Abstract
During latency, the Kaposi's sarcoma-associated herpesvirus genome is maintained as a circular episome, replicating in synchrony with host chromosomes. Replication requires the latency-associated nuclear antigen (LANA) and an origin of latent DNA replication located in the viral terminal repeats, consisting of two LANA binding sites (LBSs) and a GC-rich sequence. Here, we show that the recruitment of a LANA dimer to high-affinity site LBS-1 bends DNA by 57° and towards the major groove. The cooccupancy of LBS-1 and lower-affinity LBS-2 induces a symmetrical bend of 110°. By changing the origin architecture, LANA may help to assemble a specific nucleoprotein structure important for the initiation of DNA replication.
Kaposi's sarcoma-associated herpesvirus (KSHV, or human herpesvirus 8 [HHV-8]) is the etiological agent of KS, multicentric Castleman's disease, and primary effusion lymphoma (11, 47). As is true for all members of the gammaherpesvirus family, KSHV exploits two contrasting modes of infection: lytic or productive replication and latency. In late-stage KS or primary effusion lymphoma, the latent form of the virus predominates and drives the proliferation and survival of the tumor cells. The 140-kbp double-stranded-DNA KSHV genome is maintained as multiple circular episomes that are assembled into chromatin and retained in the nucleus (29, 35). Only a few of the 90 or so viral genes are expressed at this time and invariably include the latency-associated nuclear antigen (LANA) encoded by open reading frame 73 (12, 23, 34, 42). LANA is associated with host chromatin throughout the cell cycle, giving rise to characteristic foci in all infected cells (37, 45, 49). Based on amino acid composition, we divide LANA into three domains or regions (Fig. 1A): a proline and basic-residue-rich N terminus (LANAN), a highly acidic and repetitive central region, and a C terminus (LANAC) that is enriched in charged and hydrophobic residues (39).
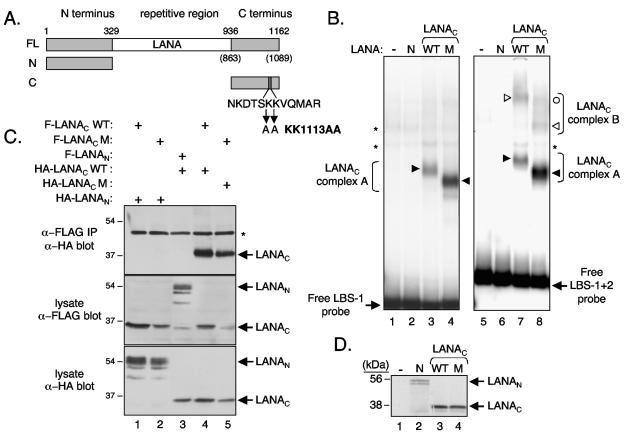
FIG. 1.
Association of wild-type and KK1113AA mutant LANAC with LBS-1 and LBS-2. (A) Schematic showing full-length LANA and fragments used in this study. The amino acid numbering corresponds to the prototype BC-1 sequence (36). The numbers in parentheses signify coordinates of the actual LANA variant (GenBank accession no. AAB62657 [31]) used here. FL, full length; N, LANAN; C, LANAC. (B) EMSA using 32P-labeled probes containing LBS-1 or LBS-1 and LBS-2. The first two lanes of each panel are controls showing the probe mixed with unprogrammed lysate (lanes 1 and 5) and lysate expressing LANAN (lanes 2 and 6; lanes labeled “N”). Lysates expressing LANAC WT (lanes 3 and 7; lanes labeled WT) or LANACKK1113AA (lanes 4 and 8; lanes labeled “M” for “mutant”) were incubated with each probe, giving rise to one or more specific complexes (filled and open arrowheads). Nonspecific complexes are indicated with an asterisk. (C) Coimmunoprecipitation assay to monitor self-association. HeLa cells were transiently transfected with expression plasmids (1.5 μg each) encoding FLAG-tagged LANAC WT (lanes 1 and 4), FLAG-LANACKK1113AA (mutant; lanes 2 and 5), or FLAG-LANAN (lane 3) and HA-tagged LANAC WT (lanes 3 and 4), LANACKK1113AA (lane 5), or LANAN (lanes 1 and 2). At 24 h posttransfection, protein extracts were prepared using high-salt extraction buffer and immunoprecipitated (IP) with anti-FLAG antibody (α-FLAG)-coupled beads (Sigma). Immunoprecipitated complexes were resolved by SDS-12% PAGE and probed by immunoblotting with anti-HA (α-HA) antibody (upper panel). One-thirtieth of the starting extracts were immunoblotted in parallel using anti-FLAG (middle panel) or anti-HA (lower panel) antibodies. Nonspecific cross-reactive polypeptides are indicated with an asterisk. Sizes of the molecular mass standards are in kilodaltons. (D) SDS-PAGE analysis of in vitro-translated 35S-labeled proteins used in panel B detected by fluorography.
LANA contributes to many aspects of latency and is required for the replication and maintenance of the viral episomes (1, 2, 18, 21, 28). Episome replication occurs during the S phase, utilizing the host DNA synthesis machinery, and requires two viral components: LANA and an origin sequence found in each of the 20 to 40 copies of the 801-bp terminal repeats (TRs) present in the KSHV genome (2, 13, 18, 21, 27). LANA is required to initiate DNA synthesis at the origins and to ensure efficient segregation of the newly replicated molecules during mitosis. The latter is achieved by the tethering of the viral DNA to components of host chromatin using a short sequence at the extreme N terminus of LANAN (3, 25, 32, 40, 49). The association of LANA with KSHV DNA is mediated by a DNA-binding/dimerization domain located in LANAC that recognizes two GC-rich sequences (LANA binding site 1 [LBS-1] and LBS-2) found in each TR (2, 9, 15, 16, 41). A single TR is sufficient for plasmid replication in transient assays, and the minimal origin comprises a 71-bp sequence containing LBS-1, LBS-2, and a GC-rich “replication element” (22).
There is no evidence for a resident helicase function in LANA, and instead, it seems likely that its initiation function involves recruiting components of the cellular DNA replication initiation machinery (52). Subunits of the origin recognition complex (ORC) and MCM3 are detected at the TR in latently infected cells by chromatin immunoprecipitation, and there is evidence that LANA interacts directly with at least two ORC subunits (28, 43). In addition to recruiting, LANA may also reorganize the chromatin around the origins and create a specific structure recognized by the ORC (43). Here, we address the architectural consequences of LANA binding to TR DNA. Using a circular permutation assay (CPA), we show that LANAC can induce a strong bend (57° or greater) in the DNA helix of LBS-1. A similar bend is introduced when LBS-2 is occupied, giving a cumulative bend angle of at least 110°. These observations were confirmed by phasing analysis, which also showed that the DNA helix is bent towards the major groove. An alanine substitution mutant within the DNA-binding domain altered the relative gel mobility of the DNA-bound complex and reduced the extent of the induced bend by 25° when both LBSs were occupied. Several other well-characterized viral and bacterial replication initiator proteins, including EBNA1 from Epstein-Barr virus (EBV), induce bends of similar magnitude in origin DNA, suggesting that this is a fundamental aspect of initiator protein function.
Mutation in the C terminus of LANA alters the mobility of the protein-DNA complex.
DNA-binding function by LANAC (Fig. 1A) was analyzed by electrophoretic mobility shift assay (EMSA) using 32P-labeled DNA probes containing either the high-affinity LBS-1 (Fig. 1B, lanes 1 to 4) or the combination of high- and low-affinity sites (LBS-1 and LBS-2 [LBS-1+2]) (Fig. 1B, lanes 5 to 8). The coding sequence for LANAC (residues 936 to 1162 in the prototype sequence) (36) was subcloned into the in vitro translation vector pTCITE, a derivative of pCITE2a+ (Novagen) that adds a T7 epitope tag to the N-terminal end of the recombinant protein. The LANAC protein was then expressed in vitro using the TNT Quick coupled transcription/translation system (Promega) in the presence of [35S]methionine. Probes were prepared by introducing oligonucleotide duplexes spanning LBS-1 (30 bp) or both LBS-1 and LBS-2 (54 bp) into the unique XhoI site of a modified pUC119 polylinker. Radiolabeled probes, with lengths of 157 bp and 181 bp were generated by PCR amplification using flanking primer pairs that had been end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Probes were gel purified on a 5% native polyacrylamide gel to remove single-stranded probe DNA and excess primer and then eluted into 10 mM HEPES (pH 7.9), 60 mM KCl. In vitro translation reaction mixtures (5 μl) were incubated with each probe at room temperature for 25 min, with the buffer adjusted to 10 mM HEPES (pH 7.9), 50 mM KCl, 1 mM EDTA (pH 8.0), 1 μg poly(dI-dC), 10 mM dithiothreitol, and 10% glycerol (total reaction volume, 20 μl). Reaction mixtures were loaded onto a 5% native polyacrylamide gel that had been prerun for at least 1 h at room temperature. After electrophoresis, gels were fixed in 10% methanol-10% acetic acid, dried, and visualized by exposure to Kodak X-ray film or a Molecular Dynamics PhosphorImager plate.
Incubation of the LBS-1 probe with wild-type LANAC (LANAC WT) gave a robust LANAC-specific complex (complex A) (Fig. 1B, lane 3) that was not found when the probe was incubated with either unprogrammed reticulate lysate (lane 1) or the N terminus of LANA (lane 2). The same translation mixtures were incubated with the 181-bp probe incorporating both LBS-1 and LBS-2 (Fig. 1B, lanes 5 to 8). Again, LANAC WT (Fig. 1B, lane 7) formed specific complexes not found when the probe was incubated with unprogrammed reticulate lysate (lane 5) or LANAN (lane 6). From the literature (15), it is likely that the faster-mobility complex (complex A) consists of one LANAC dimer bound to high-affinity LBS-1, while the slower-migrating complex (complex B) is composed of two LANAC dimers, with each dimer occupying each site. The complexes often resolve as doublets and may suggest posttranslational modifications introduced by the lysates or possibly slightly different protein confirmations.
Next, we examined complex formation by a mutant version of LANAC—named LANACKK1113AA—in which lysines 1113 and 1114 of the prototype sequence had been changed to alanine. This mutant was selected from a panel of alanine substitution mutants that will be described elsewhere and was chosen because of its increased affinity for TR DNA and altered gel mobility relative to that of the wild type. When mixed with the LBS-1 probe, LANACKK1113AA (Fig. 1B, lane 4) gave rise to a more robust complex than LANAC WT (lane 3). Both proteins were translated at similar levels as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1D), and titration experiments indicated that the complex-forming activity of the mutant was approximately threefold greater than that of the wild type (data not shown). Note that the in vitro-translated LANAC proteins are predominantly full length. More significantly, there were marked and reproducible differences in the relative mobilities of the major complexes formed by the proteins, with that of LANACKK1113AA consistently running faster than that of LANAC WT. This difference was also evident when complex formation was assayed using the larger LBS-1 and LBS-2 probes (Fig. 1B, lanes 7 and 8). Three discrete complexes were observed with LANACKK1113AA, and the two slower-migrating complexes were unique to the LBS-1 and LBS-2 probes. The nature of the slowest complex is currently unclear and may reflect the recruitment of more than two LANAC dimers to a single DNA molecule. Similar complexes are sometimes observed with LANAC WT (data not shown).
We have previously shown that LANAC contains a self-interaction domain (39), and it is possible that the altered mobility of the mutant results from a difference in oligomerization state. We, therefore, asked whether LANACKK1113AA was competent to self-associate using a coimmunoprecipitation assay (Fig. 1C). Using the lipid transfection reagent PolyFect (QIAGEN), human HeLa cells were transiently transfected with expression plasmids encoding versions of LANAC WT and the mutant tagged with either FLAG or hemagglutinin (HA) epitopes. Twenty-four hours after transfection, protein extracts were prepared in high-salt lysis buffer containing 420 mM KCl, 20 mM Tris-HCl (pH 7.9), 10% glycerol, and 0.25% NP-40 and immunoprecipitated using anti-FLAG antibody-coupled agarose beads (Sigma). The resulting immunocomplexes were resolved by SDS-12% PAGE, transferred to a nitrocellulose membrane, and immunoblotted with an anti-HA monoclonal antibody (12CA5; Covance). As expected, HA-tagged LANAC WT was recovered in the presence of FLAG-tagged LANAC WT (Fig. 1C, lane 4) but not in the presence of FLAG-tagged LANAN (lane 3). The HA-tagged LANAC mutant was similarly coprecipitated in the presence of its FLAG-tagged version (Fig. 1C, lane 5), showing that the mutation does not prevent self-association. HA-tagged LANAN was not recovered with either FLAG-tagged protein (Fig. 1C, lanes 1 and 2), confirming that the observed LANAC interactions were specific. Direct immunoblotting of the starting extracts using either anti-FLAG or anti-HA antibodies showed that LANACKK1113AA was expressed at a slightly lower level than LANAC WT, thus accounting for the slightly reduced level of immunoprecipitated LANAC mutant protein. These results indicate that the observed differences in relative gel mobility of the protein-DNA complexes are not due to the inability of the mutant to oligomerize with itself.
LANAC bends DNA when bound to LBS-1.
A number of virus-encoded initiators of DNA replication are known to bend origin DNA (4, 7, 8, 17, 24, 30, 33, 51). This raised the possibility that LANA might also bend DNA and that the KK1113AA mutation alters the extent of the induced bend. To determine if LANAC WT was capable of bending TR DNA, we adapted the EMSA for use as a CPA. In this approach, the relative mobility of a protein-DNA complex through the gel matrix is measured using a series of probes of identical lengths but in which the position of the binding site is moved relative to the probe ends (10, 50). If the binding of a protein causes the DNA to bend, then the migration rate of the resultant complex will vary depending on the position of the bend relative to the probe ends. Probes that are bent at one end exhibit a faster mobility during electrophoresis than probes bent near the middle. Furthermore, the extent and position of the bend can be calculated from the relative mobilities of the bound probes.
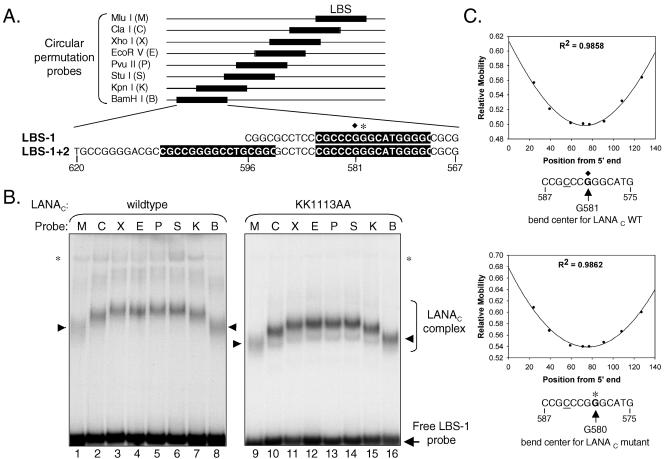
To generate an appropriate probe set, we inserted a 36-bp oligonucleotide spanning LBS-1 into the unique XbaI and HindIII sites of the plasmid pBEND2H (a gift from A. Stenlund, Cold Spring Harbor Laboratory, NY). This plasmid contains a duplicated polylinker and can be digested to release a series of probes of identical lengths (17, 48). The DNA was radiolabeled by PCR amplification in the presence of [α-32P]dCTP using the oligonucleotide primers Bend 1 (5′-TAGGCGTATCACGAGGCCCT-3′) and Bend 2 (5′-CGTTAGCAATTTAACTGTGAT-3′), which anneal to sequences on each side of the permutated polylinker region. To improve the efficiency of amplification of the highly GC-rich LBS-1 sequence, the PCR was performed in the presence of 1 M betaine (20). After PCR, the amplification product was aliquoted and digested with eight different restriction endonucleases (Fig. 2A) to give a series of probes with an overall length of 157 nucleotides. The probe fragments were purified from template DNA and cleaved arms by native gel electrophoresis and eluted from the gel slices prior to incubation with in vitro-translated protein. The results of a representative CPA using LANAC WT (lanes 1 to 8) and LANACKK1113AA (lanes 9 to 16) are shown in Fig. 2B. All eight unbound probes migrated at similar rates, indicating that the probe DNA had little or no intrinsic curvature. In contrast, the LANAC-specific complexes showed an obvious difference in mobility depending on the position of the LBS-1 site relative to the ends of the probe. This result indicates that both versions of LANAC induce a significant bend in DNA upon binding.
FIG. 2.
LANAC bends DNA. (A) Design of probes used for the CPA. Sequences of LBS-1 and LBS-2 as determined by Garber et al. (15) are boxed, and the numbering follows that of the prototype TR sequence (GenBank accession no. U86666) (26). Probes were labeled by PCR in the presence of [32P]dCTP and digested at one of eight restriction endonuclease sites in the duplicated polylinker of pBEND2H, yielding probes of identical lengths but with the LBS at different positions relative to the ends. The center of the bend induced by the binding of LANAC WT to the LBS-1 probe was at guanine-581 (filled diamond); with LANACKK1113AA, the center of the bend was at guanine-580 (asterisk). (B) EMSA using 32P-labeled probe sets containing LBS-1 incubated with in vitro-translated LANAC WT (lanes 1 to 8) or the KK1113AA mutant (lanes 9 to 16). Bands correspond to the LANAC-induced complex (filled arrowheads), and unbound probes are indicated. (C) Plot of the relative mobilities of retarded complexes (mobility of a complex divided by the mobility of the free probe) compared to the position of an arbitrary reference point (cytosine-584 [underlined]) from the 5′ end of each probe. The best fit to the cosine function curve was generated using SigmaPlot. The upper graph represents the relative mobility of LANAC WT. By extrapolation, the fastest-migrating complex corresponds to a probe with a reference nucleotide at position 75.3 within the 157-bp probe (95% confidence interval, 73.6 to 78.0). The predicted bend center should be equidistant from the ends of this probe and would therefore correspond to guanine-581. The lower graph shows the analogous mobility plot for LANAC KK1113AA. Minimal migration occurs when the reference point is at position 74.5 in the probe (95% confidence interval, 72.0 to 77.0) and thus predicts the bend center to be at guanine-580.
From this analysis, the bend angle can be estimated using the empirically derived formula (μM/μE) = cos (α/2), where μM is the slowest-migrating complex, μE is the fastest-migrating complex, and α is the estimated bend angle (46). In essence, this equation compares the mobility of the complex bound in the middle of the probe, the slowest-migrating complex, with the mobility of the complex bound at the end of the probe, the fastest-migrating complex. Using measurements derived from six independent CPA experiments, we derived an estimated bend of 57° ± 3° for LANAC WT bound to LBS-1. This confirms that the binding of LANAC to a single high-affinity site induces a significant distortion in the DNA. The analysis was also applied to LANAC KK1113AA (Fig. 2A, lanes 9 to 16) and yielded an estimated bend angle of 57° ± 1.5°. In spite of the fact that both proteins give rise to protein-DNA complexes with different relative mobilities, the DNAs are bent to similar degrees.
By plotting the mobility of each protein-DNA complex relative to the free probe (y axis) against the position of an arbitrarily chosen nucleotide from one end of the probe (x axis), we can extrapolate the position of LBS-1 that would give rise to the slowest-migrating complex (Fig. 2C, top graph). The center of the bend can then be predicted from the minimum of the cosine curve. Using measurements from six independent gel runs, we found that the center of the bend induced by the binding of LANAC WT to the LBS-1 probe corresponded to guanine-581 (Fig. 2A and C, upper graph). For LANACKK1113AA, the calculated bend center was at guanine-580 (Fig. 2A and C, bottom graph). Given the margin of error in these measurements, it is likely that the bend center for LBS-1 is the same for both proteins and lies within the core binding site and slightly to one side of the point of symmetry (9, 15, 41).
Additive increase in bend angle when LBS-1 and LBS-2 are occupied.
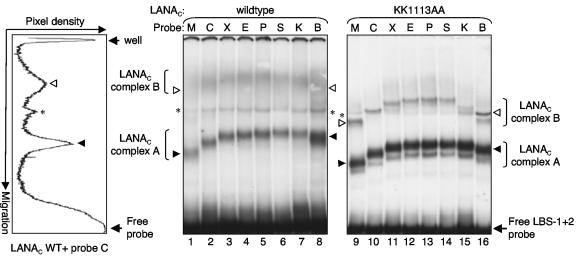
Within each copy of the TR, the two LANA binding sites are separated by 22 bp center-to-center and thus lie on approximately the same face of the DNA helix. LANA binds to the two sites in a cooperative manner such that the occupancy of LBS-1 facilitates the recruitment of a second LANA multimer (most likely a dimer) to the adjacent lower-affinity site, LBS-2 (15). To examine the extent of the induced bend when both LANA binding sites are occupied, we subcloned a 60-bp oligonucleotide spanning LBS-1 and LBS-2 into the pBEND2H vector and performed CPAs as described above. As expected, incubation with LANAC WT gave rise to two major complexes corresponding to the occupancy of LBS-1 alone or of LBS-1 and LBS-2 (Fig. 3A, lanes 1 to 8). The occupancy of two sites was reduced compared to that of the single site, a consequence of the limited amount of biologically active LANAC WT protein in the reticulocyte lysate. In spite of the weaker signal, it was clear that the upper complex migrated in a semicircular pattern similar to that of the lower complex. Using this longer probe, the bend angle for the lower complex was estimated at 65° ± 5.5° (n = 3).
FIG. 3.
CPA using LBS-1 and LBS-2 probe sets. EMSA using a set of 32P-labeled probes containing LBS-1 and LBS-2 incubated with in vitro-translated LANAC WT (lanes 1 to 8) or the KK1113AA mutant (lanes 9 to 16) is shown. Specific complexes corresponding to the occupancy of LBS-1 (LANAC complex A, filled arrowheads) or LBS-1 and LBS-2 (LANAC complex B, open arrowheads) are indicated together with the unbound probes (Free LBS-1+2). A nonspecific complex is indicated with an asterisk. A representative pixel density profile (probe C incubated with LANAC WT, lane 2) is shown on the left.
Although the upper complex formed by LANAC WT was more diffuse than that of the mutant, we were able to determine the center of the band by measuring the pixel density from the scanned autoradiogram (ImageQuant software; Molecular Dynamics) and derived an average bend angle of 110° ± 11.8° (n = 3). From comparable measurements of the well-defined upper complex formed by LANAC KK1113AA, we calculated an average bend angle of 85.4° ± 0.2° (n = 3). From these results, we conclude that LANAC bends the two binding sites in a roughly additive manner but that the cumulative angle is reduced in the case of the mutant.
LANAC bends DNA towards the major groove.
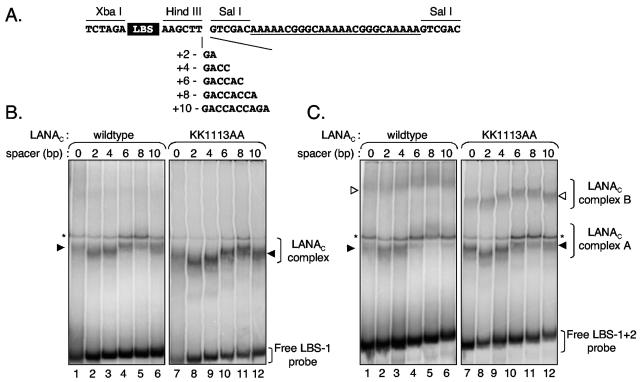
Phasing analysis offers an alternative method to identify protein-directed DNA bends and can also be used to determine the direction of the bend (38, 53). This method relies on the phase-dependent interaction between the protein-induced bend under study and an intrinsic DNA bend incorporated into each of the probes. The spacing between these two bends is varied over a single helical turn in small increments so that at a particular spacing, the two bends will either be in phase (cis isomer) or out of phase (trans isomer) with each other. In the latter, the two bends will counteract each other, and the complex will migrate through the gel with the fastest mobility. As shown in Fig. 4A, an intrinsic bend was created by the insertion of a series of three adenine (A) tracts on one side of the probe. Each copy of the A tract bends the DNA towards the minor groove by 18°.
FIG. 4.
TR DNA is bent towards the major groove. (A) Design of phasing analysis probes. Oligonucleotides containing either LBS-1 or LBS-1 and LBS-2 were inserted between unique XbaI and HindIII sites of pBEND2H. To provide the intrinsic bend, a second oligonucleotide containing three A tracts (underlined) was inserted into the unique SalI site. The size of the spacer region was increased by PCR using a series of oligonucleotides spanning the ends of the LBS and A tract was used in a PCR to clone in the spacers. Subsequently, the center of the A tract is located 40 bp (3.8 helical turns), 42 bp (4.0 helical turns), 44 bp (4.2 helical turns), 46 bp (4.4 helical turns), 48 bp (4.6 helical turns), and 50 bp (4.8 helical turns) from the molecular bend center of G581 when the spacer lengths are 0, 2, 4, 6, 8, and 10 nucleotides, respectively. (B) 32P-labeled probes containing LBS-1 were incubated with in vitro-translated LANAC WT (lanes 1 to 6) or the KK1113AA mutant (lanes 7 to 12). Specific LANAC-induced complexes and the unbound probes are indicated. The numbers indicate the linker length inserted between LBS-1 and the A tract. (C) Phasing analysis using 32P-labeled probes containing LBS-1 and LBS-2 incubated with in vitro-translated LANAC WT (lanes 1 to 6) or the KK1113AA mutant (lanes 7 to 12). Specific LANAC-induced complexes A and B plus the positions of unbound probes are indicated. Spacer lengths are listed above the lanes.
The three-part A tract oligonucleotide was subcloned into the unique SalI site of the pBEND2H constructs used in the previous CPAs (Fig. 4A). Insertions of 2, 4, 6, 8, and 10 bp between the LBS inserts and the three-part A tract were then made by PCR. Constructs were verified by DNA sequence analysis and then used as the templates for PCR amplification in the presence of [α-32P]dCTP, betaine, and the Bend 1 and 2 primers. Amplification products were digested with PvuII to remove excess vector sequences and gel purified. Each phasing probe (LBS-1 alone or LBS-1 and LBS-2) was incubated with in vitro-translated LANAC WT (Fig. 4B and C, lanes 1 to 6) or KK1113AA proteins (Fig. 4B and C, lanes 7 to 12), and the relative gel mobilities of the resultant protein-DNA complexes were measured by EMSA.
Both LANAC WT and LANAC KK1113AA gave a robust complex with each of the phasing probes containing LBS-1 alone (Fig. 4B). As expected, the relative mobility of the LANA-induced complex was dependent on the number of additional bases inserted between the LBS-1 site and the three-part A tract. Insertion of 2 or 4 bp increased the mobility of the complex, whereas insertion of 6 or 8 bp reduced the mobility. By comparison, the unbound probes showed a linear relationship of mobility to probe length, confirming the absence of an intrinsic bend in the KSHV sequence. The distinctive sinusoidal behavior of the LANAC-induced complexes shows that the binding of either protein to LBS-1 induces a significant bend in the DNA. The slowest-migrating complex (8-bp insertion) corresponds to the cis isomer, and in this probe, the bend center is located 48 bp from the middle of the A tract. Assuming a periodicity of 10.5 bp per helical turn, this corresponds to 4.6 helical turns. In this orientation, the induced and intrinsic bends distort the DNA in the same direction. The fastest-migrating complex (trans isomer) was formed on the probe with a 2-bp insertion such that the centers of the two bends were separated by 42 bp or exactly four helical turns. Based on the bending at the center of the A tracts towards the minor groove of DNA, we can conclude that the LANAC-induced bend is directed towards the major groove. A similar sinusoidal mobility profile was observed using the LBS-1 and LBS-2 probe sets (Fig. 4C) for both the lower (LANAC complex A) and upper (LANAC complex B) complexes. The linker insertions are expected to have equivalent effects on single and double complexes because the binding sites are separated by 22 bp and are therefore in helical phase.
In conclusion, we have shown that the C-terminal DNA binding and dimerization domain of LANA bends TR DNA by a minimum of ∼57° at LBS-1 and to a similar degree at LBS-2. This is a property of other origin-binding proteins from both viral and bacterial systems, and it is likely that distortion of DNA within the origin is a necessary step in the initiation of DNA replication (8, 17, 24, 30, 33, 51). As noted by others, there are interesting parallels in the organization of the KSHV and EBV latent origins (22, 43). In EBV oriP, DNA replication initiates near the dyad symmetry element DS, which contains two pairs of EBNA1 binding sites (14). Each pair is composed of one high- and one low-affinity site separated by 21 bp center-to-center, analogous to the arrangement in KSHV, in which the centers of LBS-1 and LBS-2 are separated by 22 bp or two helical turns (15). EBNA1 has also been shown to bend the DNA within each binding site, and measurements using CPAs give values that are remarkably similar to those presented here with LANA. For high-affinity site 4, a single EBNA1 dimer bends the DNA by approximately 55°, increasing to a minimum of 88° when sites 3 and 4 are occupied (4). Evidence of bending is also seen in the structures of EBNA1-DNA cocrystals, in which the distorted B-form DNA wraps around the surface of the DNA-binding domain (5, 6). It is noteworthy that the bovine papillomavirus E1 and E2 initiator proteins also bend DNA (17). The structure of the core DNA-binding domain of E2 is similar to that of EBNA1, and the dimers induce a bend of at least 50° (19). One function of the induced bend at the bovine papillomavirus origin is to bring the transactivation domain of E2 into contact with the helicase domain of E1, a rate-limiting step in initiation (17, 44). It is possible that DNA architecture also facilitates interactions between LANA and cellular replication proteins such as ORC or the MCM complex, with its resident helicase activity.
Despite an obvious difference in the relative gel mobilities between complexes formed by the wild-type and KK1113AA versions of LANAC and LBS-1, the calculated bend angles and the location of the bend centers were very close, if not identical. At face value, this suggests that the mobility difference is due to the conformation of the protein moieties rather than the DNA probe; however, the calculated bend angles differ by 25° when LBS-1 and LBS-2 are occupied simultaneously. The reason for this discrepancy is not clear but might reflect differences in the interactions between the two dimers. So far, we have not observed a measurable defect in the ability of the KK1113AA mutant to support plasmid replication in transient assays (J. Hu, L.-Y. Wong, A. C. Wilson, and R. Renne, unpublished data). It is possible that the higher affinity of the KK1113AA variant for DNA compensates for the effects of the shallower bend or that the transient replication assay is simply not sensitive enough to detect subtle differences in the frequencies of initiation. In conclusion, the shared ability of LANA and EBNA1 to bend origin DNA may be a necessary step in creating a specific platform capable of recruiting ORCs and associated replication initiation factors.
Acknowledgments
Søren Ottosen patiently guided us through the calculations and theory of these analyses, and we very much appreciate his significant help. We also thank Arne Stenlund, Jianhong Hu, and Rolf Renne for providing plasmids. Jim Borowiec, Ian Mohr, and Rolf Renne gave valuable comments on the manuscript.
This work was supported by funds from the Department of Microbiology and NIH grant GM61139-04.
REFERENCES
- 1.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 75:10603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev, A., E. Bochkareva, L. Frappier, and A. M. Edwards. 1998. The 2.2 Å structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1. J. Mol. Biol. 284:1273-1278. [DOI] [PubMed] [Google Scholar]
- 7.Borowiec, J. A., F. B. Dean, and J. Hurwitz. 1991. Differential induction of structural changes in the simian virus 40 origin of replication by T antigen. J. Virol. 65:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramhill, D., and A. Kornberg. 1988. A model for initiation at origins of DNA replication. Cell 54:915-918. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 10.Crothers, D. M., M. R. Gartenberg, and T. E. Shrader. 1991. DNA bending in protein-DNA complexes. Methods Enzymol. 208:118-146. [DOI] [PubMed] [Google Scholar]
- 11.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fejer, G., M. M. Medveczky, E. Horvath, B. Lane, Y. Chang, and P. G. Medveczky. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts preferentially with the terminal repeats of the genome in vivo and this complex is sufficient for episomal DNA replication. J. Gen. Virol. 84:1451-1462. [DOI] [PubMed] [Google Scholar]
- 14.Gahn, T. A., and C. L. Schildkraut. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58:527-535. [DOI] [PubMed] [Google Scholar]
- 15.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 16.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillitzer, E., G. Chen, and A. Stenlund. 2000. Separate domains in E1 and E2 proteins serve architectural and productive roles for cooperative DNA binding. EMBO J. 19:3069-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegde, R. S. 1995. Structure of the BPV-1 E2 DNA-binding domain bound to its DNA target. J. Nucl. Med. 36:25S-27S. [PubMed] [Google Scholar]
- 20.Henke, W., K. Herdel, K. Jung, D. Schnorr, and S. A. Loening. 1997. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 25:3957-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, J., and R. Renne. 2005. Characterization of the minimal replicator of Kaposi's sarcoma-associated herpesvirus latent origin. J. Virol. 79:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koepsel, R. R., and S. A. Khan. 1986. Static and initiator protein-enhanced bending of DNA at a replication origin. Science 233:1316-1318. [DOI] [PubMed] [Google Scholar]
- 25.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagunoff, M., and D. Ganem. 1997. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology 236:147-154. [DOI] [PubMed] [Google Scholar]
- 27.Lim, C., T. Seo, J. Jung, and J. Choe. 2004. Identification of a virus trans-acting regulatory element on the latent DNA replication of Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 85:843-855. [DOI] [PubMed] [Google Scholar]
- 28.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 77:11425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mysiak, M. E., C. Wyman, P. E. Holthuizen, and P. C. van der Vliet. 2004. NFI and Oct-1 bend the Ad5 origin in the same direction leading to optimal DNA replication. Nucleic Acids Res. 32:6218-6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1998. Human herpesvirus 8—the first human Rhadinovirus. J. Natl. Cancer Inst. Monogr. 23:73-77. [DOI] [PubMed] [Google Scholar]
- 32.Piolot, T., M. Tramier, M. Coppey, J.-C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polaczek, P. 1990. Bending of the origin of replication of E. coli by binding of IHF at a specific site. New Biol. 2:265-271. [PubMed] [Google Scholar]
- 34.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renne, R., M. Lagunoff, W. Zhong, and D. Ganem. 1996. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 70:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakakibara, S., K. Ueda, K. Nishimura, E. Do, E. Ohsaki, T. Okuno, and K. Yamanishi. 2004. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J. Virol. 78:7299-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salvo, J. J., and N. D. Grindley. 1987. Helical phasing between DNA bends and the determination of bend direction. Nucleic Acids Res. 15:9771-9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J.-I. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan, V., T. Komatsu, M. E. Ballestas, and K. M. Kaye. 2004. Definition of sequence requirements for latency-associated nuclear antigen 1 binding to Kaposi's sarcoma-associated herpesvirus DNA. J. Virol. 78:14033-14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stedman, W., Z. Deng, F. Lu, and P. M. Lieberman. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78:12566-12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stenlund, A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 4:777-785. [DOI] [PubMed] [Google Scholar]
- 45.Szekely, L., C. Kiss, K. Mattsson, E. Kashuba, K. Pokrovskaja, A. Juhasz, P. Holmvall, and G. Klein. 1999. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J. Gen. Virol. 80:2889-2900. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viejo-Borbolla, A., and T. F. Schulz. 2003. Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8): key aspects of epidemiology and pathogenesis. AIDS Rev. 5:222-229. [PubMed] [Google Scholar]
- 48.Wang, Q., and J. M. Calvo. 1993. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 12:2495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong, L.-Y., G. A. Matchett, and A. C. Wilson. 2004. Transcriptional activation by the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J. Virol. 78:10074-10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, H. M., and D. M. Crothers. 1984. The locus of sequence-directed and protein-induced DNA bending. Nature 308:509-513. [DOI] [PubMed] [Google Scholar]
- 51.Zahn, K., and F. R. Blattner. 1987. Direct evidence for DNA bending at the lambda replication origin. Science 236:416-422. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, J., C. Chau, Z. Deng, W. Stedman, and P. M. Lieberman. 2005. Epigenetic control of replication origins. Cell Cycle 4:889-892. [DOI] [PubMed] [Google Scholar]
- 53.Zinkel, S. S., and D. M. Crothers. 1987. DNA bend direction by phase sensitive detection. Nature 328:178-181. [DOI] [PubMed] [Google Scholar]