Short abstract
DNA microarrays were used to examine the temporal program of gene expression following treatment of a human prostate cancer cell line with androgen. Significant changes in levels of transcripts of more than 500 genes were observed; most were not previously known to be regulated by androgens.
Abstract
Background
Androgens are required for both normal prostate development and prostate carcinogenesis. We used DNA microarrays, representing approximately 18,000 genes, to examine the temporal program of gene expression following treatment of the human prostate cancer cell line LNCaP with a synthetic androgen.
Results
We observed statistically significant changes in levels of transcripts of more than 500 genes. Many of these genes were previously reported androgen targets, but most were not previously known to be regulated by androgens. The androgen-induced expression programs in three additional androgen-responsive human prostate cancer cell lines, and in four androgen-independent subclones derived from LNCaP, shared many features with those observed in LNCaP, but some differences were observed. A remarkable fraction of the genes induced by androgen appeared to be related to production of seminal fluid and these genes included many with roles in protein folding, trafficking, and secretion.
Conclusions
Prostate cancer cell lines retain features of androgen responsiveness that reflect normal prostatic physiology. These results provide a broad view of the effect of androgen signaling on the transcriptional program in these cancer cells, and a foundation for further studies of androgen action.
Background
Androgens are central to sexual differentiation and the maintenance of male secondary sexual characteristics and functions. Individuals with inactivating mutations of their androgen receptor (AR) (androgen insensitivity syndromes (AIS)) are phenotypically female in appearance and behavior and lack male sex accessory organs (prostate, seminal vesicles and Cowper's glands) despite the presence of testes, high levels of testosterone and a Y chromosome [1]. The effects of androgens are cell-specific and developmental stage-specific. For instance, castration of an adult male does not change the external genitalia or voice pitch, but causes a profound involution of the prostate, particularly in the epithelial compartment [2,3]. The exquisite sensitivity of prostatic epithelial cells to androgens has been exploited therapeutically in the management of prostate cancer [4].
The mechanisms underlying the temporal and tissue-specific effects of androgens are not understood. Like all steroid hormones, androgens act by binding to a nuclear hormone receptor, enacting conformational changes that allow binding of the receptor to specific regulatory elements in AR-responsive genes. The ensuing cascade of transcriptional events, unique to each cell type, underlies the dramatic and diverse effects of androgen. Understanding the molecular programs regulated by androgens in critical target tissues may lead to improvements in treatment of AIS in children, benign prostatic hyperplasia and prostate cancer in adult men, and diverse other conditions in which androgens play a key role.
Unfortunately, few model systems are available for studying the effects of androgens in human tissues. There are no practical models of early development, when androgen exerts its most striking effects on the genitalia, body habitus and brain. Only a handful of immortalized cell lines express AR and respond to its ligands. All of these cell lines have been isolated from patients with advanced prostate carcinomas that have become refractory to androgen deprivation therapy [5,6,7]. In three of these cell lines, LNCaP, MDA PCa 2a and MDA PCa 2b, mutations in the ligand-binding domain of the AR confer broad steroid-binding affinity [8,9]. A fourth cell line, LAPC-4, bears a wild-type AR.
As an initial foray into understanding androgen-mediated gene regulation, we used cDNA microarrays to characterize the transcriptional program activated by the synthetic androgen R1881 in these four human prostate cancer cell lines. The results provide a broad overview of characterized and uncharacterized genes modulated by androgens and identify many previously unknown androgen targets. The similarity of the transcriptional responses among the four androgen-responsive cell lines suggests that this expression program may be intrinsic to prostatic epithelial cell function. Indeed, most of the functionally characterized genes induced by androgen in these prostatic cells appear to have important roles in the production of seminal fluid.
Results and discussion
In our initial experiments, we focused exclusively on the well-characterized LNCaP human prostate cancer cell line. LNCaP cells were cultured in conditions of androgen deprivation for 48 hours, then exposed to 1 nM of the synthetic androgen R1881 or to ethanol control (0.01%). After treatment for intervals varying from 8 to 72 hours, poly(A)+ RNA was extracted from the cells and analyzed using DNA microarrays. Two types of array analyses were performed. In the first, mRNA from each sample of treated or control cells was reverse transcribed to produce Cys-labeled cDNA, and this was compared to a 'common reference' of Cy3-labeled cDNA prepared from mRNA extracted from several cell lines [10]. These experiments revealed changes in expression of several hundred genes in the R1881-treated LNCaP cells, but no corresponding alterations in the ethanol-treated control (data not shown). As the ethanol control treatment did not appear to affect gene expression significantly, we analyzed another similar R1881 time course in which cDNAs representing transcripts from treated (Cy-5 labeled) and control (Cy-3 labeled) cells were compared directly by hybridization to the same microarrays. Measured changes in gene-expression patterns were virtually identical to those observed in the first experiment. The remaining experiments were carried out using the direct comparative hybridization design.
The significance analysis of microarrays (SAM) procedure was used to identify genes with statistically significant changes in expression after treatment with R1881. The SAM procedure accurately identifies transcripts with reproducible changes in gene expression and is more reliable than conventional means of analyzing microarrays [11]. Hierarchical clustering of the expression patterns of 567 transcripts representing 517 unique genes with statistically significant changes in expression in response to 1 nM R1881 in LNCaP is displayed in Figure 1a. Expression levels changed more than twofold in 539 (95%) of the transcripts. The raw microarray data and full details including transcript identities can be found at [12]. Approximately one third of these transcripts represent uncharacterized and unnamed genes and were not previously known to be regulated by androgen. Of the named genes, many have not been previously recognized as responsive to androgen. Virtually all of the known androgen-regulated genes that were represented on the array and adequately measured responded as previously reported (Figure 1b).
Figure 1.

Transcriptional program activated by R1881 exposure in LNCaP cells. (a) Hierarchical cluster analysis of androgen-responsive genes in LNCaP cells treated with R1881. This is a scaled-down overview of the cluster diagram generated by querying microarray data against the list of genes generated by SAM analysis (see text). The complete image with all gene names is viewable at [12]. Each column represents data from microarray analysis of a single RNA sample from LNCaP cells corresponding to a given time; the columns under the green heading show data in which RNA from R1881-treated and control cells were hybridized on the same microarray, while those under the black heading show experiments in which RNA from treated cells was hybridized against a common reference and normalized to the '0' time point. Numbers above each column denote specific time points. The dose of R1881 was 1 nM in all experiments. Red squares indicate transcripts with expression levels higher than that of ethanol-treated control cells; green squares, levels lower than that of controls; black, levels approximately equal to those in controls; gray, data of insufficient quality. (b) Representative androgen-regulated genes measured in microarray analysis of LNCaP cells treated with 1 nM R1881. Expression profiles of transcripts representing previously published androgen target genes are depicted. Genes listed more than once indicate that the microarray contained multiple elements representing that gene. As indicated by the scale bar, color saturation reflects magnitude of expression ratio.
To further examine the expression changes induced in response to androgen, we conducted two additional sets of experiments. In the first, LNCaP cells were treated for 24 hours with dihydrotestosterone (DHT), the predominant androgen found in the prostate in vivo. A clear relationship was observed between increasing dose of DHT and increased levels of gene expression (Figure 2a and see Additional data files). Furthermore, the pattern of expression changes observed in response to DHT largely paralleled those observed after treatment with R1881 (R2 = 0.7705; Figure 2b). We conducted a second set of experiments to examine the response of LNCaP cells to androgen withdrawal. After carrying cells in androgen-replete media with charcoal-stripped fetal bovine serum (FBS) (supplemented with 1 nM R1881), cells were passaged in media without androgen and harvested at 48 and 70 hours (Figure 2a). After 48 hours, the change in gene-expression pattern was largely reciprocal to that induced by androgen treatment and these effects increased by 70 hours. For the SAM-selected gene set, expression patterns after 70 hours of androgen deprivation were inversely correlated with those seen 50 hours after R1881 treatment (R2 = 0.7119; Figure 2c).
Figure 2.

Expression patterns of androgen-responsive genes in LNCaP cells exposed to R1881, DHT, or androgen deprivation. (a) Gene-expression changes in LNCaP treated with 1 nM R1881 and with dihydrotestosterone (DHT) at 10 nM, 100 nM or 1,000 nM. Note the apparent increase in gene expression with successively higher doses of DHT. On the far right are two experiments in which LNCaP cells were deprived of androgen for 48 and 72 h and that display reciprocal expression patterns. Green, red, black and gray bands as in Figure 1. Color-saturation scale as in Figure 1. (b) Correlation of gene-expression levels for LNCaP treated with 1 nM R1881 for 24 h and 1,000 nM DHT for 24 h. (c) Inverse correlation of gene-expression levels for LNCaP after treatment with 1 nM R1881 for 50 h and androgen deprivation for 72 h.
We have previously observed that varied stimuli produce temporally complex changes in gene-expression patterns in yeast and mammalian cells in vitro [13,14,15]. In many cases, genes can be grouped by their expression patterns using hierarchical clustering, and these groups often comprise genes that are functionally related. To our surprise, genes in the SAM-generated dataset varied little in their expression between 8 and 72 hours after androgen treatment. In general, expression levels of genes induced or repressed by androgen treatment remained at a constant level between 8 and 72 hours. As the SAM procedure identified genes with significant changes in expression in androgen-treated cells compared to controls, we were concerned that genes had been excluded whose expression levels varied for only part of the time course. Analysis of the raw data by hierarchical clustering analysis alone again generally failed to identify genes whose expression varied over the time course. The lack of complexity in the temporal response to androgen in this dataset precludes useful grouping of genes based on differences in their responses by cluster analysis. Perhaps a more detailed investigation of expression changes within the first 8 hours after treatment will reveal temporal patterns of response of LNCaP to androgens and provide additional insights into androgen action.
We were curious to know whether the expression changes observed in LNCaP would also occur in other androgen-responsive human prostate cancer cell lines (MDA PCa 2a, MDA PCa 2b, and LAPC-4), or in LNCaP and LAPC-4 subclones carried in androgen-depleted conditions for extended periods. All cell lines were treated with 1 nM R1881 and at least one sample was collected between 20 and 70 hours after addition of R1881. RNA samples from treated and control cell lines were compared directly on the same microarray. Responses to androgen of the set of transcripts identified as androgen-responsive in the LNCaP experiments were examined in each of these cell lines. Many genes were modulated similarly in all of the cell lines, suggesting conservation of a specific androgen-responsive program, perhaps a vestige of the prostatic epithelial phenotype, between these cell lines (Figure 3; see also [12] and Additional data files). For example, the transcript levels of 310 of these genes changed by 1.5-fold or more in MDA PCa 2a and 2b cells following 48 hours of exposure to R1881; transcripts for 262 genes changed by 1.5-fold or greater in single treatments of all four LNCaP androgen-deprived subclones; and transcripts for 81 genes changed by 1.5-fold or greater in the three LAPC-4 and LAPC-4 (androgen-deprived) array experiments.
Figure 3.

Hierarchical clustering analysis of expression patterns of androgen-responsive genes in four prostate cancer cell lines and sublines. Details are viewable at [12]. The column labels are enlarged on the right.
There were a few noteworthy differences in expression profiles between the cell lines. When we focused our attention on transcripts whose relative abundance changed by more than 2.6-fold in at least two experiments ([12] and see Additional data files), one group of genes differed strikingly between the cell lines (Figure 4). Most of the genes in this group have previously been found to be consistently expressed at significantly higher levels in proliferating cells [10,16]. Treatment with 1 nM R1881 promotes proliferation in the MDA PCa 2a and 2b cell lines, has no proliferative effect in the LNCaP and LAPC-4 cell lines, and represses proliferation in LNCaP cell lines carried for prolonged periods in conditions of androgen deprivation (data not shown, and see references in [17,18,19]). As seen in Figure 3, the proliferation cluster is relatively induced in the MDA PCa 2a and 2b cell lines, shows little change in LNCaP and LAPC-4, and is repressed in the androgen deprived LNCaP cell lines. Thus, transcript levels of the genes in this proliferation cluster parallel the responses of these cell lines to treatment with androgens.
Figure 4.
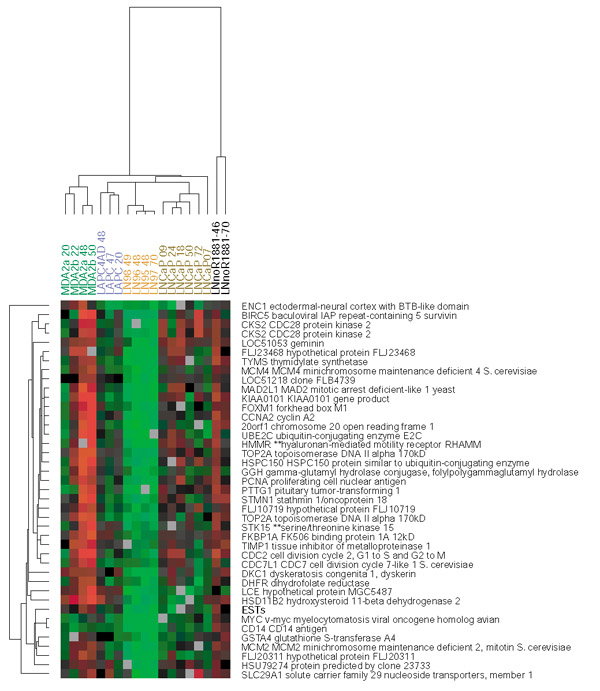
A cluster of genes with distinct patterns of modulation in LNCaP cells compared with other prostate cancer cell lines. This cluster includes many proliferation-associated genes. Column headings have been color-coded to distinguish the various cell lines as follows: Green text, MDA 2a and 2b cells; blue text, LAPC-4 cells and androgen-deprived subclone (LAPC-4AD); orange text, the four LNCaP androgen-deprived subclones; brown text, LNCaP cells + R1881; black text, LNCaP cells, R1881 deprivation. Numbers following each cell-line descriptor indicate length of exposure time in hours. Identifiers for each element shown here are gene symbol and gene name. The full cluster image is viewable at [12].
The expression patterns induced by R1881 offer possible insights into the effects of androgens in prostate physiology. Many features of the androgen-induced expression program appear to be related to activation of the cell's capacity to produce seminal fluid. After puberty, the normal prostate produces a protein- and organic solute-rich fluid that contributes approximately 30-50% of the volume of the ejaculate. The prostatic secretions are poorly characterized, but are undoubtedly critical to fertility as the prostate is the only sex accessory gland present in nearly all mammals [20]. Androgen induces several classes of genes that contribute to the production and secretion of prostatic fluid.
Perhaps the most striking change caused by treatment with R1881 occurs in the class of genes that contribute to the synthesis and modification of secretory proteins. All androgen-responsive genes in this class show increased expression in response to androgen. We have previously identified putative secretory and membrane-bound proteins based on their synthesis on membrane-associated polysomes ([21] and M.D. and P.O.B., unpublished observations) and over 200 of these are among the androgen-responsive genes (see [12] and Additional data files). Included in this set are genes encoding proteins previously known to be secreted by the prostate into the seminal plasma, including prostate-specific antigen (PSA), sorbitol dehydrogenase, apolipoprotein D [22], and vascular endothelial growth factor (VEGF) [23]. An additional set consists of 30 androgen-responsive genes with roles in protein trafficking and secretory vesicle formation and in transport of secretory vesicles (Figure 5a). This set of genes includes several with roles in membrane budding, vesicular formation, and membrane fusion, and translocation of proteins across the membrane of the endoplasmic reticulum. Androgen treatment also induced a number of genes that participate in protein folding and glycosylation (Figure 5b). One of these, FK506-binding protein 5/FKBP51, a recently described androgen target [24], was the most highly induced gene (more than 20-fold) we observed in our experiments. FK506-binding protein 5/FKBP51 is a co-chaperone protein that has peptidylprolyl isomerase activity and interacts with the progesterone receptor [25]. Whether it functions in the processing of androgen-induced secretory proteins or in other pathways induced by androgens or perhaps even in cell-cell signaling is unclear, but the degree of induction suggests it may have an important role in prostatic physiology.
Figure 5.

Androgen-responsive genes in LNCaP cells that are likely to participate in production of prostatic secretory fluid. As in Figure 1, each column represents an individual LNCaP time-point experiment. Images for these genes were grouped according to functional category: (a) protein trafficking (labeled in black text) and vesicular formation and transport (labeled in blue text); (b) protein folding (black text) and glycosylation (blue text); (c) polyamine biosynthesis, and (d) transporter and ion channels.
Prostatic fluid is also rich in small organic compounds that account for much of its osmotic pressure. For instance, prostatic secretions contain the highest levels of polyamines of all bodily fluids. The transcript encoding ornithine decar-boxylase (ODC), a known androgen target that catalyzes the rate-limiting step in polyamine biosynthesis [26], was induced coordinately with enzymes that catalyze downstream steps in polyamine synthetic pathway (spermine synthase, spermidine/spermine N1-acetyltransferase, and S-adenosylmethionine decarboxylase) (Figure 5c). Cholesterol and lipids are also found at high levels in semen and are major components of secretory vesicles referred to as prostasomes, which are thought to regulate the function of spermatozoa [27]. Note that several genes for proteins involved in cholesterol biosynthesis (HMG CoA reductase, IDI1, sterol C4 methyl oxidase) and fatty acid metabolism (ACAT2, DEGS, ACATN, FACL3, PEC1) were modulated by R1881. Choline, sorbitol, amino acids and citrate are also found in the prostatic secretions, although only a few genes (for sorbitol dehy-drogenase, phosphatidylcholine transfer protein, amino-acid transporter) related to these pathways appeared to respond directly to androgen. Citrate production is regulated by transport of aspartate into the mitochondria by aspartate amino-transferase, and indeed, the gene encoding this enzyme was induced by androgen in each of these cell lines [28]. Genes encoding several mitochondrial proteins were also induced in response to androgen, perhaps reflecting the increased production of citrate, or the increased demand for energy in these cells as they mobilize secretory pathways.
Zinc, calcium, phosphate, potassium, sodium, chloride and other ions are abundant in the ejaculate, although sodium and chloride are at levels below their serum concentrations. Several transporter molecules and ion-channel genes which may have roles in the active transfer of these ions into and out of the lumen of the prostate acini and secretory granules were modulated in response to R1881 (Figure 5d). Transport of sodium and chloride out of the lumen of the prostate acini is accompanied by the passive transfer of water and the prostatic secretions are thereby concentrated [20]. Transport of ions and small organic compounds into the acinar lumen will concentrate them in the seminal plasma. The ank gene, which codes for a transmembrane protein that secretes pyrophosphate into joint spaces [29], was strongly induced by R1881 exposure. Our data suggests that the pyrophosphate in the ejaculate maybe secreted into the prostatic fluid by the ANK protein.
The prostate undergoes dramatic morphologic changes as it acquires the ability to produce seminal fluid, and these changes can be observed at puberty or by stimulating the prostate with androgens after castration. The epithelial cells proliferate and change from stratified to cuboidal, while glands branch and acini expand. These morphologic changes follow complex reprogramming of the prostate cell that involves poorly understood alterations of the cytoskeleton and the extracellular matrix and changes in downstream signaling pathways [30,31]. Many genes encoding components of the extracellular matrix, proteins important in cell-cell interactions and cytoskeletal proteins were regulated in response to R1881, suggesting that they may participate in the morphologic changes induced by androgens (Figure 6a,6b). Many previous studies have pointed to a critical role for the cytoskeleton and extracellular matrix in transducing signals for growth, differentiation and secretion in normal and cancerous prostate cells [32]. Our data may therefore help point to one important feature of the transcriptional program that sets the stage for these signaling events.
Figure 6.

Androgen stimulation modulates signal transduction, transcriptional-regulatory, extracellular matrix and cytoskeletal genes. As in Figure 1, each column represents an individual LNCaP time-point experiment. Transcripts regulated in response to androgen grouped by functional category: (a) cell-cell interactions/extracellular matrix; (b) cellular dynamics/cytoskeleton; (c) regulation of transcription; (d) signal transduction.
We also found evidence of modulation of several genes encoding transcriptional regulatory and signaling proteins in response to androgen in the prostate cancer cell lines (Figure 6c,6d). It is possible that some of these pathways have important roles in reprogramming the normal prostate cell in response to androgen [33]. However, some of these signaling and transcriptional proteins may be modulated abnormally in these cancer-derived cell lines because of molecular genetic alterations acquired with transformation [34,35]. Yet other pathways may not be direct targets of AR activation but could be modulated in response to androgen-induced changes in the internal or external milieu of the cells. For instance, treatment of LNCaP with androgens has been shown to cause a burst of oxidative stress that may contribute to prostate carcinogenesis [19]. The observed increased expression of NFκB and several DNA repair and stress-response genes may be a secondary response to this oxidative stress rather than a direct response to AR signaling [36]. Further work will clearly be necessary to dissect the roles of these signaling pathways in the response of the prostate to AR activation.
In summary, we have provided a global characterization of the gene-expression programs triggered by androgen exposure in human prostate cancer cells. Analysis of the gene-expression program induced by R1881 strongly suggests that all of these prostate cancer cell lines retain features of prostatic epithelial cells, despite being derived from prostate cancer metastases and despite their long propagation in vitro. We recognize that the in vitro model systems used here offer at best only an approximation of normal and cancerous prostatic epithelia. Nevertheless, we believe that many of the target genes identified in these experiments are likely to be regulated similarly in vivo, just as PSA clearly is, and might serve as clinically useful diagnostic markers or therapeutic targets. These data may also offer some insight into the contribution of the prostate to the normal seminal fluid, including molecular components that may be important in fertility. Further investigation into androgen-induced changes in gene and protein expression in vitro and in vivo will undoubtedly shed light on prostatic physiology and prostatic carcinoma, and also on broader physiological functions of androgen. Beyond applications of these data to studies on the role of androgen in prostatic cells, this work provides a foundation for future investigation into androgen signaling in other model systems. Comparison of the transcriptional programs induced by androgen in other examples of the diverse cells and tissues that respond to androgen will undoubtedly contribute to a molecular understanding of the basis of the diverse and profound effects of androgens on human development and physiology and in human diseases.
Materials and methods
Cell culture and androgen treatments
The LNCaP cell line was obtained from the ATCC and maintained in RPMI media with 10% FBS and penicillin/streptomycin. The LN95, LN96, LN97, and LN98 variants of LNCaP, designated by the year in which androgen-free culture was initiated [18], were maintained in RPMI media without phenol red and with 10% charcoal-stripped, dextran-filtered FBS (Hyclone, Logan, UT) and penicillin/streptomycin. The MDA PCa 2a and MDA PCa 2b cells were generously provided by Nora Navone at the University of Texas M.D. Anderson Cancer Center and were cultured in defined BRFF-HPC1 media (Biological Research Faculty & Facility, Ijamsville, MD) and maintained on poly-lysine-coated tissue culture plates (Becton Dickinson Labware, Franklin Lakes, NJ). The LAPC-4 cells were cultured in Iscove's media with 10% FBS and penicillin/streptomycin, and the androgen-deprived variant LAPC-4 AD was maintained in Iscove's without phenol red and with 10% charcoal-stripped FBS.
Before androgen treatments, LNCaP and LAPC-4 cells were androgen-deprived by 48 h culture in media without phenol red, containing 10% dextran-filtered, charcoal-stripped FBS. For the MDA cells, media was F12K supplemented with 1% charcoal-stripped FBS, 40 mM phosphoethanolamine, 10 pg/ml hydrocortisone, 45 nM selenious acid, 5 μg/ml insulin, and 10 ng/ml epidermal growth factor (EGF) [6]. After androgen deprivation, cells were treated with either R1881 or DHT (NEN Life Science Products, Boston, MA) dissolved in ethanol, or ethanol carrier alone, and incubated for varying lengths of time. The final concentration of ethanol in media was 0.01%. Messenger RNA was prepared from cells using the Oligotex Direct kit for direct isolation of polyadenylated RNAs (Qiagen, Chatsworth, CA).
DNA microarray hybridizations and data analysis
The DNA microarrays used in this study consisted of PCR-amplified cDNAs printed on modified glass microscope slides as described previously [37]. Each array contained 24,000 cDNA spots, representing approximately 18,000 unique human genes and expressed sequence tags (ESTs). Some of the samples were also analyzed on microarrays with 9,600 cDNA elements. Microarray hybridizations were carried out according to previously published protocols [14,37,38,39]. For each hybridization, 2 μg of each purified sample of polyadenylated RNA was reverse-transcribed in the presence of fluorescently labeled nucleoside triphosphates (Cy3-dUTP for reference sample, Cy5-dUTP for experimental sample). Pairs of labeled cDNAs were hybridized to microarray slides for 16-18 h at 65°C in solution with blocking agents. After several washes, microarrays were scanned with a GenePix microarray scanner (Axon Instruments) and were analyzed with either Scanalyze or Genepix software. After visual inspection, spots of insufficient quality were flagged and excluded from further analysis. Data files were entered into the Stanford Microarray Database [40], and compiled experimental data were further analyzed with hierarchical clustering software and visualized with Treeview software [41,42,43]. Identities of genes or hypothetical genes represented by the spotted cDNAs are based on information available in UniGene cluster Build 144 (November 2001) [44].
Statistical analysis of microarray data
Genes with potentially significant changes in expression in response to androgen treatment were identified using the SAM procedure [11,45]. Changes in gene expression for any single gene as measured in several array experiments provide a statistically testable measure of robustness, regardless of the magnitude of change in expression. The SAM procedure computes a two-sample T-statistic (for example, for androgen-treated vs untreated cell lines) for the normalized log ratios of gene-expression levels for each gene. It thresholds the T-statistics to provide a 'significant' gene list and provides an estimate of the false-discovery rate (the percentage of genes identified by chance alone) from randomly permuted data.
For the experiments described in our study, the raw expression ratio dataset was filtered, using the program Cluster [43], for genes whose transcript levels differ from their median value by at least 1.5-fold in androgen-treated cells compared to controls in at least two experiments (with not more than 30% of measurements discarded because of poor data quality for each entry). To minimize noise, only genes with fluorescent intensity in each channel that was greater than 1.4 times the local background were selected. Two sets of time-course experiments were included in the analysis. The filtered dataset, containing more than 3,900 genes, was then analyzed with the SAM procedure. We used a selection threshold giving a median estimate of 0.5 false-positive genes (false detection rate of 0.088%) for further analysis. This list consists of 517 unique genes (of 568 total cDNAs) of which 421 are induced and 96 are repressed by exposure to R1881.
Additional data files
Supplemental figures showing full gene names and GenBank accession numbers for Figures 1, 2, 3, 4 and 5 are available and at [12].
Supplementary Material
Full gene names and GenBank accession numbers for Figure 1
Full gene names and GenBank accession numbers for Figure 2
Full gene names and GenBank accession numbers for Figure 3
Full gene names and GenBank accession numbers for Figure 4
Full gene names and GenBank accession numbers for Figure 5
Acknowledgments
Acknowledgements
We thank Vince Paton, Paul-Martin Holterhus and Jon Pollack for helpful discussions. We gratefully acknowledge the generous financial support of the Kovitz Foundation, the Grove Foundation, the Doris Duke Foundation, the National Cancer Institute and the Howard Hughes Medical Institute. S.E.D. was supported in part by a Stanford University School of Medicine Dean's Fellowship. M.D was supported by the National Institute of General Medical Sciences training grant GM07365. P.O.B. is an Associate Investigator of the Howard Hughes Medical Institute.
References
- Hiort O, Holterhus PM, Nitsche EM. Physiology and pathophysiology of androgen action. Baillieres Clin Endocrinol Metab. 1998;12:115–132. doi: 10.1016/s0950-351x(98)80495-3. [DOI] [PubMed] [Google Scholar]
- Bruckheimer EM, Kyprianou N. Apoptosis in prostate carcinogenesis. A growth regulator and a therapeutic target. Cell Tissue Res. 2000;301:153–162. doi: 10.1007/s004410000196. [DOI] [PubMed] [Google Scholar]
- Isaacs JT, Furuya Y, Berges R. The role of androgen in the regulation of programmed cell death/apoptosis in normal and malignant prostatic tissue. Semin Cancer Biol. 1994;5:391–400. [PubMed] [Google Scholar]
- Isaacs JT. The biology of hormone refractory prostate cancer. Why does it develop? Urol Clin North Am. 1999;26:263–273. doi: 10.1016/s0094-0143(05)70066-5. [DOI] [PubMed] [Google Scholar]
- Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- Navone NM, Olive M, Ozen M, Davis R, Troncoso P, Tu SM, Johnston D, Pollack A, Pathak S, von Eschenbach AC, Logothetis CJ. Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clin Cancer Res. 1997;3:2493–2500. [PubMed] [Google Scholar]
- Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, Lamb DJ, Marcelli M, Belldegrun A, Witte ON, Sawyers CL. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- Veldscholte J, Ris-Stalpers C, Kuiper GG, Jenster G, Berrevoets C, Claassen E, van Rooij HC, Trapman J, Brinkmann AO, Mulder E. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem Biophys Res Commun. 1990;173:534–540. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Malloy PJ, Krishnan AV, Swami S, Navone NM, Peehl DM, et al. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med. 2000;6:703–706. doi: 10.1038/76287. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supplementary data to Genome Biology. 2002. pp. research0032.1–0032.12.http://genome-www.stanford.edu/androgen
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1006/jmbi.1998.2134. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1006/abio.2000.4611. [DOI] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Boyle B, Krishnan AV, Navone NM, Peehl DM, Feldman D. Two mutations identified in the androgen receptor of the new human prostate cancer cell line MDA PCa 2a. J Urol. 1999;162:2192–2199. doi: 10.1016/S0022-5347(05)68158-X. [DOI] [PubMed] [Google Scholar]
- Pflug BR, Reiter RE, Nelson JB. Caveolin expression is decreased following androgen deprivation in human prostate cancer cell lines. Prostate. 1999;40:269–273. doi: 10.1002/(SICI)1097-0045(19990901)40:4<269::AID-PROS9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Ripple MO, Henry WF, Rago RP, Wilding G. Prooxidant-antioxidant shift induced by androgen treatment of human prostate carcinoma cells. J Natl Cancer Inst. 1997;89:40–48. doi: 10.1093/jnci/89.1.40. [DOI] [PubMed] [Google Scholar]
- Mann T, Lutwak-Mann C. Male Reproductive Function and Semen New York: Springer-Verlag; 1981.
- Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Simard J, Veilleux R, de Launoit Y, Haagensen DE, Labrie F. Stimulation of apolipoprotein D secretion by steroids coincides with inhibition of cell proliferation in human LNCaP prostate cancer cells. Cancer Res. 1991;51:4336–4341. [PubMed] [Google Scholar]
- Joseph IB, Nelson JB, Denmeade SR, Isaacs JT. Androgens regulate vascular endothelial growth factor content in normal and malignant prostatic tissue. Clin Cancer Res. 1997;3:2507–2511. [PubMed] [Google Scholar]
- Amler LC, Agus DB, LeDuc C, Sapinoso ML, Fox WD, Kern S, Lee D, Wang V, Leysens M, Higgins B, et al. Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen-independent prostate cancer xenograft model CWR22-R1. Cancer Res. 2000;60:6134–6141. [PubMed] [Google Scholar]
- Nair SC, Rimerman RA, Toran EJ, Chen S, Prapapanich V, Butts RN, Smith DF. Molecular cloning of human FKBP51 and comparisons of immunophilin interactions with Hsp90 and progesterone receptor. Mol Cell Biol. 1997;17:594–603. doi: 10.1128/mcb.17.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts AM, Waite I, Neal DE, Robson CN. Androgen regulation of ornithine decarboxylase in human prostatic cells identified using differential display. FEBS Lett. 1997;405:328–332. doi: 10.1016/s0014-5793(97)00209-3. [DOI] [PubMed] [Google Scholar]
- Carlini E, Palmerini CA, Cosmi EV, Arienti G. Fusion of sperm with prostasomes: effects on membrane fluidity. Arch Biochem Biophys. 1997;343:6–12. doi: 10.1006/abbi.1997.9999. [DOI] [PubMed] [Google Scholar]
- Costello LC, Franklin RB. The intermediary metabolism of the prostate: a key to understanding the pathogenesis and progression of prostate malignancy. Oncology. 2000;59:269–282. doi: 10.1159/000012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AM, Johnson MD, Kingsley DM. Role of the mouse ank gene in control of tissue calcification and arthritis. Science. 2000;289:265–270. doi: 10.1007/s004030050191. [DOI] [PubMed] [Google Scholar]
- Aumuller G, Seitz J. Protein secretion and secretory processes in male accessory sex glands. Int Rev Cytol. 1990;121:127–231. doi: 10.1016/s0074-7696(08)60660-9. [DOI] [PubMed] [Google Scholar]
- Partin AW, Coffery DS. In Campbell's Urology Edited by Campbell MF, Retik AB, Vaughn ED, Walsh PC 7th edn Philadelphia, PA: WB Saunders Co; 7. 1998. The molecular biology, endocrinology, and physiology of the prostate and seminal vesicles. p. 3432. [Google Scholar]
- Getzenberg RH, Pienta KJ, Ward WS, Coffey DS. Nuclear structure and the three-dimensional organization of DNA. J Cell Biochem. 1991;47:289–299. doi: 10.1002/jcb.240470402. [DOI] [PubMed] [Google Scholar]
- Peterziel H, Mink S, Schonert A, Becker M, Klocker H, Cato AC. Rapid signalling by androgen receptor in prostate cancer cells. Oncogene. 1999;18:6322–6329. doi: 10.1038/sj/onc/1203032. [DOI] [PubMed] [Google Scholar]
- Craft N, Shostak Y, Carey M, Sawyers CL. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat Med. 1999;5:280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- Yeh S, Lin HK, Kang HY, Thin TH, Lin MF, Chang C. From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc Natl Acad Sci USA. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripple MO, Henry WF, Schwarze SR, Wilding G, Weindruch R. Effect of antioxidants on androgen-induced AP-1 and NF-kappaB DNA-binding activity in prostate carcinoma cells. J Natl Cancer Inst. 1999;91:1227–1232. doi: 10.1093/jnci/91.14.1227. [DOI] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- The Brown Lab http://brownlab.stanford.edu
- Stanford Microarray Database http://genome-www5.stanford.edu/MicroArray/SMD/
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, et al. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen Lab http://rana.lbl.gov
- UniGene http://www.ncbi.nlm.nih.gov/UniGene/
- SAM: significance analysis of microarrays http://www-stat.stanford.edu/~tibs/SAM/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full gene names and GenBank accession numbers for Figure 1
Full gene names and GenBank accession numbers for Figure 2
Full gene names and GenBank accession numbers for Figure 3
Full gene names and GenBank accession numbers for Figure 4
Full gene names and GenBank accession numbers for Figure 5


