Abstract
The pathogen Candida albicans responds to amino acid starvation by activating pseudohyphal development and the expression of amino acid biosynthetic genes (GCN response). In Saccharomyces cerevisiae, the GCN response is dependent on Gcn2, which regulates the translation of the transcription factor Gcn4. Therefore, we examined the role of Gcn2 in C. albicans by using molecular, cellular, and genomic approaches. We show that C. albicans GCN2 encodes an eIF2α kinase, like its S. cerevisiae homologue. However, GCN4 appears to be regulated mainly at the transcriptional level in C. albicans. Furthermore, the inactivation of C. albicans Gcn2 only partially attenuates growth under amino acid starvation conditions and resistance to the histidine analogue 3-aminotriazole. Our comparison of the Gcn4 and Gcn2 regulons by transcript profiling reinforces the view that Gcn2 contributes to, but is not essential for, the activation of general amino acid control in C. albicans.
Several factors are thought to promote the virulence of the major systemic fungal pathogen of humans Candida albicans (10, 39). These include yeast-(pseudo)hypha morphogenesis and biofilm formation (23, 31). In C. albicans, responses to amino acid availability are intimately linked with morphogenesis and biofilm formation (7, 19, 46). Furthermore, C. albicans induces amino acid biosynthetic genes in response to phagocytosis by human neutrophils (42). Hence, the ability to regulate amino acid metabolism appears to be an important weapon in the armory of this medically important pathogen.
The GCN response (or general amino acid control) is characterized by the induction of genes in most amino acid biosynthetic pathways in response to starvation for even a single amino acid. This response has been shown to exist in C. albicans and to be involved in both morphogenesis and biofilm formation (19, 46, 49). Furthermore, this response depends on the transcription factor Gcn4 (46), which is characteristic of fungal GCN-like responses (25, 27, 48).
GCN signaling has been best characterized in Saccharomyces cerevisiae (reviewed in references 25 and 26). In budding yeast, amino acid starvation leads to the intracellular accumulation of uncharged tRNAs, which interact with the regulatory histidyl-tRNA synthetase-like domain of Gcn2, thereby stimulating its eukaryotic initiation factor 2α (eIF2α) kinase activity. Phosphorylation of eIF2α at serine-51 by Gcn2 inhibits guanine nucleotide exchange on eIF2, thereby decreasing the activity of this essential translation initiation factor. In this way, Gcn2 causes the global rate of mRNA translation to decrease in response to amino acid deprivation. However, in parallel, the decrease in eIF2 activity enhances translation of the GCN4 mRNA. This translational control is mediated by the unusually long 5′-leader sequence on the GCN4 mRNA, which carries four upstream open reading frames (ORFs). Essentially, these upstream ORFs repress translation of the main GCN4 ORF, and this repression is alleviated by Gcn2-mediated phosphorylation of eIF2α during amino acid starvation. As a result, Gcn4 levels increase, thereby leading to the transcriptional activation of amino acid biosynthetic genes via Gcn4 response elements (GCRE) (TGACTC) in their promoters (25, 37). In S. cerevisiae, GCN4 transcription increases about twofold in response to amino acid starvation (2), but Gcn4 synthesis is regulated primarily at the translational level (26). Hence, in S. cerevisiae, the GCN response is blocked by mutations that inactivate Gcn2 or Gcn4.
By analogy with S. cerevisiae, we reasoned that Gcn2 might be important for the GCN response in C. albicans. In this study we show that while Gcn2 does contribute to the GCN response in C. albicans, it plays a nonessential role. Instead, GCN4 appears to be regulated primarily at the transcriptional level in C. albicans.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains (Table 1) were grown in YPD or synthetic complete medium (SC) (45). 3-Aminotriazole (3AT) was used at the concentrations specified.
TABLE 1.
Strains analyzed in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| S. cerevisiae | ||
| BY4743HIS | MATα/MATα his3/his3 leu2/leu2 LYS2/lys2 MET15/met15 ura3/ura3; pRS303 (HIS3) | 49 |
| Y30249HIS | Like BY4743, except gcn4::kanMX4/gcn4::kanMX4; pRS303 (HIS3) | 49 |
| Y33642HIS | Like BY4743, except gcn2::kanMX4/gcn2::kanMX4; pRS303 (HIS3) | This study |
| C. albicans | ||
| SC5314 | Clinical isolate | 22 |
| CAF2-1 | URA3/ura3::λ imm434 | 18 |
| CAI-4 | ura3::λ imm434/ura3::λ imm434 | 18 |
| GTC41 | Like CAI-4, except GCN4/gcn4::hisG-URA3-hisG | 46 |
| GTC42 | Like CAI-4, except GCN4/gcn4::hisG | 46 |
| GTC43 | Like CAI-4, except gcn4::hisG-URA3-hisG/gcn4::hisG | 46 |
| GTC44 | Like CAI-4, except gcn4::hisG/gcn4::hisG | 46 |
| HTC41, HTC45, HTC49a | Like CAI-4, except GCN2/gcn2::hisG-URA3-hisG | This study |
| HTC42, HTC46, HTC50a | Like CAI-4, except GCN2/gcn2::hisG | This study |
| HTC43, HTC47, HTC51a | Like CAI-4, except gcn2::hisG-URA3-hisG/gcn2::hisG | This study |
| HTC44, HTC48, HTC52a | Like CAI-4, except gcn2::hisG/gcn2::hisG | This study |
| CAI-8 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ade2::hisG | 18 |
| GTC82 | Like CAI-8, except GCN4/gcn4::hisG | 46 |
| GTC84 | Like CAI-8, except gcn4::hisG/gcn4::hisG | 46 |
| HTC81, HTC85, HTC89a | Like CAI-8, except GCN2/gcn2::hisG-URA3-hisG | This study |
| HTC82, HTC86, HTC90a | Like CAI-8, except GCN2/gcn2::hisG | This study |
| HTC83, HTC87, HTC91a | Like CAI-8, except gcn2::hisG-URA3-hisG/gcn2::hisG | This study |
| HTC84, HTC88, HTC92a | Like CAI-8, except gcn2::hisG/gcn2::hisG | This study |
| HTC101 | Like CAI-4, except GCN4/GCN4-GFP | This study |
| HTC102 | Like CAI-4, except GCN4/gcn4-GFP | This study |
| HTC103 | Like CAI-4, except gcn4::hisG/GCN4-GFP | This study |
| HTC104 | Like CAI-4, except gcn4::hisG/gcn4-GFP | This study |
Three independent sets of congenic mutants.
C. albicans strain construction.
To disrupt GCN2, the 5′ end (−90 to +500) was PCR amplified with primers 5′-AAAAAGCTTACACGAAACATCAATTCA and 5′-GCCAGATCTTCATCCTTCTTCTGTTTC (HindIII and BglII sites underlined), and the 3′ end (+4767 to +5327) was amplified with primers 5′-GATAGATCTTTTGGTGACTGAATTATT and 5′-AAGCTAGCAAGCATGCTTGTAGGTATG (BglII and NheI sites underlined). These products were cloned into pGEM-T Easy. The hisG-URA3-hisG sequence (18) was inserted at the BglII site between the 5′ and 3′ ends of GCN2 to create the gcn2::hisG-URA3-hisG disruption cassette. This cassette was released from the plasmid backbone with NotI, and the two GCN2 alleles were disrupted sequentially in CAI-4 and CAI-8 (Table 1) in two rounds of Ura blasting (18). Disruptions were confirmed by Southern blotting and PCR diagnosis (not shown).
GCN4-GFP and gcn4-GFP fusions were constructed at the GCN4 locus with the GFP-URA3 cassette as described previously (20). For the GCN4-GFP fusion, the primers used were 5′-GAAAAGCAAGCTTTACAAGATCAAGTTGAA AGATTACAAGAATTGTTAAGAGTTAATGGTATTCAATTTGGTGGT GGTTCTAAAGGTGAAGAATTATT and 5′-TAAAAAAACAATAATAAT TTTCTAAATTTTTCTTTTTTTAAAAAAATAACGAGAGGTATATAT AGTAGTTCTAGAAGGACCACCTTTGATTG (R1-GCN4). To create the gcn4-GFP fusion, which disrupted the GCN4 locus, primer 5′-GATTTATTT GCTTCTCCAGTTAAACAACAACATCAAAAGGTTGATACTGTTGCT ACCAAAAACGAAATTGGTGGTGGTTCTAAAGGTGAAGAATTATT was used with the R1-GCN4 primer.
To create pGCN4GCRE-GFP, the wild type GCN4 5′ region (−996 to +1) was PCR amplified with primers 5′-CTCTAAAGACTCGAGAAATAGCGAAAA TGTAAATATAAATTTATGAGTCATAATTTCTCTCG and 5′-AGTAGC AAGCTTTTTATCTAATAATAATAATGGTAACAAAGCTAATTAATG TAATGTAATTTAATTTAAATAGC (XhoI and HindIII sites underlined; GCRE in italics). To generate pGCN4-GFP, the GCN4 allele (−996 to +1) was amplified from the GCN4 allele lacking the GCRE at −582 (TTATTAA) with the forward primer 5′-CTCTAAAGACTCGAGAAATAGCGAAAATGTAAATATAAATTTATAATGTATAATTTCTCTCG (XhoI site underlined; mutated GCRE at −968 in italics) and the same reverse primer. This product lacked both GCRE at −968 and −582. Both the wild-type and mutant sequences were cloned between the XhoI and HindIII sites in pGFP (3) and resequenced to generate pGCN4GCRE-GFP and pGCN4-GFP, respectively. These plasmids were integrated at the RPS1 locus (34). (The RP10/RPS10 gene has been renamed RPS1.)
Renilla reniformis LUC (RrLUC) promoter fusions were made in a plasmid containing a basal C. albicans ADH1 promoter upstream of the RrLUC reporter (46). The GCRE-RrLUC fusion was made by inserting the sequence 5′-CTG ACTCTGAGGTGACTCGGATCCTGACTCTACTGTGACTCTATAGTG ACTCT (GCRE underlined) between the BstEII and SpeI sites of this basal construct. RrLUC plasmids were linearized with HindIII and transformed into C. albicans. Single-copy integration at the ade2 locus was confirmed by PCR and Southern blotting.
Blotting and RT-PCR.
Southern (35), Northern (9), and Western (12) blotting were performed as described previously. The rabbit anti-phospho-eIF2(pSer51) antibody was from Sigma (St Louis, MO). Reverse transcription-PCR (RT-PCR) was performed by standard methods (44) with the following green fluorescent protein (GFP) primers: 5′-CACTGGTGTTGTCCC and 5′-CCATACCAT GGGTAA (677-bp product). The intron-containing EFB1 product was used to control for loading and genomic DNA contamination (44).
Reporter assays.
GFP fluorescence was visualized by microscopic analysis as described previously (3). Renilla luciferase activities (105 relative light units/20 μg protein) were measured in quadruplicate (35) in C. albicans transformants after 3 h of growth at 30°C in SC lacking histidine and containing 40 mM 3AT. Similar data were obtained in three experiments done with independent transformants.
Transcript profiling.
Transcript profiling was performed on the congenic C. albicans strains CAF2-1, GTC43, HTC43, HTC47, and HTC51 growing in SC lacking histidine after 3 h of exposure to 0 or 40 mM 3AT. Transcript profiling was performed as described previously (16, 36). Briefly, total RNA was isolated (24), Cy3- and Cy5-labeled cDNAs were prepared, and these probes were hybridized with C. albicans microarrays (Eurogentec, Seraing, Belgium). Slides were scanned at 10-μm resolution with a ScanArray Lite scanner (Perkin-Elmer Life Sciences, Beaconsfield, United Kingdom) and quantified with QuantArray software (version 2.0). GeneSpring software (Silicon Genetics, Redwood City, CA) was used for data normalization and analysis, and statistical analysis was performed with SAM (significance analysis of microarrays) (47). Data from at least three independent biological replicates were used for each analysis, and the SAM false-discovery rate was set at 10%. Expression ratios for each gene that displayed a reproducible and statistically significant change in expression are available at the Galar Fungail websites (http://www.pasteur.fr/recherche/unites /Galar_Fungail/ and http://www.galarfungail.org/data.htm). C. albicans gene annotations were obtained from CandidaDB (http://genolist.pasteur.fr/CandidaDB) (14). Functional categories for C. albicans genes were assigned mainly on the basis of the MIPS functional assignments for S. cerevisiae homologues (http://mips.gsf.de/proj/yeast/CYGD/db/index.html) (49).
RESULTS
C. albicans GCN2 encodes an eIF2α kinase.
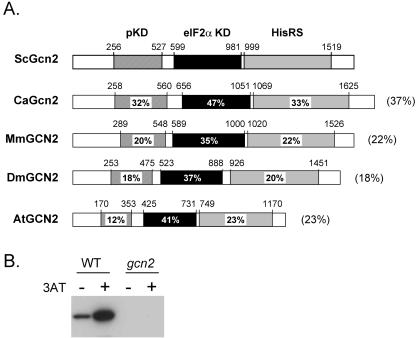
Our initial working hypothesis was that regulation of the GCN response in C. albicans would closely mirror the S. cerevisiae GCN control mechanisms that have been defined in some detail by numerous groups over many years (reviewed in references 25 and 26). As described above, Gcn2 plays a critical role in control of the GCN response in S. cerevisiae. Therefore, we screened the C. albicans genome sequence for GCN2-like loci. Only one C. albicans ORF (GCN2; orf19.6913) encodes a protein with significant sequence similarity to S. cerevisiae Gcn2 (37% identity). The C. albicans Gcn2 protein sequence contains partial kinase-, eIF2α kinase-, and histidyl-tRNA synthetase-like domains. It also displays significant sequence similarity to GCN2 eIF2α kinases from mammals, flies, and plants (Fig. 1A).
FIG. 1.
C. albicans GCN2 encodes an eIF2α kinase. (A) C. albicans orf19.6913 encodes a protein (CaGcn2) with sequence similarity to Gcn2-like proteins from other eukaryotes. Mus musculus (MmGCN2; TrEMBL accession no. Q9QZ05), Drosophila melanogaster (DmGCN2; TrEMBL Q9V9X8), and Arabidopsis thaliana (AtGCN2; TrEMBL Q8H2D3) proteins are compared with S. cerevisiae Gcn2 (TrEMBL P15442). The coordinates of the partial kinase (pKD), eIF2α kinase (eIF2α KD), and histidyl-tRNA synthetase-like (HisRS) domains are shown above each protein. The numbers within each domain represent the percentage of amino acid sequence identity with the corresponding domain in ScGcn2. The overall percentage of amino acid sequence identity with ScGcn2 is given in parentheses on the right. (B) C. albicans eIF2α kinase activity is dependent on GCN2. Equal amounts of protein extracts from wild-type (WT; CAI-4) or Δgcn2 (HTC44) C. albicans cells (see Table 1) were either left untreated (−) or treated with 40 mM 3AT (+) for 3 h and then subjected to Western blotting with an antibody that specifically recognizes the serine-51 phosphorylated form of eIF2α. Similar data were obtained from two independent experiments.
To test whether eIF2α kinase activity in C. albicans is dependent on Gcn2, we generated homozygous gcn2/gcn2 null mutants. Essentially, the two GCN2 alleles in this diploid fungus were disrupted sequentially, removing codons 167 to 1589 of the 1,764-codon ORF to create three independent homozygous mutants in two C. albicans strains (CAI-4 and CAI-8), thereby generating a total of six gcn2/gcn2 null mutants (Table 1). The eIF2α kinase activities in mutant and wild-type cells were compared by Western blotting with an antibody specific for the phosphorylated form of eIF2α (Fig. 1B). No detectable eIF2α kinase activity remained in the mutant cells, indicating that GCN2 encodes the major eIF2α kinase activity in this fungus.
The eIF2α kinase activity increased in wild-type cells following exposure to 3AT (Fig. 1B), suggesting that Gcn2 is activated in response to amino acid starvation. This is consistent with the idea that Gcn2 plays a role in the C. albicans GCN response.
GCN2 inactivation reduces the tolerance of C. albicans to 3AT.
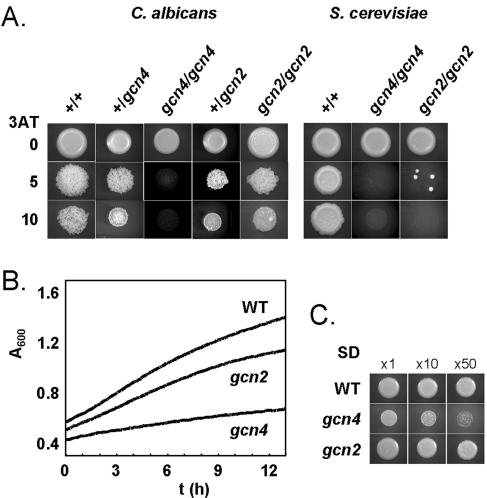
We tested whether the ability of C. albicans to respond to amino acid starvation depends on GCN2. Resistance to the histidine analogue 3AT is a classic indicator of the GCN response in S. cerevisiae and C. albicans (25, 46). Therefore, we compared the 3AT sensitivity of gcn2 cells with that of wild-type and gcn4 controls (Fig. 2A). Inactivation of GCN2 only partially attenuated the resistance of C. albicans to 3AT: gcn2 cells grew slowly on solid medium containing 10 mM 3AT. In contrast, S. cerevisiae gcn2 cells were sensitive to 5 mM 3AT. The same phenotype was observed for all six independent gcn2-null mutants (HTC44, HTC48, HTC52, HTC84, HTC88, and HTC92 [Table 1]). Therefore, C. albicans is less dependent on Gcn2 for growth in the presence of this toxic histidine analogue than S. cerevisiae. However, both yeasts require Gcn4 for 3AT resistance (Fig. 2).
FIG. 2.
Growth during amino acid starvation is only partially attenuated in C. albicans gcn2 cells. (A) Growth of C. albicans and S. cerevisiae strains was compared after 2 days on SC containing different concentrations of 3AT (mM). C. albicans strains: +/+ (GCN4/GCN4), CAF2-1; +/gcn4 (GCN4/gcn4), GTC41; gcn4/gcn4, GTC43; +/gcn2 (GCN2/gcn2), HTC41; gcn2/gcn2, HTC43. S. cerevisiae strains: +/+ (wild type), BY4743HIS; gcn4/gcn4, Y30249HIS; gcn2/gcn2, Y33642HIS. (See Table 1.) (B) Effect of 1 mM 3AT on the growth of the same strains in SC lacking histidine. Similar data were obtained for three independent gcn2 mutants. WT, wild type. (C) Growth of WT (CAF2-1), gcn2 (HTC43), and gcn4 (GTC43) C. albicans cells on SD for 2 days. Cells were diluted 1-, 10-, or 50-fold.
We compared the growth of C. albicans gcn2, gcn4, and wild-type cells in the absence of amino acids (Fig. 2B and C). As expected, gcn4 cells grew relatively slowly, confirming that Gcn4 is required for normal responses to amino acid starvation in C. albicans. However, gcn2 cells grew relatively normally under these conditions. Therefore, in contrast to the situation in S. cerevisiae, the C. albicans GCN response is not dependent on Gcn2.
Amino acid starvation stimulates morphogenesis in C. albicans but not in S. cerevisiae (46). Hence, C. albicans forms wrinkly colonies in the presence of 3AT. Interestingly, gcn2 cells also formed wrinkly colonies (Fig. 2A), indicating that Gcn2 is not required for the morphogenetic response of C. albicans to amino acid starvation.
Transcriptional activation via the GCRE is not dependent on Gcn2.
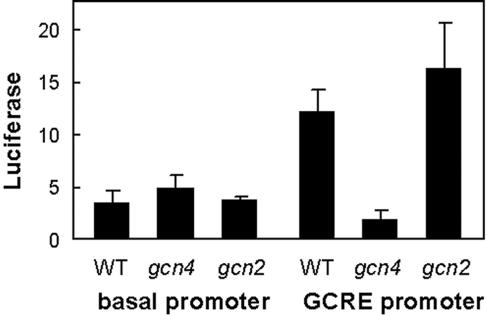
The GCN response in C. albicans is mediated by Gcn4, which activates the transcription of genes via the GCRE (46). Hence we reasoned that if Gcn2 plays a relatively minor role in this GCN response, the expression of a GCRE reporter gene in C. albicans should not depend on GCN2. To test this, the activities of basal and GCRE-containing luciferase reporters were measured in wild-type, gcn4, and gcn2 cells following exposure to 3AT. The GCRE reporter contained five copies of the GCRE upstream of a control basal reporter with the TATA box and RNA start site from the C. albicans ADH1 promoter. The expression of the control basal reporter was not affected by the disruption of Gcn2 or Gcn4. Also, the activation of the GCRE reporter was inhibited in gcn4 cells (Fig. 3), as expected (46). As predicted, inactivation of Gcn2 had no significant effect on this GCRE reporter. These data confirm that the GCRE mediates the transcriptional activation of C. albicans genes by Gcn4, and they suggest that Gcn2 does not contribute significantly to this activation.
FIG. 3.
Inactivation of GCN2 does not inhibit Gcn4/GCRE-mediated transcriptional activation. Relative luciferase activities were measured in C. albicans strains transformed with basal or GCRE-RrLUC reporters after 3 h of growth in SC with 40 mM 3AT. Wild type (WT), CAI-8; gcn4, GTC84; gcn2, HTC84. (See Table 1.)
GCN4 is transcriptionally activated.
If the activation of GCN4 is not dependent on Gcn2, how is the GCN response activated in C. albicans? Our data indicated that GCN4 mRNA levels increase in response to 3AT (46). Therefore, we tested this further by examining the responses of two types of GCN4-GFP fusion to amino acid starvation.
The first GCN4-GFP fusion contained the wild-type 5′ region (from −996 to +1) cloned upstream of the synthetic, codon-optimized yEGFP gene (12) in pGCN4GCRE-GFP. We noticed that the region upstream of the main GCN4 ORF contained two GCRE at −968 and −582 with respect to the start codon. Therefore, we made a second GCN4-GFP fusion, which lacked these putative GCREs, to test whether they are required for the transcriptional activation of GCN4. This second fusion was called pGCN4-GFP. The two fusions were integrated at the RPS1 locus, and the responses of the two fusions to 3AT were examined by Northern blotting and fluorescence microscopy (Fig. 4).
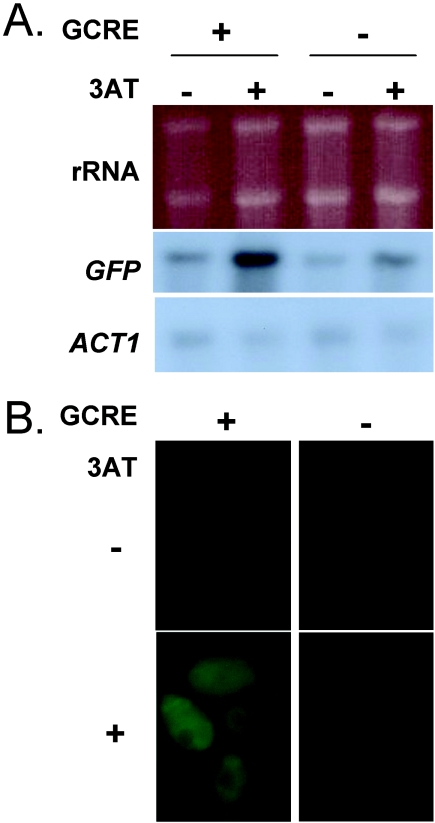
FIG. 4.
Activation of GCN4 depends on GCREs in its promoter. (A) Northern blot analysis of GFP mRNAs from cells grown in SC without (−) or with (+) 40 mM 3AT for 3 h. Wild-type (CAI-4) and gcn2 (HTC44) cells expressed GFP fusions with wild-type (GCRE +; pGCN4GCRE-GFP) or mutated (GCRE −; pGCN4-GFP) GCN4 promoters. rRNAs and ACT1 mRNAs acted as internal loading controls, and all images are from the same blot. (B) Fluorescence microscopy of GFP levels in cells from the same cultures.
In cells containing pGCN4GCRE-GFP, the GFP mRNA levels were induced in response to 3AT. Interestingly, GFP mRNA levels were not induced to the same extent in cells containing the GCN4-GFP fusion. These Northern blot data were confirmed by RT-PCR (not shown). The levels of GFP fluorescence generated by these constructs were relatively low, but this was expected, because they included the GCN4 5′ untranslated leader region. GFP fluorescence was always at background levels in cells not exposed to 3AT and in cells containing the GCN4-GFP fusion. In contrast, GFP fluorescence levels were consistently higher in cells containing the GCN4GCRE-GFP fusion. Therefore, GFP fluorescence levels correlated reproducibly with GFP mRNA levels. Hence, the data were entirely consistent with the idea that 3AT induces GCN4 transcription. Furthermore, the data indicated that the GCRE contribute significantly to this transcriptional induction.
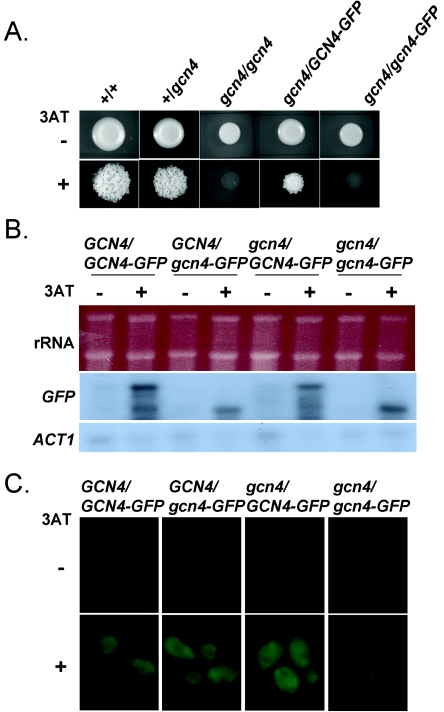
Transcriptional activation via the GCRE in C. albicans is known to be mediated by Gcn4 (46). Therefore, we tested whether Gcn4 is required for transcriptional induction of GCN4. To achieve this, we integrated GFP at the chromosomal GCN4 locus in wild-type CAI-4 cells and a heterozygous GCN4/gcn4 mutant, GTC42 (Table 1). To generate a functional fusion, the GFP ORF was fused in frame after the last codon of the GCN4 allele (strains HTC101 and HTC103). To generate a nonfunctional fusion, GFP was inserted in frame after codon 40 of GCN4 (strains HTC102 and HTC104). Strains HTC101 and HTC102 retained one wild-type GCN4 allele, whereas strains HTC103 and HTC104 carried an inactive gcn4 allele in addition to their GFP fusion (Table 1). To test the Gcn4 functionality of the GCN4-GFP and gcn4-GFP fusions, we examined the 3AT resistance of strains HTC103 and HTC104 (Fig. 5A). HTC103 was 3AT resistant, indicating that the GCN4-GFP fusion retained Gcn4 functionality. In contrast, HTC104 was 3AT sensitive, indicating that the gcn4-GFP fusion did not retain Gcn4 functionality, as predicted.
FIG. 5.
Gcn4 is not essential for the transcriptional activation of GCN4. (A) Growth of C. albicans mutants on SC-HIS containing no 3AT (−) or 5 mM 3AT (+) for 4 days at 30°C. Strains: +/+ (GCN4/GCN4), CAF2-1; +/gcn4 (GCN4/gcn4), GTC41; gcn4/gcn4, GTC43; gcn4/GCN4-GFP, HTC103; gcn4/gcn4-GFP, HTC104 (see Table 1). (B) Northern blot analysis of GFP mRNAs from cells grown in SC without (−) or with (+) 40 mM 3AT for 3 h. Strains: GCN4/GCN4-GFP, HTC101; GCN4/gcn4-GFP, HTC102; gcn4/GCN4-GFP, HTC103; gcn4/gcn4-GFP, HTC104 (see Table 1). rRNAs and ACT1 mRNAs acted as internal loading controls. (C) Fluorescence microscopy of GFP levels in cells from the same cultures.
GFP transcript levels were then examined in strains HTC101 to -104 by Northern blot analysis following exposure to 0 or 40 mM 3AT (Fig. 5B). As expected, the gcn4-GFP transcript was about 0.7 kb shorter than the GCN4-GFP transcript. GCN4-GFP and gcn4-GFP mRNA levels increased significantly in response to 3AT in HTC101 and HTC102 cells, which contained a functional GCN4 allele. Furthermore, GFP fluorescence levels were reproducibly elevated in these cells (Fig. 5C). Similar observations were made for HTC103, which contains the functional GCN4-GFP allele. However, GFP fluorescence remained at low levels in HTC104. This was consistent with the idea that Gcn4 is required for the activation of GCN4. However, gcn4-GFP mRNA levels did respond to 3AT, suggesting that Gcn4 is required at a posttranscriptional rather than a transcriptional level.
In light of this result, we examined the behavior of pGCN4GCRE-GFP in gcn4 cells. Northern blot analysis revealed that the GFP mRNA levels from this construct increased in gcn4 cells in response to 3AT, even though these cells did not express a functional Gcn4 (not shown). This was consistent with the above findings.
Taken together, our data suggest that C. albicans GCN4 is transcriptionally activated in response to 3AT and that GCRE contribute to this transcriptional activation, but that Gcn4 is not required for this transcriptional activation.
Definition of Gcn4 and Gcn2 regulons in C. albicans.
Our data suggested that Gcn2 plays a relatively minor role in the GCN response in C. albicans, whereas Gcn4 plays a major role in this response. To test this further, we examined the Gcn2 and Gcn4 regulons in C. albicans by transcript profiling. To our knowledge, this is the first time that the Gcn2 regulon has been studied in any fungus. The transcript profiles of wild-type, gcn2, and gcn4 cells were compared following exposure to 0 or 40 mM 3AT. Only reproducible and statistically significant changes observed in three independent experiments are discussed.
The transcript profiling data were consistent with our analysis of the Gcn4 proteome in C. albicans (49) and with previous Northern blot analyses of GCN-responsive mRNAs (ARO4, HIS4, HIS7, LYS2, and GCN4 [46]). Furthermore, the data revealed strong similarities between the Gcn4 transcriptomes of C. albicans and S. cerevisiae (37). For example, exposure to 3AT affected the expression of a relatively large proportion of C. albicans genes (about 13% of the ca. 6,200 ORFs). About half of these genes were induced, and half were repressed (Fig. 6A). For most genes (91%), this regulation was lost in gcn4 cells, indicating that Gcn4 is required for most transcriptional responses to 3AT. Furthermore, amino acid biosynthetic functions were significantly enriched among those genes that were induced by 3AT in a Gcn4-dependent fashion (Table 2). Genes in all amino acid biosynthetic pathways were induced by 3AT in a Gcn4-dependent fashion, and HIS1, HIS3, HIS4, HIS5, and HIS7 mRNAs were all induced more than threefold. These findings were entirely consistent with the observation that Gcn4 is essential for responses to amino acid starvation (Fig. 2) (46).
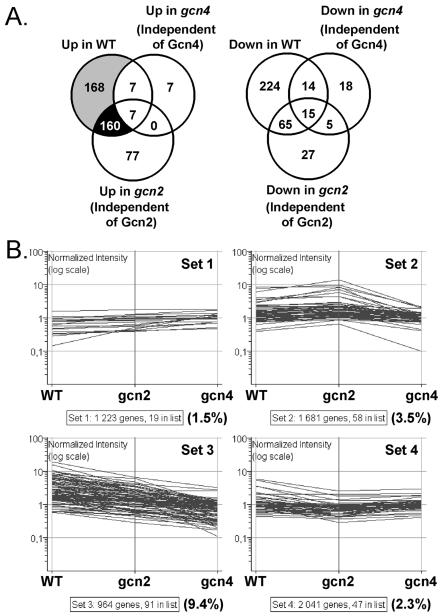
FIG. 6.
Transcript profiling of the responses of wild-type (WT), gcn4, and gcn2 C. albicans cells to 3AT. (A) Venn diagram showing the number of transcripts in each cell type that displayed reproducible and statistically significant responses to 3AT in three independent experiments. Strains: WT, CAF2-1; gcn4, GTC43; gcn2, HTC47. Gray sector, transcripts induced by 3AT in a Gcn4- and Gcn2-dependent fashion; black sector, transcripts induced by 3AT in a Gcn4-dependent but Gcn2-independent fashion. (B) Clustering of amino acid biosynthetic genes based on their responses to 3AT in WT, gcn2, and gcn4 cells. All C. albicans genes for which there were statistically significant data were clustered using K-means clustering in GeneSpring. The behavior of amino acid biosynthetic genes is shown. The total number of genes, the number of amino acid biosynthetic genes (genes in list), and the proportion of amino acid biosynthetic genes (in parentheses) in each K-means Gene Set are given below the graphs.
TABLE 2.
Functional categories that are significantly influenced by inactivation of C. albicans GCN4 or GCN2
| Functional categorya | Gcn4- and Gcn2-dependent genesb (n = 168)
|
Genes dependent on Gcn4 onlyb (n = 160)
|
||
|---|---|---|---|---|
| Fold enhancementc | Genes | Fold enhancementc | Genes | |
| Amino acid metabolism | 5.0 | AGP1, ARG8, ARO1, ARO4, BAT21, ECM42, GAP5, HIP1, HIS1, HIS3, HIS7, HOM3, HYU1, ILV1, ILV6, LEU42, LYS2, LYS4, LYS22, MET8, MXR1, PRO1, PRO2, THR1, TRP2, TRP3, TRP4, IPF8591 | 5.9 | AAT21, ARO3, ARO8, BAT22, CIS2, DIP51, ECM17, GAP6, GAP7, GCH2, GCN4, HIS4, HIS5, HOM6, ILV2, LEU41, LYS9, LYS21, MET2, MET15, SER33, THR4, THR5, TRP5, IPF1162, IPF2837, IPF8048, IPF13176, IPF14203 |
| Nitrogen and sulfur metabolism | 2.7 | MET8, NIT3, IPF8591, IPF10021, IPF11716 | 4.0 | AAT21, ECM12, GDH2, IPF3549, IPF14203, IPF20164 |
| Protein fate | 2.0 | APE3, ARP2, CDC37, CDC48, CPY1, CTM1, ELC1, HSP78, NPL4, PRC1, PRC3, PRE3, PRE6, PRE8, PUP2, RPN1, RPN2, RPN6, RPN7, RPN8, RPN10, RPT1, RPT4, RPT5, RPT6, SHR3, SRA1, UBC3, VPS1, VPS4, IPF56, IPF14031, IPF4866, IPF7556, IPF11713 | NEd | |
| Cell rescue, defense, and virulence | 2.1 | DDR48, GTT1, HAM1, HK1, HSP12, HSP78, MXR1, OSM2, PRE3, PUP2, SRA1, SSU1, TTR1, ZWF1, IPF4055, IPF9188 | NE | |
| Transport facilitation | NE | 2.3 | ARR3, CDR1, CDR11, DIP51, ENA21, FCY23, GAP6, GAP7, PXA2, SEO1, SMF12, IPF4580, IPF9670, IPF11767, IPF12884, IPF13941 | |
Functional categories are based on those of S. cerevisiae homologues as defined by MIPS (http://mips.gsf.de/genre/proj/yeast/searchCatalogFirstAction.do?db=CYGD), and C. albicans functional assignments are from CandidaDB (http://genolist.pasteur.fr/CandidaDB/) (14). Only those functional categories that show more than two fold enhancement and that also contain more than five genes are shown. Note that a gene can belong to more than one functional category.
The 168 genes that displayed significant increases in expression in response to 3AT in wild-type cells only were defined as Gcn4 and Gcn2 dependent (Fig. 6A, gray sector). The 160 genes that displayed increased expression in response to 3AT in wild-type and gcn2 cells were defined as Gcn4 dependent only (Fig. 6A, black sector).
Fold enhancement, which represents the extent to which genes in the functional category of interest were enriched, was calculated by dividing the proportion of genes within the regulatory subset that belong to that functional category by the proportion of genes in the whole genome that belong to that functional category.
NE, no significant enhancement for this functional category in this regulatory subset, whereas the other regulatory subset does show significant enhancement for this functional category.
Many putative peptidase genes were induced by 3AT (APE3, PRC1, PRC3, PRE3, PRE8, PUP2, RPN1, RPN2, RPN6, RPN7, RPN8, RPN10, RPT1, RPT4, RPT5, RPT6, and IPF4866). This suggested that protein turnover rates increase in C. albicans following amino acid starvation. Also, 43% of the 224 genes that were repressed by 3AT in wild-type cells encode protein synthesis functions. This represents a 7.5-fold enrichment compared with the genome as a whole, which is consistent with the idea that protein synthesis decreases following 3AT treatment. Hence, the transcript profiling data confirm that C. albicans activates a bona fide GCN response following exposure to 3AT. This had been suggested by previous Northern blot and proteomic analyses (46, 49) but had not been analyzed before on such a global scale.
There was significant overlap between the Gcn2 and Gcn4 regulons in C. albicans. About half of the 328 genes that were induced by 3AT in a Gcn4-dependent fashion were not induced in gcn2 cells (Fig. 6A, gray sector of Venn diagram). This indicates that the induction of these 168 genes was also Gcn2 dependent. The extent of the overlap between the regulons was even greater for 3AT-repressed genes. Genes involved in protein fate, cell rescue, defense, and virulence were significantly enriched in the subset of genes that were induced in a Gcn4- and Gcn2-dependent fashion.
Only seven mRNAs were induced by 3AT in wild-type and gcn4 cells but not in gcn2 cells (Fig. 6A). This indicated that almost all genes whose expression was induced by 3AT were dependent on Gcn4 for this induction. In contrast, 160 mRNAs were induced by 3AT in wild-type and gcn2 cells but not in gcn4 cells (Fig. 6A, black sector of the Venn diagram). The activation of these mRNAs appears to be dependent on Gcn4 but independent of Gcn2. Genes involved in transport facilitation were significantly enriched in this subset. However, it was possible that their activation was partially dependent on Gcn2. According to this scenario, the inactivation of Gcn2 would reduce, but not block, the induction of these mRNAs.
To test this, we analyzed the responses of C. albicans genes to 3AT in more depth. We performed K-means clustering to separate genes on the basis of their expression patterns in wild-type, gcn2, and gcn4 cells. C. albicans genes fell into four main K-means subsets. We then examined the behavior of the amino acid biosynthetic genes in these subsets (Fig. 6B). A small proportion of amino acid biosynthetic genes did not respond to 3AT (set 1). However, most amino acid biosynthetic genes did respond to 3AT. Some of these remained relatively unaffected in gcn2 cells (set 2), whereas the 3AT response of others was blocked by the inactivation of Gcn2 (set 4). Most amino acid biosynthetic genes fell into set 3, although this set contained the smallest number of C. albicans genes. Genes in set 3 responded to 3AT in wild-type cells; this response was all but lost in gcn4 cells, and an intermediate response was observed in gcn2 cells. This indicates that Gcn2 contributes to, but is not essential for, the activation of these genes by 3AT. These data are entirely consistent with our observation that Gcn2 is not essential for the GCN response in C. albicans.
DISCUSSION
In this study, we have shown that C. albicans has a single eIF2α kinase locus. GCN2 is the only C. albicans locus that encodes a protein with significant sequence similarity to GCN2 proteins from other organisms (Fig. 1A). Furthermore, no detectable eIF2α kinase activity is present in C. albicans cells lacking Gcn2 (Fig. 1B).
By analogy with S. cerevisiae (26), we initially predicted that Gcn2 might play a role in the C. albicans GCN response. Several of our observations suggested that this is the case. First, GCN2 inactivation attenuates the 3AT resistance of C. albicans (Fig. 2). Second, exposure to 3AT increases eIF2α kinase activity in C. albicans, and GCN2 encodes this activity (Fig. 1B). Third, transcript profiling revealed that 3AT induces the expression of some amino acid biosynthetic genes in C. albicans in a Gcn2-dependent fashion (Fig. 6; Table 2).
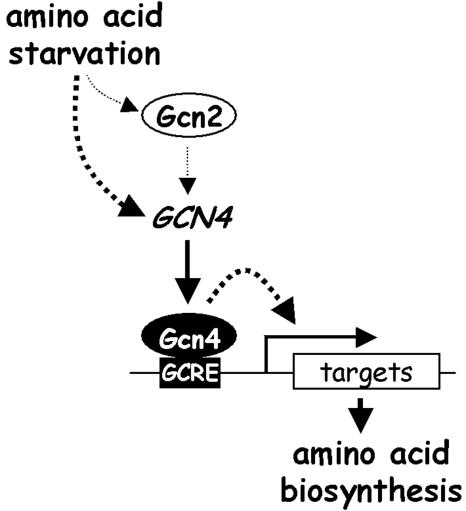
However, numerous observations suggest that the role of Gcn2 in the C. albicans GCN response is relatively minor (Fig. 7). First, Gcn2 inactivation affects C. albicans to a much lesser extent than S. cerevisiae with respect to their 3AT resistance and their growth in the absence of amino acids (Fig. 2). Second, the transcriptional activation of a GCRE reporter by Gcn4 is not dependent on Gcn2 (Fig. 3). One would have expected this GCRE reporter to be Gcn2 dependent if Gcn2 was essential for the increase in Gcn4 expression levels following 3AT exposure. Third, transcript profiling revealed that Gcn2 contributes to, but is not required for, the activation of many amino acid biosynthetic genes in response to 3AT (Fig. 6; Table 2). Consistent with this, we have observed that the GCN4 5′-leader region represses the expression of an ACT1-GFP reporter in C. albicans and that this repression is not dramatically released following exposure to 3AT (not shown).
FIG. 7.
Model illustrating the partial dependence of the C. albicans GCN response on Gcn2 (see the text).
It is conceivable that the main role of Gcn2 in C. albicans is to enhance the rate at which the GCN response is activated (for example, by accelerating the accumulation of Gcn4) rather than to significantly increase the amplitude of the response. Alternatively, the main role of Gcn2 might be control of global translation rates during amino acid starvation in C. albicans. Either of these explanations could account for our observations.
If GCN4 expression is not regulated primarily by Gcn2, how is GCN4 activated in C. albicans? Our data indicate that GCN4 is regulated mainly at the transcriptional rather than the translational level. Transcript profiling indicated that 3AT stimulates GCN4 mRNA levels in wild-type C. albicans cells (Table 2). This was confirmed by Northern blot analyses of the GCN4 mRNA and of various types of GCN4-GFP fusion (Fig. 5 and 6) (46). Furthermore, these observations have been confirmed by RT-PCR (not shown).
All of the fungal GCN4 homologues studied to date are transcriptionally regulated, at least to some extent. In S. cerevisiae, GCN4 transcription increases about twofold in response to 3AT (2), although in this yeast GCN4 is regulated primarily at the translational level (26). In contrast, C. albicans GCN4 appears to be regulated mainly at the transcriptional level. This is similar to the situation in Neurospora crassa and Aspergillus niger, where the levels of the cpc-1 and cpcA mRNAs, respectively, increase significantly in response to 3AT (40, 48). Furthermore in Aspergillus nidulans, cpcA mRNA levels are autoactivated by CPCA via CPCA response elements (CPRE) in the cpcA promoter (27). Therefore, although C. albicans is often viewed as being more closely related to budding yeast, with respect to GCN regulation this dimorphic pathogen appears to be more closely related to filamentous fungi.
Gcn4 activates C. albicans gene transcription via the GCRE (Fig. 3) (46). This GCRE is present in the upstream regions of most of the C. albicans genes that were revealed by transcript profiling to be induced by 3AT in a Gcn4-dependent fashion. We also found that the GCRE is required for transcriptional activation of GCN4 in response to 3AT (Fig. 3). Taken together, these data suggest that Gcn4 might autoactivate the GCN4 gene during the GCN response. However, transcriptional induction of GCN4 was not blocked in gcn4 cells (Fig. 5). This indicates that while GCN4 might contribute to its own activation, it is not essential for this activation (Fig. 7), thereby implicating an additional factor.
It is possible that, under normal circumstances, this additional factor contributes to the early stages of the GCN response when Gcn4 levels are low. Then, once Gcn4 levels have started to increase, Gcn4 might autoactivate GCN4 transcription via the GCRE (Fig. 4), thereby accelerating the full induction of the GCN response. Under abnormal circumstances, when no functional Gcn4 is available (Fig. 5), the additional factor might be sufficient to induce gcn4-GFP expression, albeit with relatively slow kinetics.
Several interesting questions are raised by the possible existence of a positive autoregulatory feedback loop involving Gcn4. For example, how is GCN4 activation prevented in the absence of a signal? Also, how does GCN4 expression return to normal once the amino acid starvation signal is removed? Several factors might combine to prevent inappropriate activation and timely inactivation of the GCN response. For example, the repressive effect of the 5′ leader might help to limit GCN4 expression levels in the absence of a signal. Also, the activity of the Gcn4 protein might be posttranslationally regulated. C. albicans Gcn4 contains PEST-like sequences, which by analogy with S. cerevisiae (29) appear to accelerate the degradation of the Gcn4 protein in the absence of a signal (21). Furthermore, the activation domain of S. cerevisiae Gcn4 is phosphorylated by cyclin-dependent protein kinases (11, 33). Hence, several possible posttranslational events might combine to down-regulate Gcn4 activity once C. albicans cells are released from amino acid starvation.
The fact that C. albicans and S. cerevisiae have diverged with respect to GCN regulation is not unexpected, for two main reasons. First, there are many precedents for the divergence of signaling modules in these fungi. For example, the Ras-cAMP, mitogen-activated protein kinase, and Tup1-Nrg1 modules, involved in morphogenetic signaling (5, 6, 8, 17, 32, 35, 41), the Rim101/Prr1 module, which mediates pH signaling (13, 15), and the Hog1 and Yap1/Cap1 modules, involved in stress signaling (1, 43), are conserved in C. albicans and S. cerevisiae. However, the cellular roles of some signaling modules have diverged. For example, Tup1 represses filamentous growth in C. albicans but stimulates pseudohyphal growth in S. cerevisiae (5). Rfg1 is a morphogenetic repressor in C. albicans, whereas its homologue, Rox1, is a hypoxic regulator in S. cerevisiae (28). Also, Msn2 and Msn4 mediate general stress responses in S. cerevisiae but not in C. albicans (38).
Second, S. cerevisiae and C. albicans have evolved in contrasting niches. Although Gcn4 is not required for the virulence of C. albicans during systemic infections (4), where amino acids are presumably plentiful, Gcn4 is required for biofilm formation (19), and it might be required at other infection sites. For example, the Gcn4 homologue CpcA is required for the virulence of Aspergillus fumigatus in pulmonary infections (30). Also, amino acid biosynthetic genes are up-regulated in C. albicans during phagocytosis (42). Presumably, the regulation of the C. albicans GCN response has evolved to optimize the metabolic fitness of this pathogen in the various niches it occupies within its mammalian host.
Acknowledgments
This work was supported by grants from the U.K. Biotechnology and Biological Sciences Research Council (P10256, P17124, G18883), the Wellcome Trust (063204), and the European Commission (QLK2CT-2000-00795; MRTN-CT-2003-504148).
REFERENCES
- 1.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, G., H.-U. Mosch, B. Hoffman, U. Reusser, and G. H. Braus. 1998. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:12696-12702. [DOI] [PubMed] [Google Scholar]
- 3.Barelle, C. J., C. Manson, D. MacCallum, F. C. Odds, N. A. R. Gow, and A. J. P. Brown. 2004. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 21:333-340. [DOI] [PubMed] [Google Scholar]
- 4.Brand, A., D. M. MacCallum, A. J. P. Brown, N. A. R. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277: 105-109. [DOI] [PubMed] [Google Scholar]
- 6.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in Candida albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brega, E., R. Zufferey, and C. B. Mamoun. 2004. Candida albicans Csy1 is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, A. J. P. 2001. Morphogenetic signalling pathways in Candida albicans, p. 95-106. In R. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 9.Brown, A. J. P., R. J. Planta, F. Restuhadi, D. A. Bailey, P. R. Butler, J. L. Cadahia, M. E. Cerdan, M. De Jonge, D. C. J. Gardner, M. E. Gent, A. Hayes, C. P. A. M. Kolen, L. J. Lombardia, A. M. A. Murad, R. A. Oliver, M. Sefton, J. Thevelein, H. Tournu, Y. J. van Delft, D. J. Verbart, J. Winderick, and S. G. Oliver. 2001. Transcript analysis of 1003 novel yeast genes using high-throughput Northern hybridisations. EMBO J. 20:3177-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderone, R. A. 2002. Candida and candidiasis. ASM Press, Washington, D.C.
- 11.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cormack, B., G. Bertram, M. Egerton, N. A. R. Gow, S. Falkow, and A. J. P. Brown. 1997. Yeast enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 13.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d'Enfert, C., S. Goyard, S. Rodriguez-Arnaveilhe, L. Frangeul, L. Jones, F. Tekaia, O. Bader, L. Castillo, A. Dominguez, J. Ernst, C. Fradin, C. Gaillardin, S. Garcia-Sanchez, P. de Groot, B. Hube, F. Klis, S. Krishnamurthy, D. Kunze, M.-C. Lopez, A. Mavor, N. Martin, I. Moszer, D. Onésime, J. Perez Martin, R. Sentandreu, and A. J. P. Brown. 2005. CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res. 33:D353-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Barkani, A., O. Kurzai, W. A. Fonzi, A. Ramon, A. Porta, M. Frosch, and F. A. Muhlschlegel. 2000. Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol. Cell. Biol. 20:4635-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, Q., E. Summers, B. Guo, and G. R. Fink. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Sanchez, S., S. Aubert, I. Iraqui, G. Janbon, J.-M. Ghigo, and C. d'Enfert. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 21.Gildor, T., R. Shemer, A. Atir-Lande, and D. Kornitzer. 2005. Co-evolution of the cyclin Pcl5 and its substrate Gcn4. Eukaryot. Cell 4:310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198: 179-182. [DOI] [PubMed] [Google Scholar]
- 23.Gow, N. A. R., A. J. P. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 24.Hauser, N. C., M. Vingron, M. Scheideler, B. Krems, K. Hellmuth, K.-D. Entian, and J. D. Hoheisel. 1998. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14:1209-1221. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch, A. G. 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52:248-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann, B., O. Valerius, M. Andermann, and G. H. Braus. 2001. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 12:2846-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the S. cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in C. albicans. Mol. Cell. Biol. 21:2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13:6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krappmann, S., E. M. Bignell, U. Reichard, T. Rogers, K. Haynes, and G. H. Braus. 2004. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 52:785-799. [DOI] [PubMed] [Google Scholar]
- 31.Kumamoto, C. A. 2002. Candida biofilms. Curr. Opin. Microbiol. 5:608-611. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H., J. R. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266: 1723-1726. [DOI] [PubMed] [Google Scholar]
- 33.Meimoun, A., T. Holtzman, Z. Weissman, H. J. McBride, D. J. Stillman, G. R. Fink, and D. Kornitzer. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin ligase complex. Mol. Biol. Cell 11:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murad, A. M. A., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. P. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 35.Murad, A. M. A., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. P. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A.-P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcript profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholls, S., M. Straffon, B. Enjalbert, A. Nantel, S. Macaskill, M. Whiteway, and A. J. P. Brown. 2004. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot. Cell 3:1111-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Bailliere Tindall, London, United Kingdom.
- 40.Paluh, J. L., M. J. Orbach, T. L. Legerton, and C. Yanofsky. 1988. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc. Natl. Acad. Sci. USA 85:3728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocha, C. R. C., K. Schroppel, D. Harcus, A. Marcil, D. Dignard, B. N. Taylor, D. Y. Thomas, M. Whiteway, and E. Leberer. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin-Bejerano, I., I. Fraser, P. Grisafil, and G. R. Fink. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 100:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.San Jose, C., R. A. Monge, R. Perez-Diaz, J. Pla, and C. Nombela. 1996. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 178:5850-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaller, M., W. Schafer, H. C. Korting, and B. Hube. 1998. Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol. Microbiol. 29: 605-615. [DOI] [PubMed] [Google Scholar]
- 45.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 46.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. P. Brown. 2002. CaGcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wanke, C., S. Eckert, G. Albrecht, W. van Hartingsveldt, P. J. Punt, C. A. van den Hondel, and G. H. Braus. 1997. The Aspergillus niger GCN4 homologue, cpcA, is transcriptionally regulated and encodes an unusual leucine zipper. Mol. Microbiol. 23:23-33. [DOI] [PubMed] [Google Scholar]
- 49.Yin, Z., D. Stead, L. Selway, J. Walker, I. Riba-Garcia, T. Mclnerney, S. Gaskell, S. G. Oliver, P. Cash, and A. J. P. Brown. 2004. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics 4:2425-2436. [DOI] [PubMed] [Google Scholar]