Abstract
Prp43p is a putative helicase of the DEAH family which is required for the release of the lariat intron from the spliceosome. Prp43p could also play a role in ribosome synthesis, since it accumulates in the nucleolus. Consistent with this hypothesis, we find that depletion of Prp43p leads to accumulation of 35S pre-rRNA and strongly reduces levels of all downstream pre-rRNA processing intermediates. As a result, the steady-state levels of mature rRNAs are greatly diminished following Prp43p depletion. We present data arguing that such effects are unlikely to be solely due to splicing defects. Moreover, we demonstrate by a combination of a comprehensive two-hybrid screen, tandem-affinity purification followed by mass spectrometry, and Northern analyses that Prp43p is associated with 90S, pre-60S, and pre-40S ribosomal particles. Prp43p seems preferentially associated with Pfa1p, a novel specific component of pre-40S ribosomal particles. In addition, Prp43p interacts with components of the RNA polymerase I (Pol I) transcription machinery and with mature 18S and 25S rRNAs. Hence, Prp43p might be delivered to nascent 90S ribosomal particles during pre-rRNA transcription and remain associated with preribosomal particles until their final maturation steps in the cytoplasm. Our data also suggest that the ATPase activity of Prp43p is required for early steps of pre-rRNA processing and normal accumulation of mature rRNAs.
Synthesis of eukaryotic ribosomes is an intricate process that is initiated by the synthesis of a precursor to 5S rRNA by RNA polymerase III and of a common precursor to 18S, 5.8S, and 25S rRNAs by RNA polymerase I (70, 93). Some nucleotides present in the sequences retained in the mature rRNAs are modified by site-specific enzymes or more generally by small nucleolar ribonucleoprotein particles (snoRNPs) during or immediately following synthesis of the primary pre-rRNA transcripts (50). These are then processed by a complex series of endonucleolytic and exonucleolytic digestion steps that remove so-called spacer regions to yield the mature rRNAs. These pre-rRNA processing events do not occur on naked pre-rRNAs but within preribosomal particles (96). These particles result from the association of pre-rRNAs at various stages of maturation with ribosomal proteins, which remain in the cytoplasmic ribosomes, as well as scores of nonribosomal proteins that only transiently interact with pre-rRNAs (25, 28). As they mature, preribosomal particles transit from the nucleolus to the nucleoplasm and are then transported to the cytoplasm where final maturation steps take place (88). With advancing maturation, the number of nonribosomal proteins present in preribosomal particles declines (2, 36, 63). Well over 170 nonribosomal proteins linked to ribosome biogenesis have been described previously, and the list is probably far from complete (28).
Apart from the enzymes shown to modify rRNA nucleotides or to catalyze endo- or exonucleolytic digestion steps, the precise molecular functions of most nonribosomal proteins remain a matter of speculation. For a subset of these nonribosomal proteins, potential roles can be envisaged because they belong to well-defined protein families of kinases (34, 53, 54, 90, 91), GTPases (2, 3, 35, 40, 49, 75, 76, 79, 99), ATPases (29, 30, 64), and helicases. The latter group is by far the largest. Indeed, no fewer than 18 potential helicases have been linked to ribosome biogenesis (13, 74, 85). Of these, 11 are required for the production of the large ribosomal subunit, namely, Dbp2p (4), Dbp3p (98), Dbp6p (16, 51), Dbp7p (12), Dbp9p (10), Dbp10p (7), Dob1p (15), Drs1p (72), Has1p (21, 73), Mak5p (67), and Spb4p (14), and 8 are required for the production of the small ribosomal subunit, namely, Dbp4p (56), Dbp8p (11), Dhr1p (9), Dhr2p (9), Fal1p (52), Has1p (21, 73), Rok1p (92), and Rrp3p (65). Related potential or bona fide helicases intervene in all aspects of RNA metabolism, such as RNA processing, transport, degradation, or during translation (74, 85). These enzymes are thought to drive conformational rearrangements involving RNA-RNA or RNA-protein interactions using NTP (generally ATP) hydrolysis. Potential helicases involved in ribosome biogenesis could conceivably modulate the folding of pre-rRNAs, facilitate preribosomal particle transport, assist exonucleases in the degradation of pre-rRNA spacer regions, regulate the association of small nucleolar RNAs (snoRNAs) or proteins with pre-rRNAs or their dissociation, etc. For example, the potential helicase Dob1p could assist the exosome during degradation of the 3′ end of 7S pre-rRNAs, since lack of Dob1p leads to the same 7S pre-rRNA processing defect as inactivation of the exosome (15). Dbp4p could be involved in modulating the interactions of the U14 snoRNA with pre-rRNAs, since the DBP4 gene was identified as a multicopy suppressor of a mutated allele of the gene encoding U14 (56). These ideas remain speculation, however, and in no cases have direct molecular substrates for helicases involved in ribosome synthesis been rigorously identified by experimentation. To attain a detailed understanding of ribosome biogenesis in eukaryotes, it is crucial to identify all potential helicases involved and to discover their substrates.
Recently, we have reported the characterization of the protein content of very early pre-60S preribosomal particles (17). These particles contain seven of the potential helicases listed above as well as Prp43p, a member of the family of “DEAH” potential helicases (1). This was surprising, because Prp43p had previously been shown to be involved in late steps of pre-mRNA splicing, more specifically in the release of the intron lariat from the spliceosome (1, 61). However, human Prp43p is detected both in nuclear speckles and in the nucleolus (26) whereas yeast Prp43p is mostly found at steady state in the nucleolus (45). These data suggested that Prp43p also has a direct role in ribosome biogenesis. In this paper, we show that Prp43p is a component of almost all preribosomal particles and that depletion of Prp43p strongly inhibits the synthesis of both small and large ribosomal subunits. Prp43p is thus one of the few nonribosomal proteins known to be required for the production of both ribosomal subunits and, to our knowledge, the only potential helicase required for pre-mRNA splicing and ribosome biogenesis identified so far.
MATERIALS AND METHODS
Strains, media, and plasmids.
A GAL::prp43-ZZ strain was obtained as follows. The PRP43 open reading frame flanked by BglII restriction sites was PCR amplified from plasmid pYCG_YGL120c (EUROSCARF collection) by use of oligonucleotides Prp43/5′pHA113 (5-GGGGGAGATCTAATGGGTTCCAAAAGAAGATT-3′) and Prp43/3′pHA113 (5′-GGGGGAGATCTATTTTCTTGGAGTGCTTACTCT-3′). The resulting PCR fragment was digested with BglII and inserted into the BamHI site of plasmid pHA114 between the hybrid GAL1-10/CYC1 promoter and the ZZ gene cassette (41), creating plasmid pSL1. An HpaI/PsiI fragment from pSL1 containing the PRP43 open reading frame flanked by the GAL1-10/CYC1 promoter on the 5′ side and the ZZ gene and PGK terminator on the 3′ side was inserted into centromeric vector pFL39 (5) cut with HincII, creating pSL9. This plasmid was transformed into diploid strain CBS1009 (HE) [CEN.PK MATa/α ura3-52/ura3-52 his3Δ1/his3Δ1 leu2-3,112/leu2-3,112 trp1-289/trp1-289 YGL120c(338, 2275)::kanMX4/YGL120c]. Sporulation of the transformed diploid strain was induced, tetrads were dissected, and a haploid strain [CEN.PK ura3-52 his3Δ1 leu2-3,112 trp1-289 YGL120c(338, 2275)::kanMX4 pSL9] was selected.
Strains expressing Pfa1p-tandem affinity purification tag (TAP), Rpa190p-TAP, and Net1p-TAP were purchased from Open Biosystems. Strains expressing Prp43p-TAP and Prp22p-TAP were produced as follows. Two gene cassettes flanked on the 5′ side by the last 48 or 56 nucleotides of the PRP43 or PRP22 open reading frames and on the 3′ side by a segment of the PRP43 or PRP22 terminator and containing the TAP-tag sequence followed by a URA3 marker gene from Kluyveromyces lactis were PCR amplified using plasmid pBS1539 (71) and oligonucleotides Prp43pTAPTAG5′ (5′-GAGTTGAAACAAGGTAAAAACAAAAAGAAGAGTAAGCACTCCAAGAAATCCATGGAAAAGAGAAG-3′) and Prp43pTAPTAG3′ (5′TGAATTTTCTCTCTATGAAATAGTCCTATAAATTTATATAAATCTATTTACGACTCACTATAGGG-3′) or oligonucleotides Prp22pTAPTAG5′ (5′-CATGGAGACTAAGCTCAATAAGGCAGTCAAGGGAAAGGGCATTAGGTATCAAGAGGTCCATGGAAAAGAGAAG-3′) and Prp22pTAPTAG3′ (5′GTTAAAAAATTAAATATAGGTCTATAAAACTCGATAATTATAATGCATAAAAATACGACTCACTATAGGG-3′). These cassettes were integrated into strain Y0341 (pra1-1 prb1-1 prc1-1 cps1-3 Δhis3 leu2-3,112 Δura3 Δtrp1::LEU2), creating strains expressing Prp43pTAP or Prp22pTAP.
Strains overexpressing a ZZ-tagged wild-type or dominant-negative version of Prp43p when grown on galactose-containing medium were obtained by transforming strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) or FY1679-03A (MATα ura3-52 trp1Δ63) with plasmids pFH141 or pFH142. Control strains expressing normal levels of wild-type Prp43p were obtained by transforming BY4741 or FY1679-03A with plasmid pFH140. To produce pFH140, a cassette containing the GAL1-10/CYC1 promoter, the ZZ gene, and the PGK terminator was excised from pHA114 by HpaI-PsiI double digestion and inserted into pCH32 cut with EcoRI, filled in with Klenow polymerase, and digested again with EcoRV. pCH32 is a centromeric vector conferring resistance to G418. pCH32 was constructed by inserting into the EcoRV site of pHA113 (41) the KanMX cassette, obtained from pUG6 (38) digested with NotI, the ends of which were filled in with Klenow polymerase. To produce pFH141, a cassette containing the PRP43 open reading frame flanked by the GAL1-10/CYC1 promoter on the 5′ side and the ZZ gene and the PGK terminator on the 3′ side was excised from pSL1 by HpaI-PsiI double digestion and inserted into pCH32 cut with EcoRI, filled in with Klenow polymerase, and digested again with EcoRV. pFH142 was constructed in several steps as follows. The PRP43 open reading frame flanked by BglII restriction sites was PCR amplified from plasmid pYCG_YGL120c by use of oligonucleotides Prp43/5′pHA113 and Prp43/3′pHA113. The resulting PCR fragment was digested with BglII and inserted into the BglII site of pSP72 (Promega), creating pSP72-PRP43. A mutagenized subfragment of the PRP43 open reading frame leading to the R430-to-A430 substitution was produced by PCR using pYCG_YGL120c and oligonucleotides 5′-GGGGGGAGGAGACCTGATTTGAAGATAATTATTATG-3′and 5′-GGGGGGAATGCCTCTTCAGTGTATAATCTGAAACATTTACCAGGCCTTGTAGCACCAGCACGACCAGCTCTTTGTTGGG-3′. This fragment was digested with BseRI and StuI and used to replace the corresponding wild-type fragment in BseRI/StuI-cut pSP72-PRP43, creating pSP72-PRP43R430A. The mutant prp43 open reading frame was excised from pSP72-PRP43R430A by BglII digestion and inserted into BglII-cut pHA114, generating pHA114-PRP43R430A. Finally, a cassette containing the mutant prp43 open reading frame flanked by the GAL1-10/CYC1 promoter on the 5′ side and the ZZ gene and PGK terminator on the 3′ side was excised from pHA114-PRP43R430A by HpaI-PsiI double digestion and inserted into pCH32 cut with EcoRI, filled in with Klenow polymerase, and digested again with EcoRV, generating pFH142.
Saccharomyces cerevisiae strains were grown either in YP medium (1% yeast extract, 1% peptone) supplemented with 2% galactose, 2% raffinose, 2% sucrose, or 2% glucose as the carbon source or in YNB medium [0.17% yeast nitrogen base, 0.5% (NH4)2SO4] supplemented with 2% galactose, 2% raffinose, 2% sucrose, and the required amino acids. G418 was added when required at a 0.2 mg/ml final concentration.
Immunoprecipitations.
Cells frozen in liquid nitrogen were broken with dry ice in a kitchen blender (Osterizer). Aliquots of broken cell powder corresponding to 2 × 1010 cells were resuspended in 2 ml of 20 mM Tris-HCl (pH 8.0)-5 mM MgAc-0.2% Triton X-100-200 mM potassium acetate (KAc)-1 mM dithiothreitol (DTT)-0.5 unit/μl RNasin (Promega) containing protease inhibitors (Roche). Extracts were clarified by centrifuging 10 min at 16,000 × g in a microcentrifuge (Eppendorf 5415D). Aliquots of clarified extracts corresponding to 12 mg of proteins were added to 50 μl of immunoglobulin G (IgG)-Sepharose beads (Amersham Pharmacia Biotech) in a 1 ml final volume of a buffer containing 20 mM Tris-HCl (pH 8.0), 5 mM MgAc, 0.2% Triton X-100, 200 mM KAc, 1 mM DTT, 0.5 unit/μl RNasin (Promega), and protease inhibitors. Immunoprecipitation was performed at 4°C for 1 h 30 min on a shaking table. Beads were then washed seven times with 1 ml of the buffer used for the immunoprecipitation (ice cold). A 160-μl volume of 4 M guanidinium isothiocyanate solution, 4 μl of glycogen, 80 μl of a 100 mM NaAc (pH 5)-10 mM Tris-HCl (pH 8.0)-1 mM EDTA solution, 120 μl of phenol, and 120 μl of chloroform were added to the beads. The samples were thoroughly mixed, incubated 5 min at 65°C, and centrifuged 5 min at 4°C and 16,000 × g in a microcentrifuge (Eppendorf 5415D). The aqueous phases were recovered and mixed with 120 μl of phenol-120 μl of chloroform, and the samples were centrifuged 5 min at 4°C and 16,000 × g in a microcentrifuge (Eppendorf 5415D). RNAs from the aqueous phases were then precipitated with ethanol.
Tandem affinity purification.
Prp43p-TAP purification was performed as indicated below, while analytical Pfa1p-TAP, Net1p-TAP, and Rpa190p-TAP purifications were performed using a procedure scaled down threefold. Cells frozen in liquid nitrogen were broken with dry ice in a kitchen blender (Osterizer). Aliquots of broken cell powder corresponding to 2 × 1011 cells were resuspended in 30 ml of 20 mM Tris-HCl (pH 8.0)-5 mM MgAc-0.2% Triton X-100-200 mM KAc-1 mM DTT-0.5 unit/μl RNasin (Promega) containing protease (Roche) and phosphatase (Sigma) inhibitors. Extracts were clarified by centrifuging 15 min at 25,000 rpm in a Ti 50.2 rotor (Beckman). Aliquots of clarified extracts corresponding to 1.8 g of proteins were added to 800 μl of IgG-Sepharose beads (Amersham Pharmacia Biotech). Immunoprecipitation was performed at 4°C for 1 h 30 min on a shaking table. Beads were then washed with 320 ml of the buffer used for the immunoprecipitation without RNasin, protease, and phosphatase inhibitors (ice cold) and then with 120 ml of TEV cleavage buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM DTT) and incubated 2 h at 16°C with 400 units of ActTEV enzyme (Invitrogen) in 4 ml of TEV cleavage buffer. Eluted samples were then mixed with 12 ml of calmodulin binding buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM Mg-acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 10 mM β-mercaptoethanol) to which 12 μl of 1 M CaCl2 was added and incubated with 400 μl of calmodulin beads (Stratagene) at 4°C for 1 h on a shaking table. Beads were washed with 80 ml of calmodulin binding buffer. Proteins were finally eluted by addition of 6 × 400 μl of calmodulin elution buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM Mg acetate, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10 mM β-mercaptoethanol). Eluted proteins were precipitated with trichloroacetic acid, separated on 8% or 12% polyacrylamide-sodium dodecyl sulfate (SDS) gels, and identified by Western analysis or mass spectrometry (MS).
Mass spectrometry.
Polypeptides stained with Coomassie blue (Brilliant Blue G, B 0770; Sigma) obtained after Prp43p-TAP purification were subjected to in-gel tryptic digestion using modified porcine trypsin (Promega, Lyon, France). The tryptic digest was analyzed by on-line capillary high-pressure liquid chromatography (LC Packings) coupled to a nanospray Qq-Tof mass spectrometer (QSTAR Pulsar; Applied Biosystems, Foster City, Calif.). Peptides were separated on a 75-μm inside diameter by 15-cm C18 PepMap column after loading onto a 300-μm inside diameter by 5-mm PepMap C18 precolumn (LC Packings; Dionex). The flow rate was set at 150 nl/min. Peptides were eluted using a 0 to 50% linear gradient of solvent B in 100 min (solvent A was 0.2% formic acid in 5% acetonitrile, and solvent B was 0.2% formic acid in 90% acetonitrile). The mass spectrometer was operated in positive ion mode at a 2.1 kV needle voltage. MS and MS/MS data were continuously acquired in an information-dependent acquisition mode consisting of a 10-s cycle time. Within each cycle, an MS spectrum was accumulated for 1 s over the m/z range 40 to 2,000 followed by three MS/MS acquisitions of 3 s each on the three most abundant ions in the MS spectrum. A 60-s dynamic exclusion duration was employed to prevent repetitive selection of the same ions within a preset time. MS/MS data were acquired using a 3-m/z-unit ion isolation window. Collision energies were automatically adjusted according to the charge state and mass value of the precursor ions, and the collision gas was N2. The MASCOT search engine (Matrix Science, London, United Kingdom) was used for protein identification by searching against nonredundant SwissProt and Trembl databases with MS/MS spectra.
Western analysis.
Proteins from total extracts (produced as described in reference 19) or present in fractions collected from calmodulin columns at the end of TAP were separated on 8% or 12% polyacrylamide-SDS gels and transferred to Hybond-C extra membranes (Amersham Pharmacia Biotech). ZZ- or TAP-tagged proteins and Nop1p were detected as described in reference 19. Endogenous Prp43p was detected using purified antibodies diluted 4 × 103-fold which had been raised in rabbits by Eurogentec against the following Prp43p peptides: H2N-GSKRRFSSEHPDPVEC-CONH2 and H2N-SNFQKGDVKLSLERIC-CONH2.
RNA extractions, Northern hybridizations, and primer extensions.
RNA extractions were performed as described by Tollervey and Mattaj (86). RNA fractionations by agarose or polyacrylamide gel electrophoresis were performed as described by Henras et al. (42). Primer extensions were performed as described by Bousquet-Antonelli et al. (6).
Pre-rRNA precursors, mature rRNAs, mRNAs, and various small RNAs were analyzed by Northern hybridization or primer extensions by use of 32P-labeled oligodeoxynucleotide probes. Sequences of antisense oligonucleotides used to detect these RNAs have been reported in references 17, 18, 19, 42, and 69 except those of oligonucleotides to detect U2 (snRNA) (5′-GTTACACTGAAAAGAACAGATACTAC-3′), tRNA-trp (5′-CATTACGAGTGCGATGCCTTAC-3′), actin mRNA (5′-GGTTCATTGGAGCTTCAGTC-3′ and 5′-GAAGATTGAGCAGCGGTTTGC-3′), cytoplasmic light chain dynein mRNA (5′-CTTTACTGATGGTTAAAATATCC-3′ and 5′-CAATCACATGCCAGGTATTGCCG-3′), Rps13p mRNA (5′-CTCAACAAGACACCAATTTG-3′ and 5′-CAGAGACAGCCTTCTTAATC-3′), Rps15p mRNA (5′-GAAATCTTCAGTGGACATTTCC-3′ and 5′-GTGACCCAACATTTCTGGTC-3′), Rpl25p mRNA (5′-CCGTTTGGTCTAACCAAAGTG-3′ and 5′-CTGTTGTAATGTGGAACAGCC-3′), and Rpl32p mRNA (5′-CTTCTTGGTGTGCTTCTTGAC-3′ and 5′-GACTAAGAAAGTCTTGTGACC-3′). Blots were hybridized with 5′ end-labeled oligonucleotide probes and washed as described by Henras et al. (42).
Pulse-chase analyses.
FY1679-03A/pFH141 or FY1679-03A/pFH142 cells were grown at 30°C in YP-glucose medium supplemented with G418 to an optical density of ∼0.4. Cells were washed twice with YP-galactose medium supplemented with G418, diluted in the same medium, and allowed to resume growth for 20 h. Cells were then washed twice with minimal YNB-galactose medium supplemented with G418 and grown for 4 h in the same medium, typically reaching an optical density of ∼0.4. To 9-ml culture samples, 450 μCi [3H]methylmethionine was added. After 3 min of labeling, 0.9 ml of 0.1 M methionine was added and 1-ml samples were collected at 1, 2, 5, 10, 20, 40, 60, 90, and 120 min following cold methionine addition.
Yeast two-hybrid screening.
The Prp43p two-hybrid screen was performed using a mating strategy as described previously by Fromont-Racine et al. (27). The CG1945 strain transformed with the pAS2ΔΔ-PRP43 bait plasmid was mated with strain Y187 transformed with a Saccharomyces cerevisiae genomic DNA library cloned into the pACTIIst plasmid. A total of 30 million diploids were screened, and 65 His+ LacZ+ colonies were selected. The genomic inserts cloned into the pACTIIst plasmids present in these colonies were identified by sequence. A total of 48 candidates were eliminated because they correspond to two antisense regions. The remaining 17 clones correspond to four different open reading frames. The complete set of genes selected in the screen is summarized in Table 1.
TABLE 1.
ORFs selected in Prp43p two-hybrid screen and corresponding genes
| Prey ORF | Corresponding prey gene | No. of clones | No. of different fusions | Start domain (nt)a | End domain (nt)b | ORF size (nt) | Gene function |
|---|---|---|---|---|---|---|---|
| YLR424W | SPP382 | 11 | 4 | 135 | 1450 | 2,127 | Splicing |
| YGR280C | GNO1 | 3 | 1 | 117 | End | 816 | rRNA maturation |
| YKL042W | SPC42 | 1 | 1c | −75 | 675 | 1,092 | Spindle pole body |
| YJL076W | NET1 | 1 | 1c | −56 | 1100 | 3,570 | RNA Pol I |
The 5′ position of the smallest overlapping fragment was accurately determined by sequencing.
The 3′ end of the smallest overlapping fragment was roughly determined according to PCR fragment size.
Out-of-frame fusion.
RESULTS
Depletion of Prp43p leads to a drastic reduction in the steady-state levels of pre-rRNA processing intermediates and mature rRNAs.
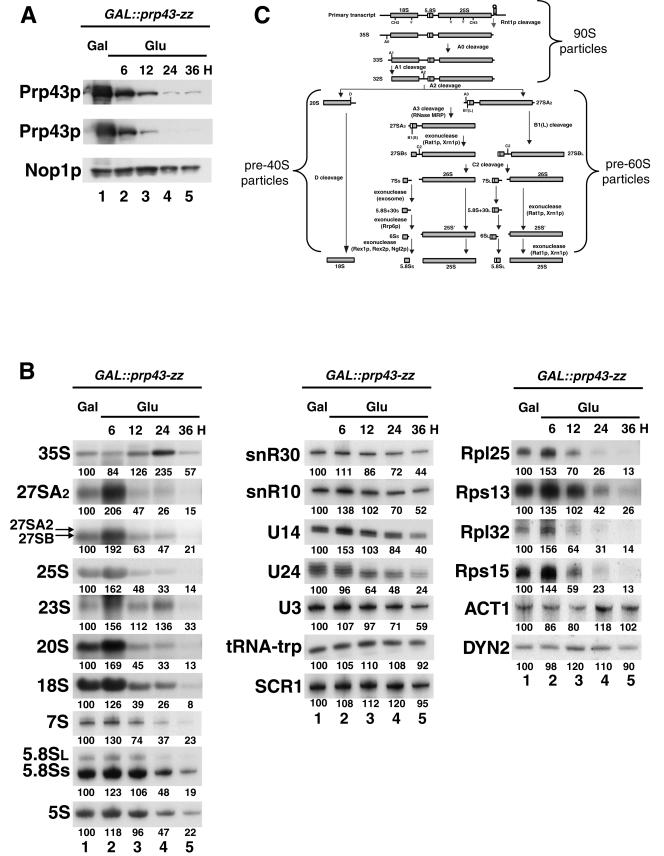
Since Prp43p has been reported to be a nucleolar protein (26, 45) and was detected among the proteins interacting with Npa1p (17), a specific component of early pre-60S preribosomal particles, we reasoned that it might be involved in ribosome biogenesis. To assess the consequences of lack of Prp43p on RNA metabolism, we constructed a yeast strain that contains a PRP43 gene transcribed from the GAL1-10-CYC1 hybrid promoter. This promoter is induced in the presence of galactose in the growth medium and is repressed when glucose is added, leading to progressive depletion of the Prp43p protein. In addition, in order to follow Prp43p depletion, a gene cassette (ZZ) encoding two IgG-binding domains derived from Staphylococcus aureus protein A was inserted in frame downstream from the PRP43 open reading frame. GAL::prp43-zz cells were grown in galactose-containing medium and transferred to glucose-containing medium for 6 to 36 h. A very severe decrease in Prp43p levels was observed after 24 h of incubation in glucose-containing medium (Fig. 1A, lane 4). Longer incubation in glucose-containing medium did not result in total disappearance of Prp43p (Fig. 1A, lane 5), presumably due to residual transcription from the GAL1-10-CYC1 promoter. As a result, even though their growth was strongly impaired, GAL::prp43-zz cells never completely ceased to divide.
FIG.1.
Prp43p depletion inhibits accumulation of mature rRNAs. A GAL::prp43-zz strain was grown in galactose-containing medium (GAL; lanes 1) and then shifted to a glucose-containing medium (Glu; lanes 2 to 5). Culture samples corresponding to the same number of cells were collected before the transfer (lanes 1) and at 6 (lanes 2), 12 (lanes 3), 24 (lanes 4), or 36 (lanes 5) h after transfer to glucose-containing medium. From these samples, total proteins (A) or RNAs (B) were extracted and subjected to Western blot (A) or Northern blot (B) analysis. In panel A, total proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), transferred to a cellulose membrane, and detected by enhanced chemiluminescence using either Dakko rabbit PAP or a polyclonal anti-Nop1p serum. Two different exposures of the film are shown for better appreciation of both the sharp decrease in Prp43p levels and the remaining quantities of that protein after 36 h of growth in glucose-containing medium. In panel B, total RNAs were separated by denaturing agarose or acrylamide gel electrophoresis and transferred to nylon membranes. Specific RNAs were detected by hybridization with antisense oligonucleotide probes. Signal intensities were measured by phosphorimager scanning. Values (indicated below each lane) are expressed as percentages of the intensities obtained in the case of galactose-grown samples. (C) Pre-rRNA processing scheme in S. cerevisiae. The primary pre-rRNA transcript is first cleaved, maybe cotranscriptionally, by the Rnt1p endonuclease in the 3′ external transcribed spacer at site B0, producing the 35S pre-rRNA. 35S is cleaved at site A0, producing 33S, which is then cleaved at site A1 corresponding to the 5′ end of mature 18S rRNA, producing 32S. Cleavage of 32S at site A2 in internal transcribed spacer 1 generates 20S, precursor to 18S rRNA, and 27SA2, precursor to 25S and 5.8S rRNAs. 20S is exported to the cytoplasm, where it undergoes endonucleolytic cleavage at site D, generating mature 18S rRNA. 27SA2 can be processed by one of two parallel pathways. A total of 90% of 27SA2 molecules are cut at site A3 by the mitochondrial RNA processing endonuclease, producing 27SA3, the 5′ end of which is digested by the Rat1p and Xrn1p exonucleases up to B1S, producing 27SBS. A total of 10% of 27SA2 molecules are processed by an as-yet-unknown mechanism at site B1L, releasing 27SBL. 27SBS and 27SBL are then processed identically. C2 cleavage releases the 26S and 7SS or 7SL molecules. The 5′ end of 26S is digested by the Rat1p and Xrn1p exonucleases up to the 5′ end of mature 25S rRNA. The ITS2 fragment remaining on 7SS or 7SL molecules is removed in successive steps by several exonucleases. The core exosome intervenes first to produce 5.8S plus 30S or 5.8S plus 30L, followed by the Rrp6p exosome component generating 6SS or 6SL molecules. The remaining ITS2 nucleotides are removed by the Rex1p, Rex2p, and Ngl2p exonucleases. The endonucleases acting at sites A0, A1, A2, and C2 are unknown. Also indicated are the names of the preribosomal particles in which these pre-rRNA intermediates are embedded.
Prp43p depletion is accompanied by a transient accumulation of the 35S and 23S pre-rRNAs and a strong decrease in the steady-state levels of the 27SA2, 27SB, 20S, and 7S pre-rRNAs and of all mature rRNAs, while levels of Scr1 RNA or tRNA-trp were little affected (Fig. 1B and 1C). This suggests that Prp43p depletion does not block pre-rRNA transcription but inhibits to some extent cleavages at the A1 and A2 sites, resulting in decreased levels of the 20S and 27SA2 pre-rRNAs. The decrease in the levels of all pre-rRNAs tested present in pre-60S particles suggests that processing steps that take place in these particles are inhibited and/or that these particles are turned over. The levels of the intronic U24 snoRNA were decreased by half after 24 h of growth in glucose. This is consistent with the findings that U24 maturation requires debranching of the intron lariat (68) and that Prp43p activity, at least in vitro, is needed to allow access of the debranching enzyme to the branch point (61). Levels of independently transcribed unspliced snoRNAs, snR30 and snR10, are only mildly decreased after 24 h of growth in glucose. Levels of mature U3 snoRNAs, which are produced following splicing of pre-U3 transcripts by the spliceosome (62), are only decreased to the same extent as those of snR10 or snR30. Strikingly, levels of mature actin or cytoplasmic light chain dynein mRNAs, produced by splicing, are not diminished following Prp43p depletion. These data suggest that splicing defects due to Prp43p depletion do not lead to strong decreases in the steady-state levels of spliced RNAs. Hence we propose that the major pre-rRNA and rRNA accumulation defects seen upon Prp43p depletion result from a direct involvement of Prp43p in ribosome biogenesis and are not the indirect consequence of reduced synthesis, due to mRNA shortage, of trans-acting factors required for the production of ribosomes. We also assessed the steady-state levels of ribosomal protein mRNAs following Prp43p depletion. Levels of all ribosomal protein mRNAs tested were strongly diminished. However, levels of ribosomal protein mRNAs generated from spliced (Rpl25, Rps13) or unspliced (Rpl32, Rps15) primary transcripts were affected equally. Hence we feel that this reduction is the result of a feedback mechanism of unknown nature, but unrelated to splicing, that shuts down ribosomal protein expression when the process of ribosome biogenesis is strongly impaired.
Prp43p is a component of multiple preribosomal particles.
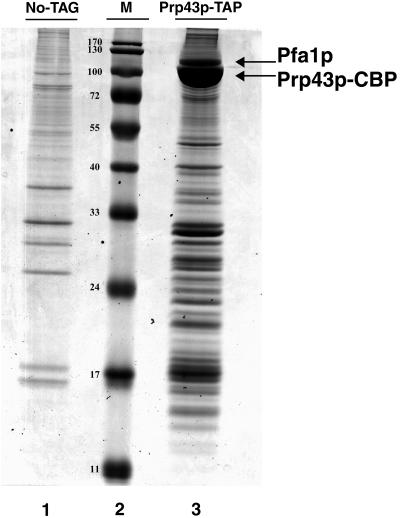
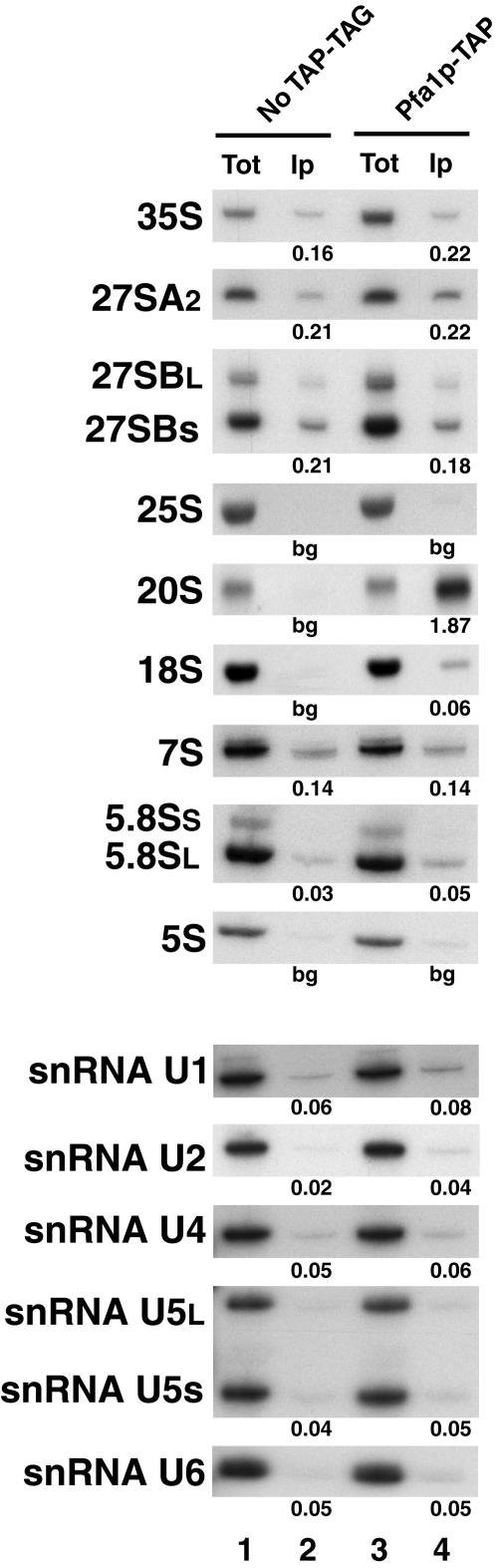
If Prp43p is directly involved in the synthesis of both small and large ribosomal subunits, we expect it to be a component of early 90S preribosomal particles and/or of both pre-60S and pre-40S preribosomal particles. With the view of identifying the RNPs which contain Prp43p, we first constructed a yeast strain expressing from the normal genomic locus Prp43p tagged with the tandem affinity tag (71), Prp43p-TAP. A control strain was also constructed that expresses tagged Prp22p, Prp22p-TAP. Prp22p is a potential helicase of the DEAH family that is required for the release of the spliced exons from the spliceosome (78, 95). Prp43p-TAP and Prp22p-TAP were precipitated from total cellular extracts using IgG-Sepharose, and the RNAs retained in the pellet were extracted and analyzed by primer extension or by the Northern blot technique (Fig. 2). Consistent with the fact that both Prp22p and Prp43p are involved in late stages of the splicing process, we find that the U2, U5, and U6 snRNAs are precipitated with both Prp22p-TAP and Prp43p-TAP (Fig. 2). In addition, several pre-rRNAs are precipitated with Prp43p-TAP but not with Prp22p-TAP. The 35S, 27SA2, and 20S pre-rRNAs are precipitated with Prp43p-TAP with efficiencies at least fourfold above background levels, while 27SB and 7S pre-rRNAs are precipitated with efficiencies about twofold above background levels. These data therefore indicate that Prp43p is a component of 90S and pre-40S as well as early pre-60S particles. They also suggest that Prp43p is a component of intermediate pre-60S particles. All snoRNAs tested were precipitated with Prp43p-TAP, most likely as a consequence of the presence of Prp43p-TAP within 90S preribosomal particles. To our surprise, mature 18S rRNA was also convincingly precipitated with Prp43p-TAP. Since the conversion of 20S pre-RNA to 18S rRNA occurs in the cytoplasm in yeast, this finding indicates that Prp43p is also present in mature or nearly mature cytoplasmic 40S ribosomal particles. Trace amounts of 25S rRNAs were also precipitated with Prp43p-TAP, while almost no background 25S rRNA precipitation was detected. Thus, Prp43p-TAP may be present in mature or nearly mature 60S particles, although we could not detect an association between Prp43p-TAP and 5.8S or 5S rRNAs (however, see below). To confirm the association of Prp43p with 90S, pre-40S, and pre-60S particles, we performed a tandem affinity purification (71) of Prp43p-TAP followed by mass spectrometry (Fig. 3 and Table 2) as well as a genome-wide double-hybrid screen using Prp43p as bait (Table 1). Consistent with our previous RNA analyses, Prp43p was found associated with splicing factors and with protein components of 90S, pre-40S, and early, intermediate, and late pre-60S preribosomal particles. The two approaches led to partially overlapping results, as both identified the putative splicing factor Spp382p and the pre-rRNA processing factor Gno1p (37) as partners of Prp43p.
FIG. 2.
Prp43p is associated with several spliceosomal snRNAs and pre-rRNAs and with mature 18S and 25S rRNAs. Immunoprecipitation experiments were carried out using IgG-Sepharose and total cellular extracts from the parental strain lacking a tagged protein (No TAP-TAG; lanes 1 and 2), or from strains expressing Prp43p-TAP (lanes 3 and 4) or Prp22p-TAP (lanes 5 and 6). RNAs were extracted from the pellets obtained following precipitation (Ip) or from an amount of input extract corresponding to 1/150 of that used for the precipitation (Tot) and analyzed by the Northern blot technique or the primer-extension procedure (to detect 35S, 27SA2, and 27SB pre-rRNAs). cDNA products obtained by primer extensions were separated on a 6% sequencing gel, and RNAs were separated and detected as described in the legend of Fig. 1B. Signal intensities were measured by phosphorimager scanning to derive the percentage of input RNAs precipitated (indicated below each Ip lane). bg, background value.
FIG. 3.
Tandem affinity purification of Prp43p-TAP. Total extracts from strains expressing Prp43p-TAP (lane 3) or devoid of tagged protein (lane 1) were subjected to the tandem affinity purification procedure. Polypeptides eluted from the calmodulin columns following EGTA addition were resolved by SDS-PAGE and stained with Coomassie blue. Stained proteins were excised, digested by trypsin, and identified by coupling liquid chromatography and mass spectrometry. Lane 2: molecular mass markers (in kilodaltons).
TABLE 2.
Proteins specifically associated with Prp43pa
| Category and protein | Accession no.b | Category and protein | Accession no.b | |
|---|---|---|---|---|
| RNA Pol I machinery | ||||
| Rpa190p | P10964 | |||
| Rpa135p | P22138 | |||
| Rpa49p | Q01080 | |||
| Rpa34p | P47006 | |||
| Rpc19p | P28000 | |||
| 90S/Pol I | ||||
| Utp10p | P42945 | |||
| Utp15p | Q04305 | |||
| 90S | ||||
| Utp9p | P38882 | |||
| Utp14p | Q04500 | |||
| Utp20p | P35194 | |||
| Utp22p | P53254 | |||
| Bms1p | Q08965 | |||
| Kre33p | P53914 | |||
| Krr1p | P25586 | |||
| Nsr1p | P27476 | |||
| 90S/60S | ||||
| Has1p | Q03532 | |||
| Nop13p | P53883 | |||
| Rrp5p | Q05022 | |||
| Rrp12p | Q12754 | |||
| 60S | ||||
| Arx1p | Q03862 | |||
| Brx1p | Q08235 | |||
| Dbp10p | Q12389 | |||
| Ebp2p | P36049 | |||
| Erb1p | Q04660 | |||
| Fpr3p | P38911 | |||
| Fpr4p | Q06205 | |||
| Lsg1p | P53145 | |||
| Mrt4p | P33201 | |||
| Noc1p | Q12176 | |||
| Nog1p | Q02892 | |||
| Nog2p | P53742 | |||
| Nop12p | Q08208 | |||
| Nop53p | Q12080 | |||
| Npa1p | P34241 | |||
| Nsa2p | P40078 | |||
| Nsa3p | P38779 | |||
| Nug1p | P40010 | |||
| Rpf1p | P38805 | |||
| Rrp1p | P35178 | |||
| Rrp14p | P36080 | |||
| Spb1p | P25582 | |||
| Ssf2p | Q12153 | |||
| Xrn1p | P22147 | |||
| YCR072cp | P25382 | |||
| 40S | ||||
| Pfa1p | P53866 | |||
| Rio1p | Q12196 | |||
| H/ACA snoRNP | ||||
| Cbf5p | P33322 | |||
| Gar1p | P28007 | |||
| Nhp2p | P32495 | |||
| C/D snoRNP | ||||
| Nop1p | P15646 | |||
| Nop56p | Q12460 | |||
| Nop58p | Q12499 | |||
| Snu13p | P39990 | |||
| snoRNP | ||||
| Srp40p | P32583 | |||
| Other ribosome biogenesis components | ||||
| Arb1p | P40024 | |||
| Dbp2p | P24783 | |||
| Gno1p | P53335 | |||
| YCR016wp | P25617 | |||
| Chromatin remodeling/RNA Pol II-associated factors | ||||
| Arp9p | Q05123 | |||
| Snf2p | P22082 | |||
| Snf5p | P18480 | |||
| Spt2p | P06843 | |||
| Spt7p | P35177 | |||
| Ssl1p | Q04673 | |||
| Swi1p | P09547 | |||
| Swi3p | P32591 | |||
| Taf6p | P53040 | |||
| Histones | ||||
| Histone H2A.1 | P04911 | |||
| Histone H2A.2 | P04912 | |||
| Htz1p (histone H2A variant) | Q12692 | |||
| Histone H2B.1 | P02293 | |||
| Histone H2B.2 | P02294 | |||
| Histone H4 | P02309 | |||
| Splicing factors | ||||
| Brr2p | P32639 | |||
| Bud31p | P25337 | |||
| Clf1p | Q12309 | |||
| Cwc2p | Q12046 | |||
| Cwc23p | Q6B1S2 | |||
| Lea1p | Q08963 | |||
| Lsm12p | P38828 | |||
| Ntc20p | P38302 | |||
| Prp19p | Q8NJV2 | |||
| Prp45p | P28004 | |||
| Prp46p | Q12417 | |||
| Prp8p | P33334 | |||
| SmBp | P40018 | |||
| SmD2p | Q06217 | |||
| SmD3p | P43321 | |||
| Snu114p | P36048 | |||
| Spp382p | Q06411 | |||
| Syf2p | P53277 | |||
| YKR022cp | P36118 | |||
| Transport | ||||
| Gsp1p | P32835 | |||
| Gsp2p | P32836 | |||
| Srp1p | Q02821 | |||
| Sxm1p | Q04175 | |||
| Unknown | ||||
| YGR130cp | P53278 | |||
| YJL122wp | P47019 | |||
| YOR252wp | Q08687 |
Ribosomal proteins have been omitted from this table. The proteins listed were not detected in the mock purification (Fig. 3, lane 1).
Swiss Prot database.
Prp43p is associated with Pfa1p, a novel component of pre-40S particles.
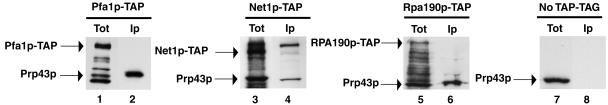
Among the proteins purified with Prp43p-TAP, the as-yet-uncharacterized polypeptide encoded by the Ynl224c open reading frame may be the most abundant, judging from the intensity of the corresponding band (Fig. 3, lane 3), the proportion of the protein covered by the tryptic peptides retrieved (sequence coverage of 61%), and the detection of numerous degradation products of this protein in several other bands. We named this polypeptide Pfa1p (Prp Forty-three Associated). To confirm the association between Pfa1p and Prp43p, we purified Pfa1p-TAP by the tandem affinity procedure and assessed the copurification of Prp43p-TAP by Western analysis using affinity-purified anti-Prp43p antibodies (Fig. 4). Prp43p could easily be detected, even in small aliquots of the final fractions eluted from the calmodulin column, both by Western analysis (Fig. 4, lane 2) and after Coomassie staining of the gel (not shown). The efficient copurification of Pfa1p with Prp43p-TAP (and vice versa) prompted us to investigate whether Pfa1p is present in all RNPs that contain Prp43p or in only a subset. Pfa1p-TAP was therefore precipitated with IgG-Sepharose from total extracts, and the RNAs retained in the pellet were extracted and analyzed by primer extension or by the Northern blot technique (Fig. 5). Only the 20S pre-rRNAs and 18S rRNAs were precipitated with efficiencies above background levels. Hence we conclude that Pfa1p is present together with Prp43p only in pre-40S preribosomal particles and in mature or nearly mature 40S small ribosomal subunits but is absent from all other preribosomal particles and from spliceosomal complexes.
FIG. 4.
Prp43p is associated with Pfa1p and with the RNA polymerase I machinery. Total extracts from cells expressing Pfa1p-TAP (lanes 1 and 2), Net1p-TAP (lanes 3 and 4), or Rpa190p-TAP (lanes 5 and 6) or devoid of tagged protein (lanes 7 and 8) were subjected to the tandem affinity purification procedure. In the case of the Pfa1p-TAP purification, 1/40th of the second fraction eluted from the calmodulin column was loaded in the immunoprecipitation (Ip) lane while in the case of the Net1p-TAP, Rpa190p-TAP, and control (No TAP-TAG) purifications, fractions eluted from the calmodulin column were pooled and all proteins present were precipitated using trichloroacetic acid and loaded in the Ip lanes. Tot: aliquot of the input extract. Proteins were subjected to SDS-PAGE, transferred to a cellulose membrane, and detected by enhanced chemiluminescence using affinity-purified anti-Prp43p polyclonal antibodies.
FIG. 5.
Pfa1p is associated with 20S pre-rRNA and 18S rRNA. Immunoprecipitations (Ip) were carried out from total extracts of cells expressing Pfa1p-TAP (lanes 3 and 4) or devoid of tagged protein (lanes 1 and 2). Immunoprecipitations and analysis of precipitated RNAs were performed as described in the legend of Fig. 2.
Prp43p is associated with the RNA polymerase I machinery.
We were surprised to discover that the two-hybrid screen conducted with the PRP43 gene as bait selected a fragment of the NET1 open reading frame as a prey. Net1p is associated with the RNA polymerase I machinery and stimulates its activity in vivo and in vitro (80). These data suggested that Prp43p interacts with the RNA polymerase I machinery. This idea is strongly supported by the detection of five subunits of RNA polymerase I among factors purified with Prp43p-TAP (Table 2). To further confirm that Prp43p interacts with the RNA polymerase I machinery, TAP-tagged Net1p and Rpa190p (the largest subunit of RNA polymerase I) were purified by the TAP procedure and the presence of Prp43p in the fractions eluting from the calmodulin column was assessed by Western analysis using anti-Prp43p antibodies (Fig. 4). These experiments confirmed that a small fraction of Prp43p is indeed associated with both Net1p and Rpa190p.
A dominant-negative form of Prp43p stalls in preribosomal particles and causes a severe reduction in the steady-state levels of mature rRNAs.
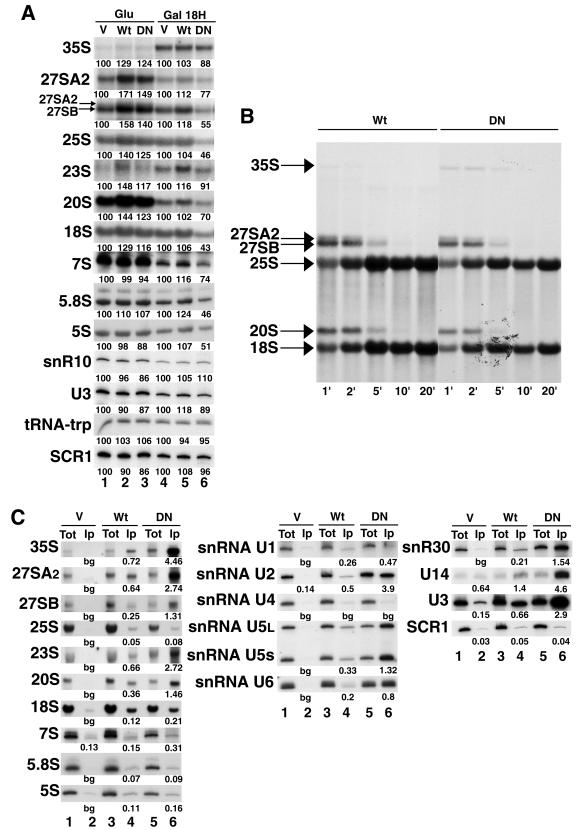
Several potential helicases, including Prp43p, display ATPase activity in vitro, and it has been proposed that conserved motif VI of these enzymes participates in ATP binding (74). Consistent with this view, Schwer and colleagues have shown that modified Prp43p proteins bearing amino acid substitutions within motif VI display severely reduced ATPase activity in vitro, cannot restore the viability of cells lacking wild-type Prp43p, and inhibit growth when overexpressed in a wild-type strain (61). We were interested in determining when and to what extent the ATPase activity of Prp43p is needed during ribosome biogenesis. Thus, we decided to analyze the phenotype induced by overexpression of a modified form of Prp43p, Prp43pR430A, containing an alanine instead of arginine at position 430 within motif VI. We confirmed that overexpression of Prp43pR430A, but not that of the wild-type protein, strongly inhibits growth (data not shown). Northern analysis demonstrates that overexpression of Prp43pR430A causes a mild decrease in the steady-state levels of 27SA2, 27SB, 20S, and 7S pre-rRNAs and a substantial drop in the amounts of all mature rRNAs (Fig. 6A). Pulse-chase analysis shows that no single pre-rRNA processing step is completely blocked by overexpression of Prp43pR430A (Fig. 6B). Nevertheless, the 35S pre-rRNA is still detected 10 min after addition of cold methionine, indicating that maturation of some 90S preribosomal particles is substantially slowed. The 35S, 27SA2, 27SB, 23S, and 20S pre-rRNAs and 18S rRNA are precipitated substantially more efficiently with Prp43pR430A than with the wild-type protein (Fig. 6C), strongly suggesting that Prp43pR430A has a reduced ability to dissociate from the ribosomal particles in which it is engaged. The increase in precipitation efficiency is most dramatic in the case of the 35S pre-rRNA (strikingly, the 35S pre-rRNA can be detected after precipitation by ethidium bromide staining of the gel) (data not shown). This latter observation is fully consistent with the slowed 35S maturation detected by pulse-chase analysis. Altogether, these data suggest that the ATPase activity of Prp43p is required already during maturation of 90S preribosomal particles and for correct production of mature ribosomes.
FIG. 6.
Phenotype induced by overexpression of Prp43pR430A. (A) Northern analysis. A wild-type yeast strain was transformed with a centromeric vector lacking an insert (V) or containing either the wild-type PRP43 gene (Wt) or a mutated prp43 allele encoding Prp43pR430A (DN) positioned downstream of the GAL1-10-CYC1 promoter and upstream of the ZZ gene cassette encoding two IgG-binding domains derived from S. aureus protein A. The transformed strains were grown in glucose-containing medium, washed, and incubated for 18 h in galactose-containing medium. Samples from cultures growing in glucose (lanes 1 to 3) or in galactose (lanes 4 to 6) corresponding to the same number of cells were collected. Total RNAs were extracted from these samples and analyzed by the Northern blot technique as described in the legend of Fig. 1. Signal intensities were measured by phosphorimager scanning. Values (indicated below each lane) are expressed as percentages of the intensities obtained in the case of V strains grown in glucose- or galactose-containing media. (B) Pulse-chase analysis. Strains transformed with the plasmid containing the wild-type PRP43 gene (Wt) or the mutated prp43 allele encoding Prp43pR430A (DN) were grown in glucose-containing medium and transferred for 24 h to a galactose-containing medium. Cells were pulse labeled with 540 μCi [3H]methylmethionine for 3 min. An excess of cold methionine was then added, and cell samples were collected at the indicated times after cold methionine addition. Total RNAs extracted from these samples were separated on a 1% agarose denaturing gel. Separated RNAs were transferred to a nylon membrane and labeled RNAs were detected by fluorography. (C) Immunoprecipitations. The V strains (lanes 1 and 2), Wt strains (lanes 3 and 4), or DN strains (lanes 5 and 6) (see above) were grown in glucose-containing medium, washed, and incubated for 24 h in galactose-containing medium. Immunoprecipitations were performed with extracts from the cultures grown in galactose as described in the legend of Fig. 2.
DISCUSSION
The yeast ATPase Prp43p has been shown to be required, at least in vitro, for the last step of splicing, that is, the dissociation of the late postsplicing complex from the spliced-out intron in the lariat form (1, 61). Consistent with this, we find that the U2, U5, and U6 spliceosomal snRNAs are precipitated with yeast Prp43p. Interestingly, we find that yeast Prp43p interacts with several components of the Prp19p-associated complex (nineteen complex or NTC) that is required for spliceosome activation (8), suggesting that yeast Prp43p may have additional, earlier roles in splicing. This seems at any rate to be the case in humans, since the likely human orthologue of yeast Prp43p, hPrp43, was detected in affinity-purified pre-spliceosome complex A (39), spliceosome complex B prior to activation (BΔU1) (60), activated B* spliceosome poised for splicing (59), and spliceosome complex C (48) that has undergone the first trans-esterification reaction. Yeast Prp43p may even associate cotranscriptionally with nascent pre-mRNAs, because tandem affinity purification of Prp43p-TAP associated with mass spectrometry analysis revealed an interaction of this protein with a surprising large number of chromatin remodeling-RNA polymerase II-associated factors (Table 2). In addition to its involvement in splicing, we propose that Prp43p also plays major, direct roles in eukaryotic ribosome biogenesis. We show in the present report that Prp43p depletion leads to a dramatic drop in the steady-state levels of all pre-rRNAs, except the 35S pre-rRNA, resulting in greatly diminished levels of all mature rRNAs.
While we cannot formally exclude the possibility that this inhibition of ribosome biogenesis is solely the indirect consequence of splicing defects, we deem it unlikely for the following reasons. In the case of both yeast and human cells, Prp43p is detected in the nucleolus (26, 45), the nuclear locale devoted primarily to early steps of ribosome biogenesis. We demonstrated by a very thorough study combining immunoprecipitations, TAP coupled to mass spectrometry, and double-hybrid analysis that yeast Prp43p is a component of almost all preribosomal particles, namely, 90S, pre-40S, and pre-60S particles. Our results extend those obtained during earlier affinity purifications (33, 43) and a double-hybrid study (46) that found Prp43p associated with a few components of 90S and pre-60S preribosomal particles. The highly significant link between Prp43p and some components of these particles has also been underscored by a recent bioinformatic analysis (55). Crucially, we note that under the experimental conditions we used, the depletion of Prp43p has little effect on the steady-state accumulation of some mature mRNAs that are produced by splicing, such as the mRNAs encoding actin or cytoplasmic light chain dynein. Thus, we think it rather unlikely that a defect in the splicing process per se did cause such a substantial drop in the steady-state levels of mRNAs for ribosome biogenesis factors, and hence a significant reduction of their de novo synthesis, that it would indirectly have led to the major perturbation of ribosome synthesis we observe. We did check the steady-state levels of ribosomal protein mRNAs and found that they declined sharply during Prp43p depletion. However, this drop is not due to splicing inhibition because levels of ribosomal protein mRNAs that are produced by splicing and of those that are not were diminished equally. This phenomenon may reflect a feedback mechanism, transcriptional and/or posttranscriptional, that shuts down ribosomal protein expression when synthesis of both small and large ribosomal subunits is severely compromised.
Prp43p is one of the few known ribosome biogenesis factors required for the synthesis of both small and large ribosomal subunits and present within pre-40S and pre-60S particles. Other examples include Rrp5p, required for the synthesis of 18S rRNA and the short form of 5.8S rRNA (22, 23, 87, 94), Rrp12p, which intervenes in the export of both small and large preribosomal subunits from nucleus to cytoplasm (66), and the DEAD-box ATPase Has1p, which is needed for 18S rRNA synthesis but is also found within pre-60S preribosomal particles (21, 73). Prp43p is also one of the rare examples of factors implicated in both splicing and ribosome biogenesis and is to our knowledge the only helicase determined as such so far. The most thoroughly studied factor shared between the splicing and ribosome biogenesis processes is Snu13p, a protein component of U4 snRNP and of all box C/D snoRNPs (97). Whether the use by the cell of Prp43p in both splicing and ribosome biogenesis contributes to a hypothetical coordination between these two processes remains to be determined.
The finding that Prp43p is associated with components of the RNA polymerase I machinery suggests that Prp43p assembles with nascent 90S preribosomal particles during pre-rRNA transcription. However, contrary to the findings with respect to the tUTP components of the U3 processome that are required for RNA polymerase I transcription (20, 31), Prp43p does not play a fundamental role in that process because the 35S pre-rRNA accumulates in Prp43p-depleted cells while it disappears in tUTP-depleted ones. We also observe a clear association of Prp43p with cytoplasmic 18S rRNA. The precipitation efficiency of 25S rRNA with tagged Prp43p is low but is nevertheless clearly above background levels. We also detect an association between Prp43p and 5.8S and 5S rRNAs when the protein is overexpressed. Moreover, Prp43p-TAP interacts with Lsg1p, a cytoplasmic GTPase required for a remodeling step of 60S ribosomal particles in the cytoplasm (40), arguing that Prp43p is associated with mature or nearly mature 60S ribosomal particles in the cytoplasm. Thus, it seems that Prp43p remains part of the ribosome biogenesis process from start (pre-rRNA transcription) to finish (production of mature 40S and 60S ribosomal particles in the cytoplasm). The prolonged association of Prp43p with preribosomal particles may seem counterintuitive for an enzyme proposed to drive transient conformational rearrangements. It is, however, a phenomenon already well documented in the case of the potential helicases Sub2p (32, 47, 57, 58, 83, 84, 101) and Dbp5p (24, 44, 77, 81, 82, 89, 100, 102), which are probably recruited to nascent pre-mRNA during transcription and remain linked to the transcript throughout splicing. Strikingly, in similarity to what we find in the case of Prp43p and preribosomal particles, Dbp5p accompanies mRNAs during export through the nuclear pore into the cytoplasm, where it drives mRNP rearrangements. The prolonged presence of Prp43p within preribosomal particles may reflect a structural role for this protein. It is also possible that Prp43p drives a succession of conformational rearrangements through several rounds of ATP hydrolysis and several association-dissociation cycles. In that respect, it is interesting that the dominant-negative forms of Prp43p, although predominantly stalled in 90S preribosomal particles, are also found in more downstream particles, suggesting that Prp43p may “enter” at several points along the pathway. The precise functions and molecular substrates of Prp43p remain to be identified. Our data suggest that the ATPase activity of this protein is required early, already within 90S preribosomal particles, to allow efficient processing of the 35S pre-rRNA. Since Prp43p associates with both pre-40S and pre-60S preribosomal particles, it must have several partners and substrates. Within pre-40S and mature or nearly mature 40S particles, and only there, Prp43p may be more closely linked to the Pfa1p protein. This link is, however, not a crucial one, because the growth disadvantage caused by lack of Pfa1p can only be detected under mixed-culture conditions. Clearly, identifying the direct partners and substrates of Prp43p constitutes a challenge for future research.
Acknowledgments
We thank J. Staley (University of Chicago) and S. Stevens (University of Texas at Austin) for communicating results prior to publication. The gifts of plasmid pBS1539 from B. Séraphin (CNRS, Gif-sur-Yvette) are gratefully acknowledged. We are thankful to members of the Ferrer laboratory for help and numerous discussions. We thank Y. de Préval for synthesis of oligonucleotides, D. Villa for art work, and A. Rivals for expert technical assistance.
S.L. is a recipient of a postgraduate fellowship from the Ministère Délégué à l'Enseignement Supérieur et à la Recherche. This work was supported by the CNRS, the Université Paul Sabatier, and grants from La Ligue Nationale contre le Cancer (“Equipe Labelisée”) to M.C.-F. and from the Région Midi-Pyrénées and the Génopole Toulouse Midi-Pyrénées to B.M.
REFERENCES
- 1.Arenas, J. E., and J. N. Abelson. 1997. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 94:11798-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 3.Becam, A. M., F. Nasr, W. J. Racki, M. Zagulski, and C. J. Herbert. 2001. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol. Genet. Genom. 266:454-462. [DOI] [PubMed] [Google Scholar]
- 4.Bond, A. T., D. A. Mangus, F. He, and A. Jacobson. 2001. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 21:7366-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonneaud, N., O. Ozier-Kalogeropoulos, G. Y. Li, M. Labouesse, L. Minvielle-Sebastia, and F. Lacroute. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7:609-615. [DOI] [PubMed] [Google Scholar]
- 6.Bousquet-Antonelli, C., E. Vanrobays, J.-P. Gélugne, M. Caizergues-Ferrer, and Y. Henry. 2000. Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA 6:826-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burger, F., M. C. Daugeron, and P. Linder. 2000. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 28:2315-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, S. P., D. I. Kao, W. Y. Tsai, and S. C. Cheng. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279-282. [DOI] [PubMed] [Google Scholar]
- 9.Colley, A., J. D. Beggs, D. Tollervey, and D. L. Lafontaine. 2000. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C + D snoRNP U3. Mol. Cell. Biol. 20:7238-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daugeron, M. C., D. Kressler, and P. Linder. 2001. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA 7:1317-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugeron, M. C., and P. Linder. 2001. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 29:1144-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daugeron, M. C., and P. Linder. 1998. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA 4:566-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 14.de la Cruz, J., D. Kressler, M. Rojo, D. Tollervey, and P. Linder. 1998. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 4:1268-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Cruz, J., D. Kressler, D. Tollervey, and P. Linder. 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 17:1128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Cruz, J., T. Lacombe, O. Deloche, P. Linder, and D. Kressler. 2004. The putative RNA helicase Dbp6p functionally interacts with Rpl3p, Nop8p and the novel trans-acting factor Rsa3p during biogenesis of 60S ribosomal subunits in Saccharomyces cerevisiae. Genetics 166:1687-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dez, C., C. Froment, J. Noaillac-Depeyre, B. Monsarrat, M. Caizergues-Ferrer, and Y. Henry. 2004. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 24:6324-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dez, C., A. Henras, B. Faucon, D. Lafontaine, M. Caizergues-Ferrer, and Y. Henry. 2001. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 29:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dez, C., J. Noaillac-Depeyre, M. Caizergues-Ferrer, and Y. Henry. 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22:7053-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emery, B., J. de la Cruz, S. Rocak, O. Deloche, and P. Linder. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 52:141-158. [DOI] [PubMed] [Google Scholar]
- 22.Eppens, N. A., A. W. Faber, M. Rondaij, R. S. Jahangir, S. van Hemert, J. C. Vos, J. Venema, and H. A. Raué. 2002. Deletions in the S1 domain of Rrp5p cause processing at a novel site in ITS1 of yeast pre-rRNA that depends on Rex4p. Nucleic Acids Res. 30:4222-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eppens, N. A., S. Rensen, S. Granneman, H. A. Raué, and J. Venema. 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5:779-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estruch, F., and C. N. Cole. 2003. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol. Biol. Cell 14:1664-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 26.Fouraux, M. A., M. J. Kolkman, A. Van der Heijden, A. S. De Jong, W. J. Van Venrooij, and G. J. Pruijn. 2002. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA 8:1428-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fromont-Racine, M., J.-C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277-282. [DOI] [PubMed] [Google Scholar]
- 28.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 29.Gadal, O., D. Strauss, J. Braspenning, D. Hoepfner, E. Petfalski, P. Philippsen, D. Tollervey, and E. Hurt. 2001. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galani, K., T. A. Nissan, E. Petfalski, D. Tollervey, and E. Hurt. 2004. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 279:55411-55418. [DOI] [PubMed] [Google Scholar]
- 31.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gatfield, D., H. Le Hir, C. Schmitt, I. C. Braun, T. Kocher, M. Wilm, and E. Izaurralde. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11:1716-1721. [DOI] [PubMed] [Google Scholar]
- 33.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Séraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 34.Geerlings, T. H., A. W. Faber, M. D. Bister, J. C. Vos, and H. A. Raué. 2003. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S pre-rRNA in Saccharomyces cerevisiae. J. Biol. Chem. 278:22537-22545. [DOI] [PubMed] [Google Scholar]
- 35.Gelperin, D., L. Horton, J. Beckman, J. Hensold, and S. K. Lemmon. 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7:1268-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 37.Guglielmi, B., and M. Werner. 2002. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J. Biol. Chem. 277:35712-35719. [DOI] [PubMed] [Google Scholar]
- 38.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henras, A., C. Dez, J. Noaillac-Depeyre, Y. Henry, and M. Caizergues-Ferrer. 2001. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res. 29:2733-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henras, A., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J.-P. Gélugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 44.Hodge, C. A., H. V. Colot, P. Stafford, and C. N. Cole. 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 18:5778-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 46.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen, T. H., J. Boulay, M. Rosbash, and D. Libri. 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11:1711-1715. [DOI] [PubMed] [Google Scholar]
- 48.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 51.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1998. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1855-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:7283-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 54.LaRonde-LeBlanc, N., and A. Wlodawer. 2004. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure (Cambridge) 12:1585-1594. [DOI] [PubMed] [Google Scholar]
- 55.Lee, I., S. V. Date, A. T. Adai, and E. M. Marcotte. 2004. A probabilistic functional network of yeast genes. Science 306:1555-1558. [DOI] [PubMed] [Google Scholar]
- 56.Liang, W. Q., J. A. Clark, and M. J. Fournier. 1997. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol. Cell. Biol. 17:4124-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Libri, D., N. Graziani, C. Saguez, and J. Boulay. 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 15:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 59.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Lührmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 60.Makarova, O. V., E. M. Makarov, H. Urlaub, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23:2381-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin, A., S. Schneider, and B. Schwer. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277:17743-17750. [DOI] [PubMed] [Google Scholar]
- 62.Myslinski, E., V. Ségault, and C. Branlant. 1990. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science 247:1213-1216. [DOI] [PubMed] [Google Scholar]
- 63.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nissan, T. A., K. Galani, B. Maco, D. Tollervey, U. Aebi, and E. Hurt. 2004. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol. Cell 15:295-301. [DOI] [PubMed] [Google Scholar]
- 65.O'Day, C. L., F. Chavanikamannil, and J. Abelson. 1996. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 24:3201-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oeffinger, M., M. Dlakic, and D. Tollervey. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 18:196-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohtake, Y., and R. B. Wickner. 1995. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol. Cell. Biol. 15:2772-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petfalski, E., T. Dandekar, Y. Henry, and D. Tollervey. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qu, L.-H., A. Henras, Y.-J. Lu, H. Zhou, W.-X. Zhou, Y.-Q. Zhu, J. Zhao, Y. Henry, M. Caizergues-Ferrer, and J.-P. Bachellerie. 1999. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19:1144-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raué, H. A. 2003. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine. .In M. Olson (ed.), The nucleolus. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 71.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 72.Ripmaster, T. L., G. P. Vaughn, and J. L. Woolford, Jr. 1992. A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl. Acad. Sci. USA 89:11131-11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rocak, S., B. Emery, N. K. Tanner, and P. Linder. 2005. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 33:999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell. Biol. 5:232-241. [DOI] [PubMed] [Google Scholar]
- 75.Saveanu, C., D. Bienvenu, A. Namane, P.-E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saveanu, C., A. Namane, P.-E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmitt, C., C. von Kobbe, A. Bachi, N. Pante, J. P. Rodrigues, C. Boscheron, G. Rigaut, M. Wilm, B. Séraphin, M. Carmo-Fonseca, and E. Izaurralde. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 18:4332-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 80.Shou, W., K. M. Sakamoto, J. Keener, K. W. Morimoto, E. E. Traverso, R. Azzam, G. J. Hoppe, R. M. Feldman, J. DeModena, D. Moazed, H. Charbonneau, M. Nomura, and R. J. Deshaies. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8:45-55. [DOI] [PubMed] [Google Scholar]
- 81.Snay-Hodge, C. A., H. V. Colot, A. L. Goldstein, and C. N. Cole. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strahm, Y., B. Fahrenkrog, D. Zenklusen, E. Rychner, J. Kantor, M. Rosbach, and F. Stutz. 1999. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 18:5761-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 84.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 85.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 86.Tollervey, D., and I. W. Mattaj. 1987. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 6:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Torchet, C., C. Jacq, and S. Hermann-Le Denmat. 1998. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA 4:1636-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 89.Tseng, S. S., P. L. Weaver, Y. Liu, M. Hitomi, A. M. Tartakoff, and T. H. Chang. 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 17:2651-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vanrobays, E., J.-P. Gélugne, P.-E. Gleizes, and M. Caizergues-Ferrer. 2003. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:2083-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vanrobays, E., P.-E. Gleizes, C. Bousquet-Antonelli, J. Noaillac-Depeyre, M. Caizergues-Ferrer, and J.-P. Gélugne. 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 20:4204-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Venema, J., C. Bousquet-Antonelli, J.-P. Gélugne, M. Caizergues-Ferrer, and D. Tollervey. 1997. Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol. 17:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 94.Venema, J., and D. Tollervey. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15:5701-5714. [PMC free article] [PubMed] [Google Scholar]
- 95.Wagner, J. D., E. Jankowsky, M. Company, A. M. Pyle, and J. N. Abelson. 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17:2926-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Warner, J. R. 2001. Nascent ribosomes. Cell 107:133-136. [DOI] [PubMed] [Google Scholar]
- 97.Watkins, N. J., V. Ségault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Lührmann. 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 98.Weaver, P. L., C. Sun, and T. H. Chang. 1997. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol. Cell. Biol. 17:1354-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wegierski, T., E. Billy, F. Nasr, and W. Filipowicz. 2001. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA 7:1254-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weirich, C. S., J. P. Erzberger, J. M. Berger, and K. Weis. 2004. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16:749-760. [DOI] [PubMed] [Google Scholar]
- 101.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao, J., S. B. Jin, B. Bjorkroth, L. Wieslander, and B. Daneholt. 2002. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 21:1177-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]