Abstract
The PilB protein of Neisseria gonorrhoeae has been reported to be involved in the regulation of pilin gene transcription, but it also possesses significant homology to the peptide methionine sulfoxide reductase family of enzymes, specifically MsrA and MsrB from Escherichia coli. MsrA and MsrB in E. coli are able to reduce methionine sulfoxide residues in proteins to methionines. In addition, the gonococcal PilB protein encodes for both MsrA and MsrB activity associated with the repair of oxidative damage to proteins. In this work, we demonstrate that the PilB protein of Neisseria gonorrhoeae is not involved in pilus expression. Additionally, we show that wild-type N. gonorrhoeae produces two forms of this polypeptide, one of which contains a signal sequence and is secreted from the bacterial cytoplasm to the outer membrane; the other lacks a signal sequence and is cytoplasmic. Furthermore, we show that the secreted form of the PilB protein is involved in survival in the presence of oxidative damage.
Pathogenic bacteria that contact the human host at the mucosal surface are subjected to host responses including localized membrane defenses such as the production of mucus, lysozyme, secretory antibodies, lactoferrin, reactive oxygen species (ROS), and reactive nitrogen intermediates (1, 2). The obligate human pathogen, Neisseria gonorrhoeae, is the causative agent of the disease gonorrhea. The hallmark of symptomatic gonorrhea is an influx of polymorphonuclear leukocytes (PMNs) into the urethra (3, 4). PMNs possess a variety of mechanisms for destroying bacteria; of particular importance is the respiratory burst of the phagocytes. This process generates a variety of ROS, including superoxide, hydrogen peroxide, hypochlorite, hydroxyl radical, and peroxynitrite (5–8). To exist as an effective pathogen, N. gonorrhoeae possesses a number of defenses to resist the host's oxidative burst, including both catalase and peroxidase activities (9, 10). Additionally, N. gonorrhoeae possesses a Mn(II) uptake system involved in resistance to oxidative damage through the accumulation of manganese (11). N. gonorrhoeae is unusual in that, despite the inevitable encounter with ROS, resistance to oxidative killing is independent of superoxide dismutases that destroy certain ROS (11). Considering the high level of toxic oxygen radicals likely encountered inside the host, it seemed probable that this pathogen has additional, and as yet unidentified, factors that permit it to survive in the presence of ROS.
Monomeric pilin, encoded by the pilE gene, is the major component of the pilus, a hair-like appendage involved in attachment of the bacterium to the host epithelium (12, 13). The PilA and PilB proteins of N. gonorrhoeae were originally reported to comprise a two-component system that regulates pilE transcription (14). The PilA protein has since been shown to be an FtsY homologue capable of hydrolyzing GTP, likely involved in protein transport and not in the regulation of pilin transcription in the gonococcus (15, 16). Additionally, controversy surrounds the involvement of PilB in the regulation of pilin production. A strain containing a transposon insertion in the pilB gene was shown to exhibit increased levels of both piliation and pilin mRNA (14). More recently, the predicted PilB protein of N. gonorrhoeae was found to have considerable homology to both the MsrA (17, 18) and MsrB proteins of Eschericia coli (19). Although encoded by two genes in most bacteria, a small subset of microorganisms, including Helicobacter pylori, Haemophilus influenza, Streptococcus pneumonia, and N. gonorrhoeae, possess both MsrA and MsrB homologues as a fused protein in a single ORF (19). MsrA and MsrB are highly conserved proteins in eukaryotes and prokaryotes, and MsrA has been identified as part of the minimum gene set required for life (20). The MsrA and MsrB proteins of E. coli are members of the peptide methionine sulfoxide reductase (PMSR) family of enzymes that catalyze the reduction of methionine sulfoxide residues in proteins (21–23). The oxidation of methionine results in the formation of two epimers of Met(O) (R and S). MsrA is specific for the S epimer of methionine sulfoxide, Met-S-(O), whereas MsrB reduces the R epimer, Met-R-(O) (19, 24, 25). The formation of methionine sulfoxide residues has been implicated in conformational changes in proteins, possibly leading to inactivation or modulation of activity (26–29). Consistent with its homology to MsrA and MsrB, the PilB protein of the pathogenic Neisseria has been shown to possess both MsrA and MsrB activity (30, 31). In the present study, we address how a protein reported to be involved in the regulation of piliation possesses both homology and enzyme activity associated with survival in the presence of ROS. We demonstrate that the PilB protein is not involved in pilus expression but is involved in survival to agents that can generate ROS. We show that the full-length gonococcal PilB protein is secreted from the bacterial cytoplasm to the outer membrane, and this extra-cytoplasmic localization is required for survival in the presence of oxidative damage. In addition, cellular fractionation experiments show the presence of a truncated cytoplasmic form of PilB in wild-type cells that does not confer protection against damage by oxidative agents.
Materials and Methods
Bacterial Strains, Bacterial Growth, and Molecular Genetics.
Gonococcal strain MS11, variant VD300, was used in all experiments, and grown on GC Medium Base (GCB; Difco) plus Kellogg supplements (12) at 37°C in 5% CO2. Transformants were selected with 10 μg/ml chloramphenicol. All pilB alleles were transferred to an MS11 VD300 strain carrying an isopropyl-β-d-thiogalactopyranoside (IPTG)-regulatable gonococcal recA allele, recA6, which has been described (32). All strains encoded the same starting pilE sequence, as described (33).
Adherence Assays.
The standard colony forming unit (CFU) adherence assay was performed as described (34) with the following differences. Gonococci (1 × 108 CFU per well) were incubated with Chang cell monolayers for 1 h at 37°C in 5% CO2. After washing, monolayers were incubated with 1% saponin (Sigma)-PBS for 10 min to disrupt the monolayers. The cell suspensions were diluted and plated on GCB for CFUs.
Analysis of Piliation.
Total pilin levels were determined by Western analysis (34) by using pilin mAb 1E8/G8 (35) at a dilution of 1:1,000. Pili were purified as described (36) and analyzed by SDS/PAGE followed by Coomassie staining to detect the pilin subunit. For electron microscopy, gonococci were grown for 18 to 20 h on plates, and poly-l-lysine-treated (1 μg/ml) formvar-coated grids (Ladd Research Industries, Burlington, VT) were used to lift cells from colonies. The grids were fixed and negatively stained as follows: once in 1% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer for 2 min, twice in sterile water for 3 s, and once in 1% (vol/vol) phosphotungstic acid (pH 6.0) for 1 min (37). Grids were viewed in a Jeol JEM-100 CX II transmission electron microscope at 60 kV.
Cloning of the Truncated Forms of PilB.
Truncated forms of PilB were cloned downstream of an IPTG-inducible promoter in pQE32 (Qiagen, Chatsworth, CA). PCR was performed on a plasmid containing the pilB gene as template (18). The forward primer used with both reverse primers shown below was 5′-TCGGATCCCATGAACACGCGCACCAT. The two reverse primers used were 5′-TAAGCTTTTATTTCACTTCGCCCTTCAAC and 5′-TAAGCTTTAGAAGCCTTTGCCTTGCG. One set of primers yielded DNA coding from amino acid 196 to the end of the protein (331 amino acids), encoding both MsrA and MsrB homology. The second primer pair produced DNA encoding a shorter protein (a termination codon has been inserted in the primer) stopping after the Phe residue at position 331 (135 amino acids) that contains only MsrA homology. Both products putatively initiate from an internal methionine residue at amino acid 196 (15). E. coli XL-1 blue cells were grown with IPTG, and the PilB derivatives were purified on Ni-NTA agarose beads. The purified proteins demonstrated the predicted masses of about 15 and 35 kDa.
Enzyme Activity.
MsrA and/or MsrB activity were determined as described (38) by using 5 nmol n-acetyl-[3H]methionine-R,S-sulfoxide as a substrate. Cell-free extracts of N. gonorrhoeae were prepared by sonication of a suspension of mid log-phase cells. The extract was centrifuged at 32,000 × g for 20 min, and the supernatant was assayed for enzymatic activity.
Oxygen Sensitivity Assays.
Hydrogen peroxide sensitivity was measured by using a disk inhibition assay. Approximately 109 CFUs were added to 3 ml of GCB-Top Agar (29 g/liter GCB dissolved in 800 ml of H2O plus 200 ml of GCBL for a final agar concentration of 0.8%), warmed to 48°C, and poured onto GCB plates containing Kellogg's supplements (12). Filter discs impregnated with 400 mM H2O2 were placed on solidified top agar. Plates were incubated overnight at 37°C with 5% CO2, and zones of inhibition were scored. The xanthine/xanthine oxidase assay was modified from that described by Wilks et al. (39). N. gonorrhoeae cells were harvested, washed once in PBS, and the concentration of cells was estimated by lysing a proportion of the suspension in 1% (vol/vol) SDS + 0.1 M NaOH and measuring A260 (A260 of 1 = 108 cells). Cells (104 to 105) were added to a solution of GCBL with Kellogg's supplements containing 43 μM xanthine (Sigma). Xanthine oxidase (3 milliunits/ml; Sigma) was added and cultures were incubated at 37°C with shaking at 180 rpm. Throughout 0–60 min, 10 μl of the cell suspension was withdrawn, plated onto GCB plates, and incubated at 37°C in 5% CO2. Experiments were done in triplicate and repeated 3 or 4 times.
Isolation of Membranes.
A modification of the procedure of Osborn and Munson (40) was used for isolation of inner and outer membranes. Pellets of whole cells were resuspended in 6.25 ml (per 250 ml of culture) of 200 mM Tris⋅HCl, pH 8.0 and diluted with an equal volume of ice cold 1 M sucrose/200 mM Tris⋅HCl, pH 8.0. The following prechilled reagents were added sequentially with vortex mixing: 25 μl of 250 mM EDTA, 100 μl of n-acetylmuramidase (5 mg/ml in H2O; mutanolysin, Sigma), 12.5 ml of 4°C H2O (added by pipetting). After 16 h shaking at 4°C, cells were sonicated (below) and centrifuged at 12,000 × g for 10 min. The supernatant then was centrifuged at 240,000 × g for 2 h at 4°C in a Ti80 rotor (Beckman Coulter). Pellets were resuspended in 1 ml of 18% (wt/wt) sucrose.
One milliliter of the membrane fraction was layered on a step gradient of 60%, 55%, 50%, 45%, 35%, 25%, and 20% sucrose (wt/wt) containing 1 mM EDTA and 200 μM DTT and centrifuged at 80,000 × g for 48 h at 4°C. After centrifugation, 1 ml of the fractions were collected and analyzed by refractometry and absorbance at 280 nm. Absorbing fractions in the density range of ≈1.1–1.2 were pooled and dialyzed. Pooled fractions were used for SDS/PAGE and Western blotting studies.
To confirm separation and outer membrane identity, the proteins of all fractions were separated by SDS/PAGE and stained with Coomassie brilliant blue, or blotted, and probed with antibodies to known cytoplasmic and periplasmic markers (EFTu; ref. 41) and outer membrane markers (Por, Omp85; refs. 42 and 43, and data not shown).
Isolation of Cytoplasm.
Cells were resuspended in 100 μl of Dulbecco's PBS and subjected to four 20-s bursts at full power using a Fisher 550 Sonic Dismembranator (Fisher Scientific). Cellular debris was removed by centrifugation at 12,000 × g for 10 m. The supernatant was aspirated and ultracentrifuged at 130,000 × g for 1 h. The supernatant was used in SDS/PAGE and Western blotting studies (44).
Alkaline Phosphatase Assay.
Alkaline phosphatase (AP) assays were performed as described (45) by using Sigma 104 phosphatase substrate (Sigma) at 28°C. Gonococci were grown on GCB for 18–24 h and swabbed into 1 M Tris pH 8.0 to approximately 108 CFU/ml. Gonococcal cell suspension (0.9 ml) in 1.0 M Tris pH 8.0 was added to 0.1 ml of 10 mM Tris pH 8.0 and used directly in alkaline phosphatase assays. AP units were standardized to the A600 of the bacterial culture.
Results
The Transposon Insertion Used in All Previous Analyses of PilB Results in Overexpression of a Truncated Form of PilB.
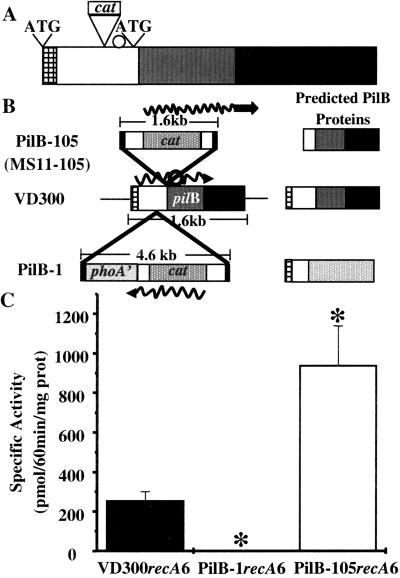
The initial account of the N. gonorrhoeae PilA and PilB described these proteins as being part of a two-component regulation system. This work demonstrated that a transposon insertion into the pilB gene of gonococcal strain MS11 (strain MS11–105) resulted in an increase in piliation and pilin mRNA (14). These results suggested that the gonococcal PilB acts as the negative regulator of a two-component system controlling pilin transcription and adherence. This observation was extended by a report of increased adherence to epithelial cells of MS11–105 compared with wild type (17). This result was particularly interesting because of the noted similarity between the PilB protein and the PMSR family of enzymes. In addition, the Neisserial protein encodes for both MsrA and MsrB activity (31). Searching the complete N. gonorrhoeae genome sequence has identified PilB as the only MsrA and MsrB homologue in this organism. Analysis of the MsrA/B activity of the original MS11–105 transposon mutant strain (14) unexpectedly revealed that this strain produced a three- to fourfold increase in MsrA/B activity compared with wild type (data not shown). We investigated how a transposon mutant of a gonococcal MsrA/B homologue might lead to an increase in MsrA/B activity and pilus expression.
For the sake of clarity, we will refer to MS11–105 as PilB-105 throughout this article, although these two strains are identical. We sequenced out of the transposon and located it to the 5′ region of the pilB gene, just upstream of the MsrA domain (Fig. 1A), with the cat gene of the transposon in the same transcriptional orientation as pilB. There is a candidate ribosome-binding site just 3′ of the site of transposon insertion, and an in-frame AUG in register to the ribosome-binding site (Fig. 1 A and B). We postulated that strain PilB-105 was producing a truncated form of PilB that expressed increased levels of MsrA/B activity (Fig. 1C) because of read-through transcription from the cat gene (Fig. 1B). In support of this, a band migrating at the size predicted for the truncated PilB protein was recognized by anti-PilB antiserum in PilB-105 (for example, see Fig. 4C). To determine if the transposon insertion in PilB-105 disrupted the MsrA activity of PilB because of the proximity of the transposon insertion to the MsrA domain, we tested the ability of the putative PilB-105 truncated protein to reduce both the S and R epimers of methionine sulfoxide. We cloned the predicted truncated form of PilB and showed that it possesses both MsrA and MsrB activity (data not shown). Additionally, we observed that a shorter truncated form of PilB that contains only the MsrA domain (see Methods) is stereospecific for the S form of methionine sulfoxide (data not shown). These results demonstrate that the transposon insertion in PilB-105 does not disrupt the reductase function of the MsrA or MsrB domains, and that the MsrA and MsrB domains possess stereospecificity. We concluded that the transposon insertion in strain PilB-105 was producing a truncated, overexpressed form of the protein, leading to increased amounts of MsrA/B activity. Therefore, we needed a PilB loss-of-function mutant to investigate the exact role of PilB in piliation.
Figure 1.
Maps of pilB alleles and their MsrA/B activity. (A) Location of transposon insertion in PilB-105 relative to protein domains. Checkered box represents signal sequence, white box represents putative disulfide oxidoreductase domain, gray box represents MsrA domain, and the black box corresponds to the MsrB domain. Transposon insertion is shown just upstream of the putative ribosome binding site (white circle). Putative start codons are marked with ATG. (B) Transposon insertions and predicted protein products. Squiggly line is the predicted mRNA transcript, white circle represents the predicted internal ribosome-binding site, and the checked bar is the predicted N-terminal signal sequence. Strain MS11–105 contains the same pilB allele as shown in PilB-105. VD300 is wild type, and PilB-1 contains the N-terminal signal sequence of PilB fused in frame to phoA'. (C) Levels of MsrA/B activity. *, P < 0.05, as compared with VD300recA6 in Student's t test.
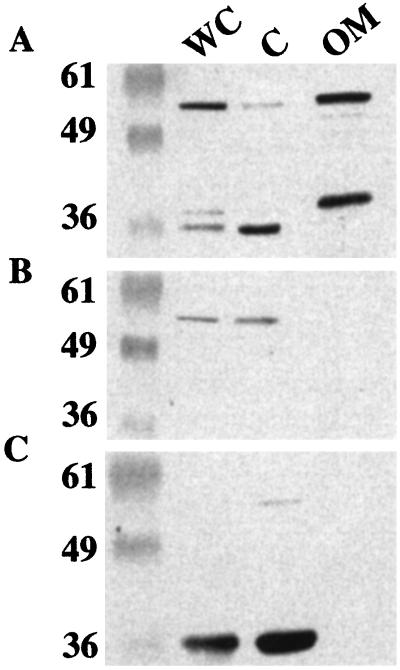
Figure 4.
PilB-1recA6 and PilB-105recA6 lack an extra-cytoplasmic form of PilB. Western blots of cellular fractions of VD300recA6, PilB-1recA6, and PilB-105recA6 using polyclonal anti-PilB antiserum raised in rabbits injected with purified PilB. Marker values are listed in kDa. Whole cell (WC), cytoplasm (C), and outer membrane (OM) lanes are marked. (A) Strain VD300recA6. (B) PilB-1recA6 loss-of-function mutant strain. Bands visible are cross-reactive bands running similarly but not exactly at the size of full-length PilB. (C) PilB-105recA6 truncated overexpressed mutant strain. Blots in A and C were exposed for shorter times than blot in B, which explains the decreased intensity of the cross-reactive bands in A and C.
Previously, a strain was isolated containing a minitransposon, carrying both the phoA' allele and a cat marker, inserted into the pilB gene (strain PilB-1; ref. 46). PilB-1 contains the cat gene in the opposite transcriptional orientation to the pilB gene and is predicted to produce a protein containing the N-terminal signal sequence of PilB fused to PhoA'. MsrA/B activity assays revealed that PilB-1 lacked detectable MsrA/B activity, whereas PilB-105 had approximately four times the activity of wild-type strain VD300 (Fig. 1C). These results provided us with a panel of three strains (VD300, PilB-1, and PilB-105) to address the role of PilB and its associated MsrA/B activity in pilus expression of N. gonorrhoeae.
PilB Is Not Involved in Piliation, Pilin Production, or Adherence.
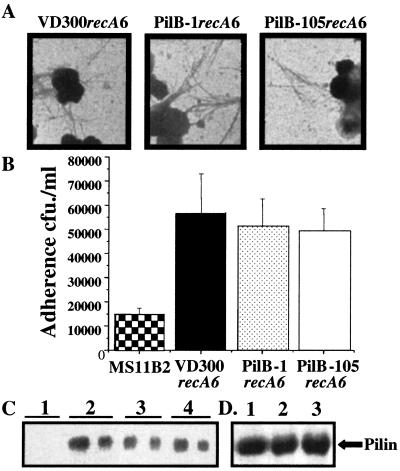
To ascertain the role of PilB in pilus expression, we transformed the two pilB alleles into MS11recA6 to allow IPTG regulation of transcription of recA and the control of antigenic variation at pilE. Transformants containing the same pilE sequence and either the wild-type pilB allele (VD300recA6), pilB-null mutant allele (PilB-1recA6), or the truncated overexpressed pilB allele (PilB-105recA6) were isolated. Comparison of piliation by transmission electron microscopy showed no substantial difference in the number of pili, or number of cells expressing pili, on VD300recA6, PilB-1recA6, or PilB-105recA6 (Fig. 2A). This observation was supported by the ability of all three strains to mediate equivalent levels of pilus-dependent adherence to Chang conjunctival cells, all of which adhered significantly better than the nonpiliated control MS11-B2 (Fig. 2B).
Figure 2.
Mutations in pilB do not affect piliation, pilin production, or adherence. MS11B2 is a pilin-null mutant strain (55) used as a negative control. (A) Negatively stained whole-mount electron micrographs. (B) Adherence assays to Chang conjuctival cells. (C) Western blot of total cellular protein using anti-Pilin antibody. Lane 1, ΔpilE; lane 2, VD300recA6; lane 3, PilB-1recA6; lane 4, PilB-105recA6. Two dilutions of protein sample loaded per strain to compare at different levels. (D) Total extracellular pilin. Lane 1, VD300recA6; lane 2, PilB-1recA6; lane 3, PilB-105recA6.
Anti-pilin Western blots demonstrate that the levels of total pilin do not vary between VD300recA6, PilB-1recA6, and PilB-105recA6 (Fig. 2C). In addition, purification of pili did not reveal any differences in the level of total extracellular pilin produced between VD300recA6, PilB-1recA6, and PilB-105recA6 (Fig. 2D). Finally, quantitative reverse transcription-PCR analysis of pilE mRNA showed no difference in the level of pilin message between the three strains (data not shown). Therefore, we conclude that, contrary to previously published reports (14, 17), the PilB protein of N. gonorrhoeae is not involved in piliation, adherence, pilin production, or pilE transcription.
PilB Affects the Survival of N. gonorrhoeae to ROS.
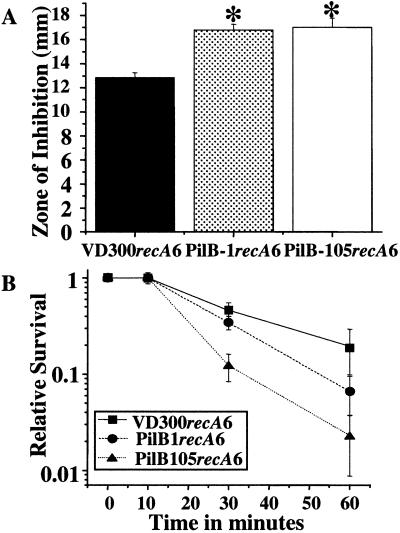
The observation that the gonococcal PilB protein has homology to proteins of the PMSR family of enzymes, combined with the MsrA/B activity of the gonococcal PilB, suggested that PilB might be involved in survival of N. gonorrhoeae to oxidative damage. To determine whether inactivation or overexpression of the truncated PilB affected growth in the presence of hydrogen peroxide, a hydrogen peroxide disk diffusion assay was carried out. Both strains—PilB-1recA6 and PilB-105recA6—were more sensitive to hydrogen peroxide than the wild-type strain (Fig. 3A). This finding demonstrates that mutations in the N. gonorrhoeae PilB lead to an increased sensitivity to the ROS generated by hydrogen peroxide.
Figure 3.
PilB mutant strains show increased sensitivity to oxygen radical generators. (A) Hydrogen peroxide disk inhibition assay. *, P < 0.05, as compared with VD300recA6 in Student's t test. (B) Xanthine/xanthine oxidase survival assay.
The effect of the superoxide ion on survival of the PilB mutant strains was determined by incubating the cells in the presence of xanthine/xanthine oxidase (39). Both PilB-1recA6 and PilB-105recA6 demonstrated a decreased survival rate, as compared with VD300recA6 in the presence of xanthine/xanthine oxidase. Notably, PilB-105recA6 survived less well than PilB-1recA6 in these assays (Fig. 3B). These results allow us to conclude that wild-type PilB is involved in survival in the presence of both hydrogen peroxide and superoxide.
Decreased Survival of the Truncated Overexpressed PilB Mutant in the Presence of Reactive Oxygen Radicals Is Due to a Loss of Secreted Forms.
PilB-105recA6 expresses more MsrA/B activity than wild type (Fig. 1C), yet survives less well than wild type when exposed to ROS (Fig. 3 A and B). Because the transposon insertion in PilB-105recA6 is predicted to eliminate the wild-type signal sequence from PilB (Fig. 1 A and B), we hypothesized that the increased sensitivity to ROS of PilB-105recA6 might be caused by inappropriate localization of PilB. We confirmed that PilB-1recA6 produces levels of alkaline phosphatase consistent with secretion of this fusion protein from the bacterial cytoplasm (data not shown; ref. 47), supporting our hypothesis that in strain PilB-105recA6, the truncated form of PilB is unable to leave the bacterial cytoplasm because of the absence of the N-terminal signal sequence.
To identify the cellular locale of the gonococcal PilB protein, cellular fractionation experiments were performed. These fractionations revealed the presence of two proteins that are recognized by anti-PilB polyclonal antisera in the outer membrane fraction of VD300recA6 (Fig. 4A). The larger protein migrates at the mass consistent for the full-length PilB protein, whereas the faster migrating band is likely a truncated form of PilB. The mutations in both strains PilB-1recA6 and PilB-105recA6 lead to the disappearance of these bands in the outer membrane (Fig. 4 B and C), demonstrating that both proteins are encoded by the pilB gene. In addition to the two proteins observed in the outer membrane fraction of VD300recA6, additional truncated forms of PilB are present in the whole-cell fraction of VD300recA6. The truncated form that migrates the fastest of the three bands specifically localizes to the cytoplasmic fraction (Fig. 4A). This band appears to have the same mobility as the truncated form of PilB observed in PilB-105recA6 (Fig. 4C). This result is not surprising, as VD300recA6 encodes the same internal RBS and AUG initiation codon that is still intact in PilB-105recA6 (Fig. 1A). The bands visible in PilB-1recA6 are cross-reactive bands running similarly but not exactly at the size of full-length PilB. No PilB or MsrA/B activity was found in the supernatant of PilB-105 (data not shown), showing that the loss of detection in the outer membrane is not caused by secretion into the medium. These results, in combination with the alkaline phosphatase activity assays, support the hypothesis that the oxygen-sensitive phenotype of PilB-105recA6 is due to the loss of the extra-cytoplasmic forms of PilB. The observation that the truncated form of PilB present in PilB-105recA6 is also present in VD300recA6 suggests that this truncated form has a physiological role in wild-type N. gonorrhoeae.
Discussion
PilB was initially designated as a regulator of gonococcal pilin expression. This work was initiated to determine (i) the level of involvement of the PilB protein in pilin expression, (ii) the physiological significance of the MsrA/B activity of PilB, and (iii) if PilB is involved in pilin production, does this involvement depend on the MsrA/B activity of PilB. The PilB-1recA6-null mutant strain and the PilB-105recA6 truncated, overexpressed mutant strain are not substantially affected in piliation, pilin production, adherence, or levels of pilin mRNA. Therefore, we conclude that PilB has no role in pilus expression or piliation. Inactivation or a lack of extra-cytoplasmic localization of the PilB protein resulted in increased sensitivity to ROS, suggesting a role for secreted PilB in protection against an oxidative burst. We propose that the name of the gonococcal PilB protein be changed to MsrA/B to conserve the nomenclature used in other organisms.
The initial descriptions of the role of the gonococcal MsrA/B protein in piliation, using the same pilB allele present in PilB-105recA6, suggested that this protein, in cooperation with PilA, regulates transcription of pilE (14, 48). Subsequent work reported an increased ability of the same strain to adhere to epithelial cells (17). Pilin-associated phenotypes, including adherence, are differentially affected by alterations in pilE sequences (34). We noticed that the strains being compared in these analyses were not matched for starting pilE sequences. Thus, because the starting pilE sequences were matched in our work, we suggest that these different results are due to the fact that strains compared in this study lack any intrinsic differences in pilin-associated phenotypes.
We show that MsrA/B is involved in the survival of N. gonorrhoeae in the presence of ROS, consistent with what would be expected from a protein with MsrA/B activity. Previous studies in E. coli (49, 50) and yeast (51) have shown that a knockout of MsrA results in the organism being more susceptible to oxygen radicals. The gonococcal PilB protein contains a candidate N-terminal signal sequence followed by two consecutive Msr domains with homology to both MsrA and MsrB, the latter in the C-terminal portion of the protein (30). The transposon insertion in PilB-105recA6 is predicted to produce a protein containing both reductase domains, but lacking the N-terminal signal sequence and a putative reductase domain (19). We have shown that the cloned and expressed truncated form possesses both MsrA- and MsrB-specific activities. We identified a similarly truncated form of MsrA/B in the cytoplasm of wild-type cells, indicating that this smaller form of MsrA/B might have some physiological significance. However, the oxygen sensitivity assays reveal that this truncated form of MsrA/B does not afford protection against oxidative damage, possibly because the critical methionine sulfoxide targets for MsrA/B are located in the outer membrane in this organism. Previous work on MsrA has speculated that the methionine residues in proteins can act as a unique antioxidant system, scavenging ROS that enter the cell (50, 52). In this mechanism, exposed Met residues in proteins act catalytically to destroy ROS through oxidation, followed by reduction by MsrA and MsrB. Also, ROS that are not inactivated by antioxidant systems such as glutathione, thioredoxin, glutaredoxin, or protein methionines can activate additional systems that protect against oxidative damage (53, 54). This mechanism could potentially lead to an increase in oxygen sensitivity of an overexpressing MsrA/B strain as compared with wild type because of a lack of the critical threshold of ROS buildup in the cells necessary to activate these additional defensive systems.
A hallmark of gonococcal infection is a purulent exudate consisting of bacteria and host neutrophils (3, 4). Upon interaction, the neutrophil produces toxic products directed at the bacterial cell, including hydrogen peroxide, hydroxyl radicals, hypochlorite, superoxide anion, and nitric oxide (6–8). Despite this inhospitable environment, N. gonorrhoeae is quite successful at colonizing the human urogenital tract. Although N. gonorrhoeae produces copious amounts of catalase and peroxidase, this organism produces limited amounts of superoxide dismutase. Additionally, the superoxide dismutase produced by the gonococcus is not required for survival in the presence of ROS (11). However, the accumulation of manganese protects against superoxide-mediated killing (11). The present work has identified a new way by which this pathogen might survive in the presence of the oxidative burst of host cells. We have shown that the gonococcal MsrA/B protein is involved in survival in the presence of hydrogen peroxide and superoxide anions. Recent work has shown that MsrA in E. coli is also important for survival in the presence of nitric oxide (50). If this protection is conserved in the gonococcus, it would demonstrate a method of protection against the different forms of ROS produced by neutrophils. Attempts to determine if the PilB mutant strains survive less well than wild type in a PMN killing assay provided inconclusive results (data not show). We believe this is due to the effectiveness of PMN killing being too great to reveal differences in survival between these three strains. Although the action of the gonococcal MsrA/B might not be sufficient to mediate survival against PMNs, the results of our in vitro assays suggest that MsrA/B is protecting against certain components of neutrophil-mediated killing in vivo. This study demonstrates that secretion of this protein from the bacterial cytoplasm is essential for complete protection against oxidative damage, underscoring the importance of proper protein localization as it relates to biological function. The fact that the gonococcal MsrA/B protein requires extracytoplasmic localization to provide protection against ROS suggests that the outer membrane requirement for certain proteins may present new targets for novel antimicrobial therapy for the disease gonorrhea.
Acknowledgments
We thank Kimberly Kline for critical reading of this manuscript. This work was supported by Public Health Service Grants R01AI33493 (to H.S.S.) and RO1AI47254 (to R.C.J.). Additionally, R.C.J. was supported by IBS-CORE (University of Montana). E.P.S. was partially supported by Public Health Service training Grant T32GM08061.
Abbreviations
- ROS
reactive oxygen species
- PMN
polymorphonuclear leukocytes
- CFU
colony forming unit
References
- 1.Dejong A R, Ros S, Smith K. Ped Emerg Care. 1994;10:238–240. doi: 10.1097/00006565-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 2.Cohen M S, Sparling P F. J Clin Invest. 1992;89:1699–1705. doi: 10.1172/JCI115770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafer W M, Rest R F. Ann Rev Microbiol. 1989;43:121–145. doi: 10.1146/annurev.mi.43.100189.001005. [DOI] [PubMed] [Google Scholar]
- 4.Rest R F, Shafer W M. Clin Microbiol Rev. 1989;2,Suppl.:S83–S91. doi: 10.1128/cmr.2.suppl.s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathan C, Shiloh M U. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitznagel J K. Prog Clin Biol Res. 1977;13:103–131. [PubMed] [Google Scholar]
- 7.Thomas E L, Lehrer R I, Rest R F. Rev Infect Dis. 1988;10, Suppl. 2:S450–S456. doi: 10.1093/cid/10.supplement_2.s450. [DOI] [PubMed] [Google Scholar]
- 8.Clifford D P, Repine J E. Mol Cell Biochem. 1982;49:143–149. doi: 10.1007/BF00231175. [DOI] [PubMed] [Google Scholar]
- 9.Archibald F S, Duong M N. Infect Immun. 1986;51:631–641. doi: 10.1128/iai.51.2.631-641.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson S R S B, Cruce D D, Perkins G H, Arko R J. Infect Immun. 1993;61:1232–1238. doi: 10.1128/iai.61.4.1232-1238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng H J, Srikhanta Y, McEwan A G, Jennings M P. Mol Microbiol. 2001;40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- 12.Kellogg D S, Jr, Peacock W L, Deacon W E, Brown L, Pirkle C I. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson J. J Exp Med. 1973;137:571–589. doi: 10.1084/jem.137.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taha M K, So M, Seifert H S, Billyard E, Marchal C. EMBO J. 1988;7:4367–4378. doi: 10.1002/j.1460-2075.1988.tb03335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arvidson C G, So M. J Biol Chem. 1995;270:26045–26048. doi: 10.1074/jbc.270.44.26045. [DOI] [PubMed] [Google Scholar]
- 16.Arvidson C G, Powers T, Walter P, So M. J Bacteriol. 1999;181:731–739. doi: 10.1128/jb.181.3.731-739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wizemann T M, Moskovitz J, Pearce B J, Cundell D, Arvidson C G, So M, Weissbach H, Brot N, Masure H R. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowther W T, Brot N, Weissbach H, Honek J F, Matthews B W. Proc Natl Acad Sci USA. 2000;97:6463–6468. doi: 10.1073/pnas.97.12.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimaud R, Ezraty B, Mitchell J K, Lafitte D, Briand C, Derrick P J, Barras F. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 20.Mushegian A R, Koonin E V. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brot N, Weissbach H. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 22.Brot N, Weissbach L, Werth J, Weissbach H. Proc Natl Acad Sci USA. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrams W R, Weinbaum G, Weissbach L, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1981;78:7483–7486. doi: 10.1073/pnas.78.12.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharov V S, Ferrington D A, Squier T C, Schoneich C. FEBS Lett. 1999;455:247–250. doi: 10.1016/s0014-5793(99)00888-1. [DOI] [PubMed] [Google Scholar]
- 25.Moskovitz J, Poston J M, Berlett B S, Nosworthy N J, Szczepanowski R, Stadtman E R. J Biol Chem. 2000;275:14167–14172. doi: 10.1074/jbc.275.19.14167. [DOI] [PubMed] [Google Scholar]
- 26.Vogt W. Free Radical Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 27.Sigalov A B, Stern L J. FEBS Lett. 1998;433:196–200. doi: 10.1016/s0014-5793(98)00908-9. [DOI] [PubMed] [Google Scholar]
- 28.Gao J, Yin D H, Yao Y, Sun H, Qin Z, Schoneich C, Williams T D, Squier T C. Biophys J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciorba M A, Heinemann S H, Weissbach H, Brot N, Hoshi T. Proc Natl Acad Sci USA. 1997;94:9932–9937. doi: 10.1073/pnas.94.18.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowther W T, Weissbach H, Etienne F, Brot N, Matthews B W. Nat Struc Biol. 2002;9:348–352. doi: 10.1038/nsb783. [DOI] [PubMed] [Google Scholar]
- 31.Olry A, Boschi-Muller S, Marraud M, Sanglier-Cianferani S, Van Dorsselear A, Branlant G. J Biol Chem. 2002;277:12016–12022. doi: 10.1074/jbc.M112350200. [DOI] [PubMed] [Google Scholar]
- 32.Seifert H S. Gene. 1997;188:215–220. doi: 10.1016/s0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- 33.Stohl E A, Seifert H S. Mol Microbiol. 2001;40:1301–1310. doi: 10.1046/j.1365-2958.2001.02463.x. [DOI] [PubMed] [Google Scholar]
- 34.Long C D, Hayes S F, van Putten J P, Harvey H A, Apicella M A, Seifert H S. J Bacteriol. 2001;183:1600–1609. doi: 10.1128/JB.183.5.1600-1609.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards M, McDade R L, Schoolnik G, Rothbard J B, Gotschlich E C. J Exp Med. 1984;160:1782–1791. doi: 10.1084/jem.160.6.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfgang M, Lauer P, Park H-S, Brossay L, He'bert J, Koomey M. Mol Microbiol. 1998;29:312–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 37.McGee Z A, Gross J, Dourmashkin R R, Taylor-Robinson D. Infec Immun. 1976;14:266–270. doi: 10.1128/iai.14.1.266-270.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brot N, Werth J, Koster D, Weissbach H. Anal Biochem. 1982;122:291–294. doi: 10.1016/0003-2697(82)90283-4. [DOI] [PubMed] [Google Scholar]
- 39.Wilks K E, Dunn K L, Farrant J L, Reddin K M, Gorringe A R, Langford P R, Kroll J S. Infect Immun. 1998;66:213–217. doi: 10.1128/iai.66.1.213-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborn M J, Munson R. Methods Enzymol. 1974;31:642–653. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- 41.Porcella S F, Belland R J, Judd R C. Microbiology. 1996;142:2481–2489. doi: 10.1099/00221287-142-9-2481. [DOI] [PubMed] [Google Scholar]
- 42.Judd R C. Clin Microbiol Rev. 1989;2,Suppl.:S41–S48. doi: 10.1128/cmr.2.suppl.s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manning D S, Reschke D K, Judd R C. Microb Pathog. 1998;25:11–21. doi: 10.1006/mpat.1998.0206. [DOI] [PubMed] [Google Scholar]
- 44.Judd R C. Anal Biochem. 1988;173:307–316. doi: 10.1016/0003-2697(88)90194-7. [DOI] [PubMed] [Google Scholar]
- 45.Brickman E, Beckwith J. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 46.Boyle-Vavra S, Seifert H S. Gene. 1995;155:101–106. doi: 10.1016/0378-1119(94)00890-5. [DOI] [PubMed] [Google Scholar]
- 47.Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taha M K, Dupuy B, Saurin W, So M, Marchal C. Mol Microbiol. 1991;5:137–148. doi: 10.1111/j.1365-2958.1991.tb01834.x. [DOI] [PubMed] [Google Scholar]
- 49.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. Proc Natl Acad Sci USA. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moskovitz J, Berlett B S, Poston J M, Stadtman E R. Proc Natl Acad Sci USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine R L, Mosoni L, Berlett B S, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim H S, Park Y W, Kasai H, Nishimura S, Park C W, Choi K H, Chung M H. Mutat Res. 1996;363:115–123. doi: 10.1016/0921-8777(96)00006-7. [DOI] [PubMed] [Google Scholar]
- 54.Wu J, Weiss B. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal E, Billyard E, So M, Storzbach S, Meyer T F. Cell. 1985;40:293–300. doi: 10.1016/0092-8674(85)90143-6. [DOI] [PubMed] [Google Scholar]