construction: TT probe 5′-AGTCGCAAGTTAACACGGAC-3′ and TT-1 probe 5′-GTCGCAGGTACCACGGACTA-3′; 3TG construction: 3TG probe 5′-TATCACTTTGCACACTAAACGT-3′ and 3TG+3 probe 5′-TCA CTTGTCGACACACTAAACG-3′; 3CG construction: 3CG probe 5′- GTTTTCATACCCGATACATC-3′ and 3CG+3 probe 5′-GTTTTC ATACCCGGGATAACATC-3′. All colonies hybridizing with the probe specific for the target strand were scored as TLS events (Figure 1B). For the (6–4)TT photoproduct, the plasmid progeny present in TLS-positive yeast colonies was analyzed following re-transformation in E.coli and sequencing in order to determine the extent and molecular nature of the mutagenic versus error-free bypass. For the G-AAF adducts, in both 3TG and 3CG sequence contexts, the extent of mutagenic TLS was determined phenotypically. Indeed, the single G-AAF adduct is located within the bacterial lacZ coding sequence expressed from the yeast LEU2 promoter (Baynton et al., 1999). The constructions are such that the lacZ gene is out of frame by –1 and +1 nucleotide in constructions pKB-3TG and pKB-3CG, allowing +1 and –1 frameshift mutations to be scored on indicator plates, respectively. The β-galactosidase activity in lacZ+ revertant yeast colonies is detected by overlaying yeast plates with X-gal-containing agarose (Boy-Marcotte et al., 1993). Plasmids isolated from independent colonies were sequenced and found to contain +1T or –1C frameshift events for pKB-3TG and pKB-3CG, respectively.
Acknowledgments
Acknowledgements
We would like to collectively acknowledge the members of the laboratory for their constructive reading of the manuscript. Special thanks are due to Adeline Martz for technical help during her internship in the laboratory.
References
- Baynton K. and Fuchs,R.P.P. (2000) Lesions in DNA: hurdles for polymerases. Trends Biochem. Sci., 25, 74–79. [DOI] [PubMed] [Google Scholar]
- Baynton K., Bresson-Roy,A. and Fuchs,R.P. (1998) Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol. Cell. Biol., 18, 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynton K., Bresson-Roy,A. and Fuchs,R.P. (1999) Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol. Microbiol., 34, 124–133. [DOI] [PubMed] [Google Scholar]
- Becherel O.J. and Fuchs,R.P. (1999) SOS mutagenesis results from up-regulation of translesion synthesis. J. Mol. Biol., 294, 299–306. [DOI] [PubMed] [Google Scholar]
- Becherel O.J. and Fuchs,R.P. (2001) Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc. Natl Acad. Sci. USA, 98, 8566–8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belguise-Valladier P., Maki,H., Sekiguchi,M. and Fuchs,R.P.P. (1994) Effect of single DNA lesions on in vitro replication with DNA polymerase III holoenzyme: comparison with other polymerases. J. Mol. Biol., 236, 151–164. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E., Buu,A., Soustelle,C., Poullet,P., Parmeggiani,A. and Jacquet,M. (1993) The C-terminal part of the CDC25 gene product has Ras-nucleotide exchange activity when present in a chimeric SDC25–CDC25 protein. Curr. Genet., 23, 397–401. [DOI] [PubMed] [Google Scholar]
- Fuchs R.P.P. and Daune,M.P. (1972) Physical studies on deoxy ribonucleic acid after covalent binding of a carcinogen. Biochemistry, 11, 2659–2666. [DOI] [PubMed] [Google Scholar]
- Fuchs R.P.P., Schwartz,N. and Daune,M.P. (1981) Hot spots of frameshift mutations induced by the ultimate carcinogen N-acetoxy-N-2-acetylaminofluorene. Nature, 294, 657–659. [DOI] [PubMed] [Google Scholar]
- Fuchs R.P., Koffel-Schwartz,N., Pelet,S., Janel-Bintz,R., Napolitano,R., Becherel,O.J., Broschard,T.H., Burnouf,D.Y. and Wagner,J. (2001) DNA polymerases II and V mediate respectively mutagenic (–2 frameshift) and error-free bypass of a single N-2-acetylaminofluorene adduct. Biochem. Soc. Trans., 29, 191–195. [DOI] [PubMed] [Google Scholar]
- Gibbs P.E., Borden,A. and Lawrence,C.W. (1995) The T–T pyrimidine (6–4) pyrimidinone UV photoproduct is much less mutagenic in yeast than in Escherichia coli. Nucleic Acids Res., 23, 1919–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Prakash,S. and Prakash,L. (2000) Replication past O(6)-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol., 20, 8001–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Unk,I., Johnson,R.E., Johansson,E., Burgers,P.M., Prakash,S. and Prakash,L. (2001) Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev., 15, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999a) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999b) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999c) Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem., 274, 15975–15977. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Haracska,L., Prakash,S. and Prakash,L. (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature, 406, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Haracska,L., Prakash,S. and Prakash,L. (2001) Role of DNA polymerase η in the bypass of a (6–4) TT photoproduct. Mol. Cell. Biol., 21, 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.K. and Choi,B.S. (1995) The solution structure of DNA duplex-decamer containing the (6–4) photoproduct of thymidylyl(3′→5′) thymidine by NMR and relaxation matrix refinement. Eur. J. Biochem., 228, 849–854. [DOI] [PubMed] [Google Scholar]
- Kim J.K., Patel,D. and Choi,B.S. (1995) Contrasting structural impacts induced by cis–syn cyclobutane dimer and (6–4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity. Photochem. Photobiol., 62, 44–50. [DOI] [PubMed] [Google Scholar]
- Koehl P., Burnouf,D. and Fuchs,R.P.P. (1989) Construction of plasmids containing a unique acetylaminofluorene adduct located within a mutation hot spot. J. Mol. Biol., 207, 355–364. [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Verdier,J.M., Bichara,M., Freund,A.M., Daune,M.P. and Fuchs,R.P.P. (1984) Carcinogen induced mutation spectrum in wild type, uvrA and umuC strains in E.coli: strain specificity and mutation prone sequences. J. Mol. Biol., 177, 33–51. [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Coin,F., Veaute,X. and Fuchs,R.P.P. (1996) Cellular strategies for accommodating replication-hindering adducts in DNA: control by the SOS response in E.coli. Proc. Natl Acad. Sci. USA, 93, 7805–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriek E., Miller,J.A., Juhl,W. and Miller,E.C. (1967) 8-(N-2-fluorenyl acetamide) guanosine and arylamidation reaction product of guanosine and the carcinogen N-acetoxy-N-2-fluorenylacetamide in neutral solution. Biochemistry, 6, 177–182. [DOI] [PubMed] [Google Scholar]
- Kuraoka I., Robins,P., Masutani,C., Hanaoka,F., Gasparutto,D., Cadet,J., Wood,R.D. and Lindahl,T. (2001) Oxygen free-radical damage to DNA. J. Biol. Chem., 276, 49283–49288. [DOI] [PubMed] [Google Scholar]
- Lambert I.B., Napolitano,R.L. and Fuchs,R.P.P. (1992) Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc. Natl Acad. Sci. USA, 89, 1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Hwang,G.S. and Choi,B.S. (1999) Solution structure of a DNA decamer duplex containing the stable 3′ TG base pair of the pyrimidine(6–4)pyrimidone photoproduct [(6–4) adduct]: implica tions for the highly specific 3′ T→C transition of the (6–4) adduct. Proc. Natl Acad. Sci. USA, 96, 6632–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley J.E. and Fuchs,R.P.P. (1994) Use of single-turnover kinetics to study adduct bypass by T7 DNA polymerase. Biochemistry, 33, 764–772. [DOI] [PubMed] [Google Scholar]
- Masutani C. et al. (1999) The XPV (xeroderma pigmentosum variant) gene encoded human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I.G., Washington,M.T., Prakash,L., Prakash,S. and Lloyd,R.S. (2001) Translesion DNA synthesis by yeast DNA polymerase η on templates containing N2-guanine adducts of 1,3-butadiene metabol ites. J. Biol. Chem., 276, 2517–2522. [DOI] [PubMed] [Google Scholar]
- Napolitano R.L., Lambert,I.B. and Fuchs,R.P.P. (1997) SOS factors involved in translesion synthesis. Proc. Natl Acad. Sci. USA, 94, 5733–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R., Janel-Bintz,R., Wagner,J. and Fuchs,R.P. (2000) All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J., 19, 6259–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.R., Gibbs,P.E., Nowicka,A.M., Hinkle,D.C. and Lawrence,C.W. (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol., 37, 549–554. [DOI] [PubMed] [Google Scholar]
- Ohmori H. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- Roy A. and Fuchs,R.P. (1994) Mutational spectrum induced in Saccharomyces cerevisiae by the carcinogen N-2-acetylamino fluorene. Mol. Gen. Genet., 245, 69–77. [DOI] [PubMed] [Google Scholar]
- Veaute X. and Fuchs,R.P.P. (1993) Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science, 261, 598–600. [DOI] [PubMed] [Google Scholar]
- Wagner J., Etienne,H., Janel-Bintz,R. and Fuchs,R.P.P. (2002) Genetics of mutagenesis in E.coli: various combinations of translesion polymerases (Pol II, IV and V) deal with lesion/sequence context diversity. DNA Repair, 1, 159–167. [DOI] [PubMed] [Google Scholar]
- Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.S. and Wang,Z. (2000) Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]
- Zhang H. and Siede,W. (2002) UV-induced T→C transition at a TT photoproduct site is dependent on Saccharomyces cerevisiae polymerase η in vivo. Nucleic Acids Res., 30, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yuan,F., Wu,X., Rechkoblit,O., Taylor,J.S., Geacintov,N.E. and Wang,Z. (2000) Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res., 28, 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]