Abstract
The sexual development and virulence of the fungal pathogen Cryptococcus neoformans is controlled by a bipolar mating system determined by a single locus that exists in two alleles, α and a. The α and a mating-type alleles from two divergent varieties were cloned and sequenced. The C. neoformans mating-type locus is unique, spans >100 kb, and contains more than 20 genes. MAT-encoded products include homologs of regulators of sexual development in other fungi, pheromone and pheromone receptors, divergent components of a MAP kinase cascade, and other proteins with no obvious function in mating. The α and a alleles of the mating-type locus have extensively rearranged during evolution and strain divergence but are stable during genetic crosses and in the population. The C. neoformans mating-type locus is strikingly different from the other known fungal mating-type loci, sharing features with the self-incompatibility systems and sex chromosomes of algae, plants, and animals. Our study establishes a new paradigm for mating-type loci in fungi with implications for the evolution of cell identity and self/nonself recognition.
Self/nonself recognition events underlie the function of the major histocompatibility locus in defense against infection and organ transplant rejection, the self-incompatibility systems that prevent inbreeding in plants, and the production of offspring by sexual reproduction. During sexual reproduction, specialized genomic regions promote self/nonself interactions. Sex-determining regions include the mating-type loci in fungi and the sex chromosomes in plants and animals. Dimorphic sex chromosome systems independently evolved in animals, mosses, and dioecious plants. A related but distinct sexual incompatibility system is found in many lower eukaryotes, including algae, protozoans, monoecious plants, and fungi. In these organisms, multiallelic mating-type (MAT) loci monitor cell interactions for sexual compatibility, and if inbreeding is detected, mating is aborted (17, 18, 50, 56).
A common theme of sex determinants is the need to be transmitted as a single unit, and recombination within sex-determining regions is suppressed to avoid generating self-fertile or sterile offspring. Several mechanisms operate to suppress recombination. In the fungal MAT loci, extensive sequence divergence prevents recombination between different alleles. In the case of sex chromosomes, both sequence divergence and chromosomal rearrangements suppress recombination. These rearrangements affect nearly the entire sex chromosome in humans or mice, whereas in lower vertebrates and certain dipterous insects, only a limited region of the sex chromosomes is rearranged. These findings suggest that the dimorphic sex chromosomes evolved via accumulation of chromosomal aberrations.
Fungal mating-type loci serve as paradigms for understanding gene regulation during sexual development and the determination of cell fate and identity (7, 16, 32, 34, 37, 40, 54). Sexual development of ascomycetous fungi is commonly controlled by a bipolar mating system involving a single mating-type locus. In these cases, the MAT locus spans only a few thousand base pairs and exists in two unrelated alleles that control cell identity by encoding transcription factors that act on distant target genes (16, 40, 54).
In contrast to mating in ascomycetes, mating in basidiomycetes is commonly regulated by two independent, unlinked loci, resulting in tetrapolar mating systems (7, 37, 40). Both mating-type loci can be multiallelic, giving rise to thousands of different mating types in some mushroom fungi (38). The structure of mating-type loci in basidiomycetes has been determined for several model systems, including the mushrooms Coprinus cinereus (41, 52, 53) and Schizophyllum commune (66-68, 70) and the maize pathogen Ustilago maydis (4, 27, 39, 62). Similar to ascomycete mating-type loci, one locus encodes a pair of homeodomain transcription factors that controls a subset of developmental processes involved in sexual reproduction (A locus in C. cinereus and S. commune and b locus in U. maydis). The second locus (B in C. cinereus and S. commune and a in U. maydis) encodes a G protein-coupled pheromone receptor linked to one or more pheromone genes.
The opportunistic human fungal pathogen Cryptococcus neoformans is an encapsulated yeast that is distributed worldwide in association with pigeon guano and trees (6). This pathogen has increased in medical importance over the past several decades because of its ability to cause fatal meningioencephalitis in immunocompromised hosts. In contrast to many basidiomycetes, C. neoformans has a bipolar mating system with two opposite mating types, MATα and MATa.
A portion of the C. neoformans MATα locus was initially identified by a difference cloning approach and was found to contain the MFα1 pheromone gene (49). Subsequent work revealed that the C. neoformans MAT locus is unusual in size and gene composition, spanning an ∼55-kb region (35) and containing several additional α-specific genes, including STE12α and STE20α (10, 46, 69, 71, 73). Recent studies have revealed the following: (i) the MAT locus is larger than previously suspected (C. M. Hull, R. C. Davidson, and J. Heitman, submitted for publication), (ii) the α and a alleles encode divergent alleles of related genes (9, 46), and (iii) the architectures of the two MAT alleles may differ. Here we present our study on the C. neoformans mating-type locus that establishes a novel paradigm for the structure of mating-type loci with implications for fungal evolution and the evolution of specialized sex chromosomes.
MATERIALS AND METHODS
Strains.
The strains used for construction of bacterial artificial chromosome (BAC) libraries and analysis of the mating-type loci were C. neoformans serotype A var. grubii strains H99 (MATα) and 125.91 (MATa) and C. neoformans serotype D var. neoformans congenic strains JEC21 (MATα) and JEC20 (MATa) (31, 42, 46). The serotype D strains used to analyze recombination in the MAT region (Fig. 6) were derived from crosses between MATα strain DSF51 (znf1α::NAT1 ste12α::URA5 ade2) or RDC20-5 (ste12α::URA5 ade2) and MATa strain JEC53 (ura5 lys1). The stability of the MAT loci through several crosses (see Fig. 4) was analyzed using the serotype D strains NIH12, NIH433, B3501, B3502, JEC20, and JEC21 (31, 42). Structural analysis of MAT loci in the population of C. neoformans var. neoformans was conducted using the unrelated serotype D strains CDC92-18, CDC92-27, MMRL760 (all MATα), and #11 (MATa).
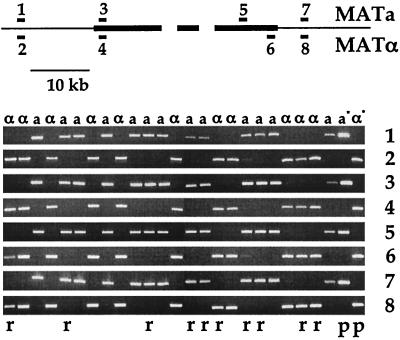
FIG. 6.
Recombination between the MAT alleles is suppressed. Twenty-four progeny from two defined crosses were tested by PCR for recombination events between mating-type alleles. Primer pairs were either mating-type specific (pairs 3, 4, 5, and 6) or strain specific (pairs 1, 2, 7, and 8). Fragments amplified within the mating-type region by the different primer pairs are indicated as thick black bars. No recombination events were observed, and the PCR-amplified fragments all cosegregated with the corresponding mating types (α or a), as determined by backcrosses. The control strains were the serotype D strains JEC20 (MATa) and JEC21 (MATα), indicated by a* and α*, respectively. The meiotic progeny were also tested for segregation of parental markers. Ten of 24 strains showed a recombinant pattern (r), whereas the remaining 14 strains exhibited a parental genotype (p), which is indicated only for the control strains.
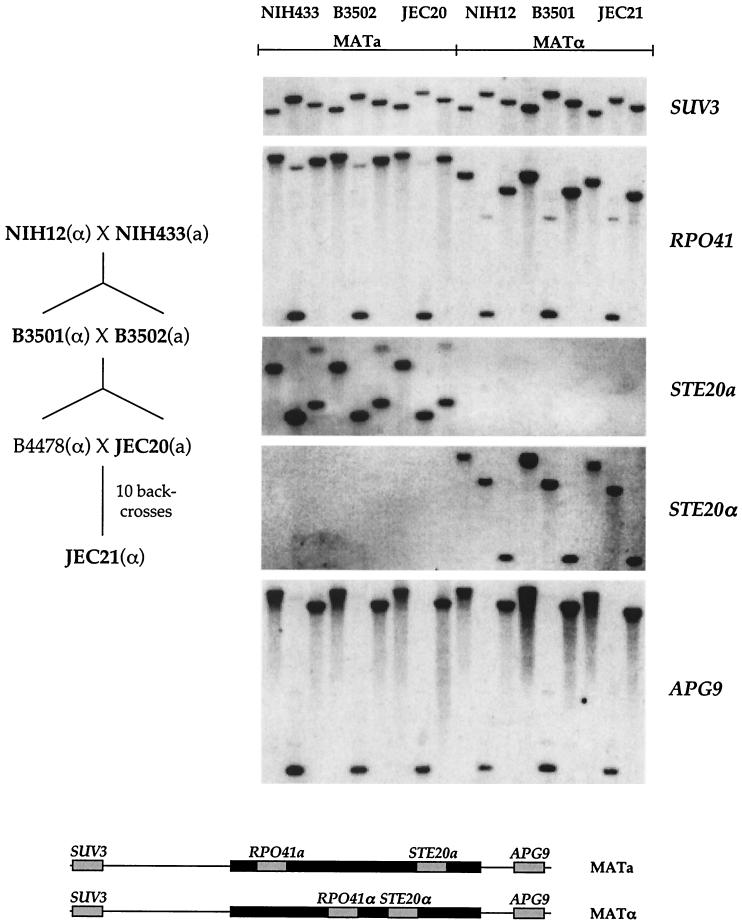
FIG. 4.
Mating-type locus is stable through several genetic crosses. The strains and backcrossing scheme used during the construction of the congenic pair of serotype D strains JEC21 and JEC20 are shown on the left. The strains used in the analysis are indicated in boldface type. Mating type is indicated as α or a. Strains in boldface type were subjected to restriction enzyme digestion (BamHI, HindIII, and PstI) and Southern blotting using the mating-type-specific and nonspecific probes indicated. No differences were apparent between JEC21 and JEC20 and the ancestral strains NIH12, NIH433, B3501, and B3502. The relative positions of the probes used are indicated below in the corresponding mating-type locus.
BAC and subgenomic libraries
To obtain high-quality chromosomal DNA from C. neoformans, protoplasts were isolated as described previously (46). In the present study, the lysis of protoplasts was prolonged to 48 h and proteinase K was added to the lysis buffer at a final concentration of 4 mg/ml. Fresh buffer was added after a 24-h incubation. Plugs containing lysed protoplasts were washed twice with ice-cold Tris-EDTA (TE) buffer containing 0.2 mM phenylmethylsulfonyl fluoride and subsequently washed four to five times with ice-cold TE buffer for 1 h each. Plugs could be stored indefinitely at 4°C in 50 mM EDTA. In collaboration with Research Genetics (Huntsville, Ala.), the chromosomal DNA was partially digested with HindIII, and ∼100-kb fragments were isolated after separation of digested DNA via pulsed-field gel electrophoresis. Genomic fragments were cloned into the BAC vector pBeloBAC11, and clones with inserts were identified using standard blue/white screening techniques. BAC clones of interest were identified by Southern hybridization with mating-type-specific gene probes by using colony lift membranes (Research Genetics). To close the remaining gaps in strains H99 and 125.91 not covered by the analyzed BAC clones (Fig. 1), desired fragments were identified by Southern blotting and isolated from subgenomic libraries or generated by PCR.
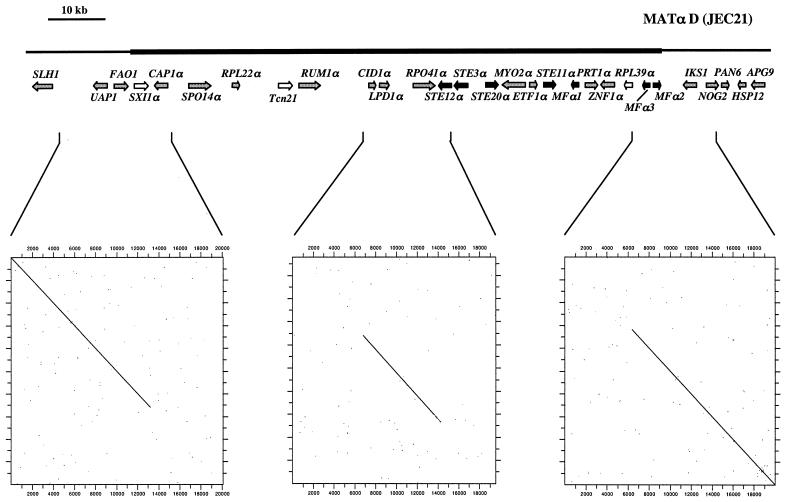
FIG. 1.
Structures of the serotype D (MATa of JEC20 and MATα of JEC21) and serotype A (MATa of 125.91 and MATα of H99) α and a mating-type alleles and adjacent genomic regions. The mating-type-specific regions are shown as thick bold lines, and flanking regions are shown as thinner black lines. Sequences were analyzed using BLASTX, and identified genes are shown as arrows in the direction of transcription. Genes encoding pheromone response pathway elements are shown as black arrows, locus-specific genes are shown as white arrows, and all other genes are shown as grey arrows. Bars above the mating-type alleles represent the BAC clones, genomic fragments, and PCR products analyzed.
BAC sequencing strategy
To generate high-quality BAC DNA, plasmid DNA was subjected to cesium chloride equilibrium centrifugation. The plasmid DNA of individual BAC clones was isolated from 500 ml of cultures by using alkaline lysis. The DNA was resuspended in 15 ml of TE buffer containing ethidium bromide (400 μl of a 10-mg/ml stock), and cesium chloride was added to a final density of 1.4 mg/ml. The DNA-CsCl solution was transferred into a 15-ml ultracentrifuge tube, and the sample was centrifuged in an NVT65.1 rotor at 65,000 rpm for ∼24 h at room temperature. The lower, plasmid-containing DNA band was removed using a 5-ml syringe with a 20-gauge needle and introduced into a 4-ml ultracentrifuge tube, which was then filled with CsCl solution to a final density of 1.4 mg/ml. The sample was centrifuged for an additional 24 h at 70,000 rpm. The DNA was removed from the second gradient, ethidium bromide was extracted several times with salt-saturated isopropanol, and the DNA was dialyzed against 5 liters of TE buffer for several hours. After adding 1/10 volume of 3 M sodium acetate, BAC DNA was precipitated with 0.6 volume of isopropanol (−20°C), washed with 70% EtOH, and resuspended in TE buffer.
Three to five micrograms of BAC DNA was sheared with a Hydroshear device (Gene Machines) to generate ∼1.5- to 3-kb DNA fragments. Sheared fragments were subsequently subjected to standard blunting and fill-in reactions, and double-stranded adapters, provided by the Duke Center for Genome Technology (CGT), were ligated in 100-fold excess to the blunted DNA fragments. Fragments were separated from free adapters by agarose gel separation, and the DNA was cloned into a special, pUC18-based linearized vector provided by the CGT containing ends compatible to the adapters ligated onto the BAC DNA fragments (51). Before large-scale sequencing, the percentage of clones with inserts and the average insert size were carefully checked. Clones were picked automatically into 384-well plates containing Luria-Bertani Hogness medium by using a Genomic Solutions Flexis robot, and the plates were heat sealed and stored at −80°C. H99 genomic shotgun libraries were prepared accordingly, starting with CsCl-purified genomic DNA. For sequencing, clones were grown in 96-well plates containing Terrific broth medium in a Higro orbital shaker (Gene Machines), and DNA was isolated using a RevPrep robot (Gene Machines). Sequencing reactions were performed with a Hydra workstation (Robbins) and an MJ Research thermal cycler using standard BigDye chemistry (Applied Biosystems). After removal of unincorporated dye, samples were analyzed on a PE3700 96-capillary sequencer, and sequence data were automatically transferred to the UNIX-based CGT server. The resulting sequence data were analyzed and assembled with the Pare/Phrased sequence package (19, 20), and assemblies were examined using Consed (29). Sequences from the C. neoformans Genome Project that were added during the assembly of the serotype D MATα mating-type locus were provided by the Stanford Genome Technology Center and The Institute for Genomic Research, funded by the National Institute of Allergy and Infectious Diseases and the National Institutes of Health under cooperative agreements U01 AI47087 and U01 AI48594, respectively. The genes were identified by comparing BAC sequences to sequences in the GenBank database by using the BLASTX algorithm (1).
PCR
Recombination within the mating-type region of the serotype D MATα and MATa loci was analyzed using mating-type and strain-specific primers designed for the corresponding sequences generated in this study (see Fig. 6). The primer sequences and primer combinations are listed in Table 1. Fragments of ∼500 bp were amplified using a synthesis time of 30 s and an annealing temperature of 66°C. Primers used in the structural analysis of mating-type loci from unrelated serotype D strains (Fig. 7) were initially designed for sequence analysis of the serotype D MAT locus from strain JEC21 and are also listed in Table 1. Depending on the primer combination, an annealing temperature between 50 and 64°C was used, whereas the synthesis time was 5 min for each reaction.
TABLE 1.
Primer pairs used in recombination and structural analysis of the C. neoformans MAT locus
| Primer pair | Sequence | Tm (°C)a |
|---|---|---|
| Pairs used in recombi- nation analysisb | ||
| JOHE7607 (6) | ATAGACATCCTCAACTTGTCCAC | 66 |
| JOHE7608 | AAGTTCAGCTGCTGAACGATCG | |
| JOHE7609 (5) | TTGAGCGTCATATTGGTCATGAC | 66 |
| JOHE7610 | GAAGACCGTCATCACACACAAG | |
| JOHE7611 (8) | GGACGACACTGTCACAATCATC | 66 |
| JOHE7612 | GAAATCGCACCGTGAGCTGAG | |
| JOHE7613 (7) | GAACGACACTGTCACAATCATG | 66 |
| JOHE7614 | GAAATCGCACCGTGAGCTTCC | |
| JOHE7615 (3) | GGTGTGCGAGGATGTAGTATGG | 66 |
| JOHE7616 | ATCCGCTCCTTCTATCAGTTCC | |
| JOHE7617 (4) | TTCGACCTGTGATAGCTCTTCC | 66 |
| JOHE7618 | TGCTTGACTCGGAAGAGGAGC | |
| JOHE7619 (1) | GATTCCATTCCACTTGCATTACG | 66 |
| JOHE7620 | GTGAGGAAGGTAGGGGAGTAGT | |
| JOHE7621 (2) | CCATTCCACTTGCATTATTATTACG | 66 |
| JOHE7622 | GTGAGGAAGGTAGGGGAGTATC | |
| Pairs used in struc- tural analysisc | ||
| JOHE3069 | GATTTATCTCAGCAGCCACG | 60 |
| JOHE5299 | ACAGCTAGTTCAACCATGGC | |
| JOHE5935 | CCTTCTACATCATCATATGTCACTTC | 64 |
| JOHE5795 | CGACGCAACAAGTCTGCTCG | |
| JOHE5724 | TTACCCACGTGGAGACAAC | 60 |
| JOHE5727 | CGATGGAATGTACATGTCTTG | |
| JOHE5867 | CGACGATCTCCAAGTTCTCG | 50 |
| JOHE5598 | ATCACCTAGAATGCAGC | |
| JOHE5870 | ATCCGCATGCTCGTCTATCC | 62 |
| JOHE5875 | CGTTACGAATACCTGATCACG | |
| JOHE5874 | TTCAGTGTATGAACTGCCCAC | 62 |
| JOHE5801 | TACCTTGGTCCGATCGAGACG | |
| JOHE5653 | TTTCGTCCTGAACAACCCAC | 60 |
| JOHE5877 | TTCTGCTGACCTTTAGTAGTAC | |
| JOHE5561 | TTGGTTCGACGCCAATGACG | 60 |
| JOHE5740 | CAACACAACCATCTGTAATGC | |
| JOHE5567 | GCATTACAGATGGTTGTGTTG | 60 |
| JOHE5570 | TCGTTCCCTGTTCTATTATCTG | |
| JOHE5741 | CAGATAATAGAACAGGGAACGA | 62 |
| JOHE5910 | GTTGATACACCGTTGTTGTACC |
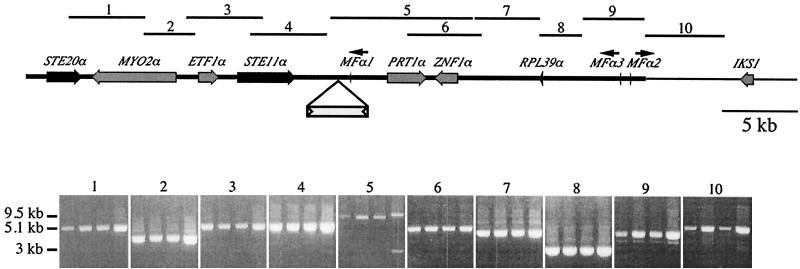
FIG. 7.
The MATα mating-type locus is conserved in the population. Using one primer combination, overlapping fragments (3 to 10 kb) spanning part of the mating-type locus were PCR amplified from (lanes from left to right in panel 1) the control strain JEC21 and the unrelated serotype D clinical isolates CDC92-18, CDC92-27, and MMRL760. Fragments of identical sizes were amplified from all four strains with nine primer combinations (panels 1 to 4 and 6 to 10). For descriptions of the 10 primer pairs, which are also represented by numbered bars indicating their positions on the MAT locus, see Table 1. One primer pair yielded a larger PCR product for strain MMRL760 (panel 5, lane 4), indicative of an ∼4-kb insertion between the STE11α and MFα1 genes. DNA sequence analysis revealed that the insertion of a novel mariner-related transposable element resulted in the addition of 4,006 bp relative to the JEC21 serotype D α allele and the creation of a TA target site duplication.
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequences reported here are as follows: JEC21 serotype D MATα AF542531; JEC20 serotype D MATa, AF542530; H99 serotype A MATα, AF542529; 125.91 serotype A MATa, AF542528; Tcn760, transposable element, AF542532.
RESULTS
Cloning and sequencing the mating-type locus of C. neoformans.
We set out to clone and sequence the mating-type locus of C. neoformans to test the following hypotheses. First, does the MAT locus encode homeodomain transcription factors that govern cell identity as in other fungi? Second, did the α and a alleles of the MAT locus diverge from a common ancestral region of DNA, which we proposed earlier based on the identification of the divergent STE20α and STE20a genes (46, 69)? Third, has the MAT locus been conserved or rearranged during the evolution of this pathogen?
To determine the complete structure of the α and a mating-type alleles of C. neoformans, genomic BAC libraries were generated from a congenic pair of serotype D α and a strains (JEC21 and JEC20) as well as from the serotype A α and a strains H99 and 125.91. Probes to known mating-type-specific genes were used to identify BAC clones spanning each allele of the mating-type locus. Shotgun libraries were produced from two or three BAC clones encompassing each allele and sequenced (Fig. 1). In the case of the α locus from serotype D, the ∼38-kb mating-type region defined by Moore and Edman was analyzed using PCR products and existing cosmid clones (49).
To establish gene order in this region and to rule out possible rearrangements of the BAC clones chosen for sequencing, hybridization-based BAC maps were generated (Fig. 2). An initial screen was conducted using probes to the STE12α and STE20α genes and sequences flanking the previously identified right end of the MAT locus. Hybridizing BAC clones were subjected to further analysis with additional probes, including the SXI1α and RUM1α genes (Fig. 2). The resulting BAC map confirmed the gene order for the initial ∼38- and ∼55-kb mating-type regions previously analyzed (35, 49) and extended this map to include numerous additional genes. No aberrant recombination events appear to have occurred during the construction of the BAC library. This was further confirmed by comparison of the sequences of the MATα loci generated in this study with BAC fingerprint maps for the α strains JEC21 and H99 generated at the Genome Center of the University of British Columbia (61).
FIG. 2.
Mapping of the serotype D (A) and serotype A (B) MATα loci by hybridization. The serotype D and serotype A MATα BAC libraries (strains JEC21 and H99) were screened with probes specific for several MAT-specific genes, the right end (RE) of the locus, and the flanking gene APG9. BAC clones that hybridized to these probes were analyzed by dot blot hybridizations using probes to the underlined genes. Additional hybridizations were conducted with a high-density BAC clone filter. These hybridization data were used to generate a linkage map and establish the gene order of the MAT locus. For all BAC clones depicted here, end sequences from the University of British Columbia database were incorporated to define endpoints that lie between hybridization probes. BACs sequenced to completion are marked with asterisks. The sizes of the BAC clones and BAC contigs depicted were determined at the University of British Columbia Genome Center. Clones used to span a gap in the H99 MAT locus are depicted as short grey lines above the locus. For additional details, see the legend to Fig. 1.
Data generated during both mapping processes revealed that the MATα locus of strain H99 was not completely covered in a library of ∼6,000 available BAC clones. This finding was confirmed during our sequencing efforts. The ∼10-kb gap in the H99 MAT locus was closed by identifying and sequencing the following: (i) genomic clones spanning MAT-specific genes, (ii) MAT-specific plasmid clones from the H99 shotgun sequencing project, and (iii) a 1.6-kb PCR product spanning the final remaining gap that proved recalcitrant to recovery in Escherichia coli (Fig. 2B). This ∼10-kb region spans three pheromone genes and the ZNF1α and PRT1α genes and includes several large inverted repeats that may render this region difficult to clone. To provide deeper sequence coverage, sequences for strains H99 (serotype A) and JEC21 (serotype D) that were available from GenBank and public genome sequencing projects were entered in the assembly. The overall region of double-stranded DNA that was bidirectionally sequenced was 245 and 210 kb for the serotype D α and a alleles, respectively, and 225 and 150 kb for the serotype A α and a alleles, respectively, for a total of 830 kb of genomic sequence.
Mapping the borders of the mating-type locus.
A portion of the serotype D α mating-type allele was identified by Moore and Edman in 1993, and subsequent work in several labs has contributed to further define the structure of the MAT locus (46, 49, 69, 71). Karos and coworkers reported a map of the serotype D MATα locus that spans an ∼55-kb region between the RPO41 and NOG2 genes (35). Here we present evidence that redefines the left border of the mating-type locus, demonstrating that the MAT locus spans an additional ∼50-kb region upstream of the RPO41 gene and defining the authentic left junction between genomic DNA and the MAT locus.
The junctions between the MAT locus and neighboring genomic DNA were identified by comparing the serotype D α and a allele sequences. The serotype D α strain analyzed (JEC21) was generated by backcrossing an α strain 10 times to an a strain (JEC20), resulting in a congenic strain pair that should differ only at the MAT locus (31, 42). Hence, sequences bordering the MAT locus should be identical or nearly so, whereas sequences within MAT should be distinct. A DNA sequence comparison of the α and a alleles by dot plot analysis revealed that flanking sequences on one side and within the UAP1-FAO1 and IKS1-NOG2 genes are nearly identical, whereas on the other side, the sequences diverge, defining the left and right junctions between the MAT locus and surrounding genomic DNA (Fig. 3, left and right panels). In addition, the order of genes surrounding the UAP1-FAO1 and IKS1-NOG2 genes in the flanking regions is identical between the α and a alleles up to the proposed junctions and then diverges in the opposite mating-type alleles (Fig. 1). In Southern analysis, probes specific to the sequences outside the predicted junctions yielded identical restriction patterns in α and a strains (Fig. 4), whereas MAT-specific probes yielded mating-type specific patterns (Fig. 4). Our findings indicate that the mitochondrial RNA polymerase gene RPO41 originally reported to define the left border of the MAT locus is in fact part of the mating-type locus and not a flanking gene. The finding that the gene order and sequence both diverge on either side of the RPO41 gene further supports this conclusion (Fig. 1 and 3, middle panel). An RPO41-specific probe also yielded different restriction patterns for the α and a strains JEC21 and JEC20 (Fig. 4).
FIG. 3.
Mapping of the ends of the mating-type locus by sequence comparison. Twenty kilobases of the sequences surrounding the proposed junctions between the MATα and MATa mating-type alleles and flanking DNA and surrounding the RPO41 mitochondrial RNA polymerase genes of the serotype D MATα and MATa strains JEC21 and JEC20 were compared using the DNA Strider program. Corresponding sequences were subjected to a pairwise comparison using a window size of 11 bp. In the graphical outputs, sequence identity is indicated by dots and stretches of sequence identity appear as diagonal lines. For additional details, see the legend to Fig. 1.
The sequences flanking the MAT loci in the α and a strains JEC21 and JEC20 share ∼99% identity for several kilobases before reaching 100% identity, reflecting the position of the most recent recombination between the mating-type junctions and surrounding genomic DNA. A small ∼100-bp region just upstream of the left MAT locus junction shares limited similarity between the α and a strains and may reflect an ancient recombination event between the alleles.
Gene order is nearly identical in the regions flanking the mating-type locus in the serotype A and D strains, whereas the central region spanning the MAT locus itself is extensively rearranged. In the left flanking regions, all four mating-type alleles share synteny. In contrast, in the right flanking region, the NOG2, PAN6, HSP12, and APG9 genes all share synteny but the gene that immediately flanks the MAT locus in serotype D (IKS1) is an integral component of the mating-type locus in both the α and a alleles in serotype A (Fig. 1). The IKS1 gene does not appear to be a component of the MAT locus in serotype D, as it is embedded in sequences that share ∼99% identity between strains JEC21 and JEC20. As discussed further below, the IKS1 gene may have entered the MAT locus in serotype A (gene capture model) or exited the locus in serotype D (gene egress model). Comparison of the IKS1 gene sequences reveals that the serotype A IKS1α and IKS1a alleles are dramatically divergent (52% identity), whereas the IKS1 genes flanking the serotype D MAT alleles share 99% identity with each other and significant identity (85%) with the serotype A IKS1α allele. These findings support a model in which the IKS1 gene was lost from the locus and fixed in the flanking region by inversion and recombination events, with concomitant loss of the IKS1a gene in the serotype D lineage.
In conclusion, based on synteny and sequence comparisons, our study demonstrates that the mating-type locus of C. neoformans is significantly larger than previously proposed, spanning an ∼105- to 130-kb region that lies between the FAO1 and IKS1-NOG2 genes in serotype D and the FAO1 and NOG2 genes in serotype A. The serotype D α and a alleles span 105,656 and ∼117,308 bp, respectively, whereas the serotype A α and a alleles span ∼102,764 and ∼127,082 bp, respectively. Thus, the a alleles of the MAT locus are larger than the α alleles.
Genes contained in the mating-type locus.
Approximately 20 genes contained in the MAT locus were identified when the BLASTX algorithm was used to compare the MAT locus sequence with sequences in GenBank (Fig. 1). Table 2 summarizes the genes identified within the mating-type alleles and flanking sequences. Transcripts corresponding to several of these genes are present in expressed sequence tags derived from cDNA from strain H99 and the serotype D strain B3501, a precursor to strain JEC21 (University of Oklahoma Health Science Center [http://www.genome.ou.edu/cneo.html]). The four alleles of the mating-type locus have been annotated with respect to the exon-intron structure of the genes contained and expressed sequence tags corresponding to genes within the MAT locus. This information is available electronically (http://cneo.genetics.duke.edu/mating-type/). Repetitive sequences and transposon remnants have also been annotated (Fig. 5).
TABLE 2.
Genes within and flanking the mating-type alleles of C. neoformans
| Gene(s) | Description or product(s) | Known or predicted cellular function(s) |
|---|---|---|
| Mating-type-specific genes | ||
| MFα/a1-3 | Pheromone precursor genes | Mating, haploid fruiting |
| STE3α/a | Pheromone receptor | Mating, haploid fruiting |
| SXI1αa | Homeodomain transcription factor | Mating, diploid filamentation |
| STE20α/a | p21-activated protein kinase | Pheromone response pathway |
| STE11α/a | Mitogen-activated protein kinase kinase kinase | Pheromone response pathway |
| STE12α/a | Homolog of S. cerevisiae Ste12 transcription factor | Pheromone response pathway |
| ZNF1α/a | Putative Zn-finger/PHD-finger transcription factor | Pheromone response pathway |
| CAP1α/a | Similar to C. neoformans Cap10 (capsule-associated protein) | Capsule biosynthesis |
| NAD4αb | NADH dehydrogenase subunit 4L | Respiratory chain |
| SPO14α/d | Similar to S. cerevisiae Spo14 (phospholipase D) | Lipid metabolism; meiosis and differentiation |
| RPL22α/a | Ribosomal protein | Protein synthesis |
| RPL39α/ac | Ribosomal protein | Protein synthesis |
| RUM1α/a | Similar to U. maydis Rum1; retinoblastoma binding protein 2 | Transcriptional repression |
| CID1α/a | Similar to C. albicans caffeine-induced death protein Cid1 | Putative nucleotidyltransferase, cell cycle |
| LPD1α/a | Dihydrolipoamide dehydrogenase | Amino acid metabolism |
| RPO41α/a | Mitochondrial RNA polymerase | Energy generation, mitochondrial transcription |
| MYO2α/a | Myosin heavy chain, class V myosin | Polar growth and secretion |
| ETF1α/a | Putative electron transport flavoprotein | Unknown |
| PRT1α/a | Translation initiation factor eIF3 beta subunit | Protein synthesis |
| IKS1α/ad | Probable serine/threonine protein kinase | Unknown |
| NCP1ae | Similar to N. crassa protein of unknown function | Unknown |
| Genes flanking the MAT locus | ||
| SLH1 | Putative member of the Snf2 family of DNA helicases | Unknown |
| UAP1 | Similar to A. nidulans UapA | Uric acid transporter |
| FAO1 | Putative iron/ascorbate oxidoreductase | Secondary metabolism |
| NOG2 | Putative nucleolar GTPase | Unknown |
| PAN6 | Pantothenate synthetase | Pantothenate synthesis |
| HSP12 | Similar to S. cerevisiae Hsp12 and C. albicans Wh11 | Protein folding, cell stress response |
| APG9 | Similar to S. cerevisiae Apg9 | Vesicular transport, autophagy |
Present only in MATα mating-type alleles.
Present only in the serotype A MATα strain H99.
Missing in the serotype D MATa strain JEC20.
Within the MAT locus in serotype A; flanking the MAT locus in serotype D.
Present only in the serotype D MATa strain JEC20.
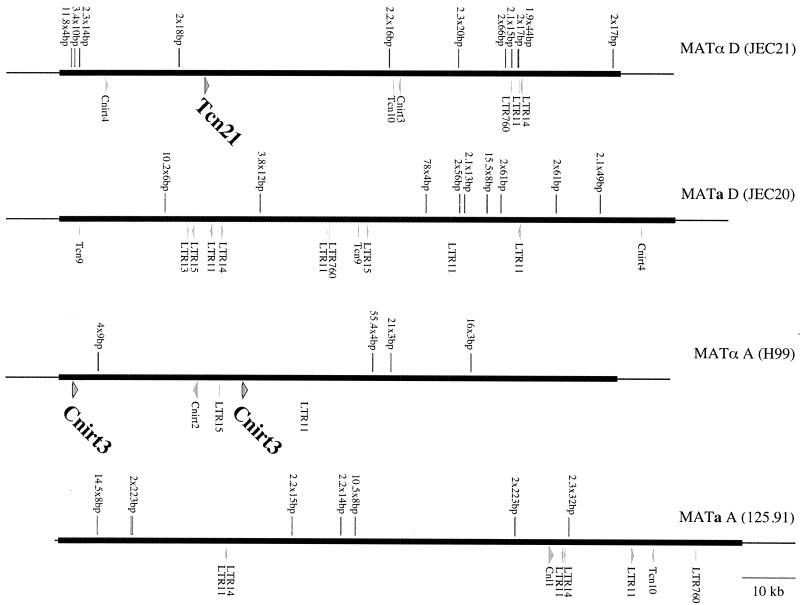
FIG. 5.
Multiple transposon remnants and repetitive sequences are embedded in the MAT locus. Transposable element-related sequences are depicted for the four alleles of the MAT locus. Complete element copies are indicated in a larger font size and boldface type. In addition, local sequence repeats were identified and annotated for each allele.
Previous studies on the C. neoformans mating-type locus had suggested that key regulators of sexual differentiation found in other basidiomycetous fungi might be missing. However, in the newly defined MAT locus, we identified homologs of both key mating-type components from model basidiomycetes. First, a pheromone receptor (STE3α/a) and several pheromone precursor genes (MFα/a) were identified in the locus, similar to those present in the a mating-type loci of U. maydis and Ustilago hordei and the B loci of C. cinereus and S. commune and in accord with several recent reports (14, 47, 63). In contrast to those in the model basidiomycetes, the genes for the pheromones and pheromone receptor are not tightly linked to one another but are instead dispersed throughout the C. neoformans MAT locus (Fig. 1). Second, a gene encoding a novel homeodomain protein (SXI1α) was identified in both the serotype A and D α mating-type alleles but not within the a mating-type alleles. This homeodomain homolog is analogous to the components found in the b mating-type loci of U. maydis and U. hordei and the A loci of C. cinereus and S. commune. The pheromones, pheromone receptor, and Sxi1α transcription factor have all been linked to roles in the sexual development of C. neoformans (14, 63; Hull et al., submitted).
The sexual development of fungi is regulated by a pheromone-activated mitogen-activated protein (MAP) kinase signaling cascade. In C. neoformans, several elements of the MAP kinase pathway are encoded by genes in the MAT locus and exist in divergent forms in the α and a alleles. These include homologs of the p21-activated protein kinase Ste20, the MEK kinase Ste11, and the transcription factors Ste12 and Znf1, which function in the sexual development and virulence of this organism (9, 10, 14, 15, 69, 73). The link between the components of the pheromone response pathway and the MAT locus is novel, and the biological importance of this unusual gene clustering for the organism is unknown but may involve the unique properties associated with the MATα locus that promote haploid fruiting and virulence.
In addition to mating-specific genes, several other genes are contained within the MAT locus that have no obvious role in sexual development (Table 2). However, based on their similarity to genes identified in other organisms, the functions of the products of a few of these genes can be predicted. For example, Rum1 is a retinoblastoma binding protein 2-like coregulator that corepresses genes regulated by the MAT locus-encoded homeodomain transcription factors bE and bW in U. maydis (55). By analogy, the Rum1α and Rum1a proteins may play similar roles in sexual differentiation in C. neoformans. Other proteins that might be involved in differentiation are the phospholipase D homolog Spo14, which is involved in meiosis and sporulation in Saccharomyces cerevisiae (59, 60), and the class V myosin heavy-chain homolog Myo2, which plays an important role in polarized growth and secretion in S. cerevisiae (3, 36, 72). Another interesting protein is Cap1, which shares amino acid identity (163 of 574 [28%] amino acids) with the product of a previously characterized gene of C. neoformans, CAP10, which is involved in capsule biosynthesis (8). Cap1 might therefore play a role in the synthesis of the capsular polysaccharide that is essential for virulence.
The MAT locus contains transposon remnants and many repetitive sequences.
Multiple transposon-related sequences were identified in each allele of the MAT locus (Fig. 5). Most of these sequences represent decayed versions of long terminal repeats associated with a ubiquitous family of retrotransposable elements that inhabits the genome of C. neoformans (28). These include several copies of LTR11, LTR14, Cnirt3, and Cnirt4. In general, the position of these elements was not conserved with respect to the locus borders or neighboring genes, suggesting that these elements were recently acquired by each allele. There are three examples of particular interest. First, the region between the SXI1α and CAP1α genes in strain JEC21 differs from that in the serotype A strain H99 in which a Cnirt3 element has inserted and replaced intervening sequences. A second complete copy of the Cnirt3 element is also present between the STE3α and MFα4 genes. The two Cnirt3 elements are in a direct orientation, but because each is flanked by ∼50-bp inverted repeats, homologous recombination events could occur between the distal or internal ends of the two elements and transpose the intervening genes. A second interesting case is the serotype D a-specific gene NCP1a, which shares homology with a Neurospora crassa protein of unknown function but is missing sequences homologous to the N-terminal region and instead contains a fragment of the Cnirt4 transposable element. Finally, several different transposase-related genes were identified in the α alleles, implying that one or more copies of Tc1/mariner-type transposons were present in the locus and might have contributed to structural rearrangements during the evolution of the MAT locus. For example, local transposition of an inserted element could create inverted sequence repeats and promote inversions by homologous recombination.
The mating-type locus was also found to contain a surprising number of repetitive sequences, including simple sequence repeats. As shown in Fig. 5, all four alleles contained multiple copies of several different simple tri- and tetranucleotide repeats. A particularly notable example was 71 imperfect copies of a tetranucleotide repeat contained within the serotype D STE11a gene that were not present in any of the other STE11 genes. In addition, the pheromone precursor genes are often encoded by divergent pairs of genes that were embedded in regions that constitute large inverted repeats. This may give rise to unique mechanisms by which the genes are duplicated, rearranged, and lost as the alleles of the mating-type locus diverged from their common ancestors. For example, inversions between the identical MFα1 and MFα2 genes in the serotype D α allele would transpose the order and direction of the intervening genes (PRT1α, ZNF1α, RPL39α, and MFα3).
Recombination is suppressed in the mating-type region.
An important feature of mating type is stable inheritance as a single unit, and recombination is suppressed in these regions to avoid generation of sterile or self-fertile offspring. We used PCR analysis with α and a allele-specific primers to test whether the sequences we defined as the MAT locus faithfully cosegregate with mating type and whether recombination occurs in this locus (Fig. 6). Twenty-four progeny derived from two defined crosses between multiply marked strains were tested by PCR to test whether recombination occurred between the ends of the MAT locus. In addition, mating type was scored by genetic backcrosses. No recombination was observed in the mating-type region, and mating-type-specific sequences faithfully cosegregated with the corresponding mating types as determined by mating assays with tester strains. Forche and coworkers recently reported an amplified fragment length polymorphism-based physical map for C. neoformans and established the recombination frequency for the mating-type chromosome at ∼24 kb/centimorgan, demonstrating that recombination readily occurs elsewhere on this chromosome (24). In addition, the CNB1 gene resides on the mating-type chromosome but is completely unlinked to the MAT locus in genetic crosses (data not shown) (25), providing additional evidence for recombination events distal to MAT.
Sequences flanking the mating-type locus are nearly identical, but a few sequence polymorphisms between the α strain JEC21 and the a strain JEC20 are present immediately upstream and downstream of the MAT locus. We used primers designed for these sequences and the same set of meiotic progeny to test whether recombination occurred just outside of the borders of the MAT locus. No recombination events were observed, providing additional evidence for the integrity of the locus and its correct assignment (Fig. 6).
The α and a alleles of the MAT locus are stably inherited.
The α and a congenic pair of serotype D strains JEC21 and JEC20 was generated by a series of 10 backcrosses (Fig. 4) (31, 42). One concern was whether the mating-type alleles might have rearranged during the process of strain construction, possibly as a result of increased recombination during meiosis. Southern analysis was used to compare the genomic structure of the α and a alleles of strains JEC21 and JEC20 with those of their ancestors by using probes to sequences within and flanking the MAT locus (Fig. 4). No differences in restriction patterns were observed for any of the genes analyzed. Thus, the structure of the mating-type locus has been stably inherited through multiple generations, providing additional evidence that recombination is suppressed in this genomic region.
Structure of the serotype D MATα mating-type locus is conserved in nature.
The serotype D α and a strains JEC21 and JEC20 and their derivatives are widely used because of their congenic background and because of the ability to conduct classical genetic experiments with them (31). This was one of the major reasons why these strains were chosen to determine the structure of the mating-type locus in serotype D. However, an important issue is whether the structure of the mating-type locus of these lab strains is representative of unrelated serotype D strains. We addressed this by a PCR-based approach using primers that amplify overlapping fragments spanning the original ∼38-kb serotype D MATα mating-type locus proposed by Moore and Edman. Fragments of identical sizes were obtained with 9 of the 10 primer pairs using as templates DNA from the unrelated serotype D strains JEC21, CDC92-18, CDC92-27, and MMRL760 (Fig. 7). Only one primer combination produced a larger, ∼14-kb PCR product from strain MMRL760 (Fig. 7), compared with an ∼10-kb PCR product from JEC21 and the two CDC strains. Further PCR analysis revealed that an insertion of ∼4 kb had occurred between the STE11α and MFα1 genes of the MAT locus of this atypical yet still fertile strain (Fig. 7).
Sequence analysis of this region of the MAT locus of strain MMRL760 revealed that a novel mariner-related transposable element had inserted into the locus. Compared with that of strain JEC21, an additional 3,906 bp are present in the MAT locus of strain MMRL760, and this novel sequence is flanked by 136-bp inverted repeats that are identical at 135 of 136 positions. In addition, the element is inserted at a TA sequence and created a TA-TA duplication at the insertion site. The right half of this element encodes an open reading frame that might represent a transposase gene. Importantly, by comparison with the results of the ongoing genome project, this region of the element was found to share significant sequence identity with five distinct regions of the genome of serotype D strain JEC21. Curiously, the MAT locus of strain JEC21 contains a region of several hundred base pairs that shares identity with the left end of this element. Thus, this element may have either transposed into a remnant of itself, or strain JEC21 contains a fragment of the element as a result of a previous excision event.
Structural rearrangements during evolution and divergence of the MAT alleles.
Our findings reveal that the C. neoformans mating-type locus is significantly larger than previously suspected. In serotype D, the α allele spans ∼105 kb and the a allele spans ∼117 kb, whereas in serotype A, the α and a alleles span ∼103 and ∼127 kb, respectively. Thus, in both serotypes, the a allele is larger than the α allele. The number of genes identified within the locus ranges from 19 (MATa in serotypes A and D) to 23 (MATα in serotype A). While some of the genes encoded by the MAT locus have already been shown or predicted to function in the pheromone response pathway that regulates mating (Fig. 1, black arrows), other genes have no obvious function with respect to sexual development.
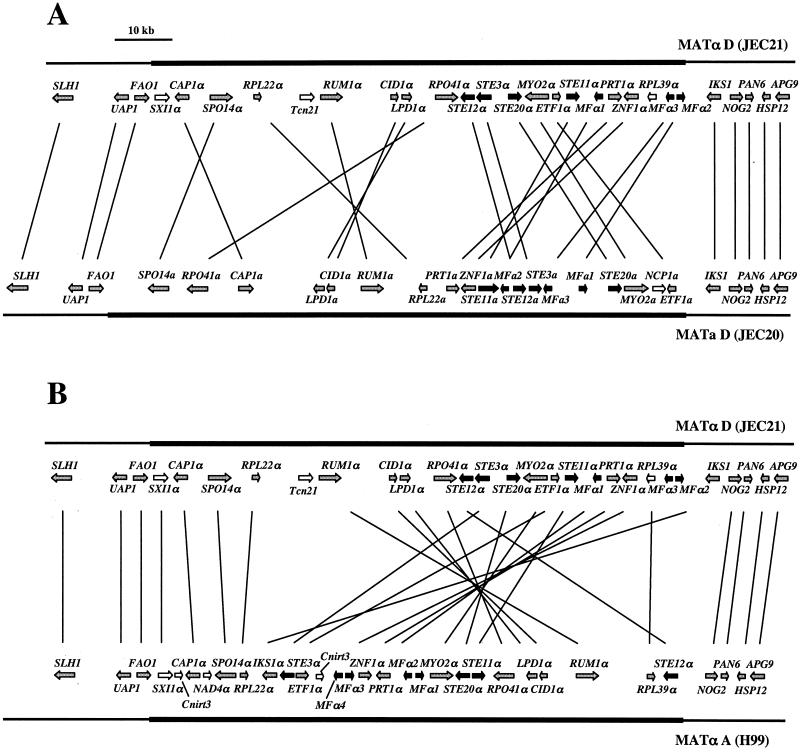
The gene order is strikingly different between different alleles of the mating-type locus. Genes outside the mating-type locus exhibit synteny in both serotype A and serotype D (Fig. 1 and 8A), whereas gene order inside the mating-type locus has been dramatically remodeled (Fig. 8A and data not shown). In addition, a few genes are present in either the α or the a mating-type allele but not in both, including SXI1α, RPL39α, and NCP1a in serotype D and SXI1α and NAD4α in serotype A (Fig. 1 and 8A) (Table 2). When the α or a mating-type alleles were compared between serotypes A and D, the rearrangement of the locus was even more striking (Fig. 8B). Furthermore, several genes identified are unique to the mating-type allele of only one serotype (Table 2). Interestingly, the IKS1 gene flanks the mating-type locus in serotype D but is located within the mating-type locus in serotype A, possibly as the result of a DNA inversion. With this exception, the order of the genes outside the MAT locus is conserved between the two mating types and varieties. Interestingly, the orders of the genes just inside the left ends of the MATα mating-type loci of serotype A strain H99 and serotype D strain JEC21 are similar, with the exception of one inversion (SPO14α) and two small insertions (Cnirt3 and NAD4α) (Fig. 8B).
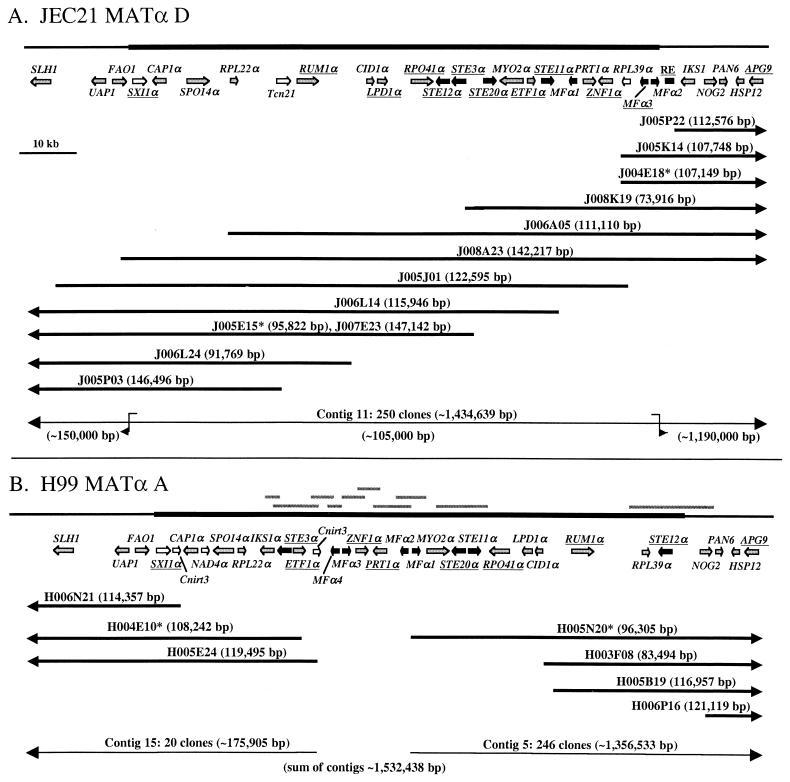
FIG. 8.
Structural comparison of the C. neoformans mating-type alleles. (A) Comparison of the α and a mating-type alleles of serotype D for analysis of the relative positions of the genes found within and adjacent to the MAT locus. Vertical and diagonal lines connect diverged gene alleles present in both alleles and illustrate substantial gene rearrangements within the MAT locus, whereas gene order outside the MAT locus has been conserved. In addition, a few genes that are present in only one of the two MAT alleles were identified (white arrows). (B) Comparison of the α mating-type alleles between serotypes A and D. Similar to what occurred in the α and a alleles from serotype D, significant rearrangements of gene order have occurred in the α allele during strain divergence (vertical and diagonal lines), and some genes are unique to one serotype (white arrows). Interestingly, the IKS1 gene flanks the MAT locus in serotype D but is located within the locus in serotype A. For additional details, see the legend to Fig. 1.
In summary, our findings reveal that the α and a mating-type alleles diverged from a common ancestral region of DNA by a process involving rearrangements, inversions, and nucleotide substitutions. Moreover, the α and a alleles have both undergone extensive rearrangements as the serotype A and serotype D strains evolved into varieties or even distinct species.
DISCUSSION
We have analyzed the structure of four mating-type alleles of the fungal pathogen C. neoformans, including the α and a alleles of the serotype A and D varieties grubii and neoformans. The MAT locus of C. neoformans is considerably larger than previously reported (35) and spans ∼105 to 130 kb. While the gene order outside the locus is largely conserved, even between the two serotypes, genes inside the mating-type locus have been subject to extensive rearrangements. This is true not only for the two opposite mating-type alleles in a given serotype but also for a single MAT allele compared between serotypes. In addition, a few genes were identified that are present in only one or the other allele. Recombination in the mating-type locus and the surrounding genomic region is suppressed, and the mating-type alleles are stable through multiple genetic crosses without rearrangement. The basic structure of the mating-type locus in C. neoformans is largely conserved within the population of this organism, but the detection of a transposable element in the α locus of an atypical serotype D strain reveals that genetic alterations can occur in the population.
When compared to those of other model ascomycetes or basidiomycetes, the mating-type locus of C. neoformans has a unique structure in terms of both size and gene composition. The MAT locus in most ascomycetes is limited in size and encodes transcription factors that determine mating type and cell identity. In basidiomycetes with tetrapolar mating systems, one mating-type locus resembles ascomycete mating-type loci in size and gene composition. The second locus encodes pheromone and pheromone receptor systems and can extend up to 20 kb via gene duplications. Only a few other fungal mating-type loci have been found to contain genes lacking an obvious function in mating (33, 54, 64). The mating-type locus of C. neoformans is the largest single-copy MAT locus known, and the locus contains a striking number of genes, including ones that function in mating and others with no predicted role in sexual differentiation.
In contrast to what occurs in other basidiomycete mating-type loci, the C. neoformans genes encoding pheromones (MFα1-3 and MFa1-3) and pheromone receptors (STE3α and STE3a) are not adjacent to each other but rather dispersed throughout the locus (Fig. 1). The MFα1-3 pheromone and STE3α pheromone receptor genes were identified in the previously published C. neoformans MAT locus (35), but no transcriptional regulators of the homeodomain or HMG domain family were previously known. In our studies, BLASTX analysis of the complete α and a mating-type sequences identified a gene close to the left end of the α mating-type alleles that exhibited weak similarity to other homeodomain transcription factors involved in mating and cell identity in other fungi (Fig. 1). As will be presented elsewhere, deletion analysis of the SXI1α (sex inducer 1 α) gene reveals a role for Sxi1α in sexual development (Hull et al., submitted). No SXI1α-related gene is present in either a allele studied, and no cross-hybridizing genes are present in a-specific DNA by Southern analysis (Hull et al., submitted). The identification of the mating-type-specific homeodomain transcription factor Sxi1α brings the mating-type system of C. neoformans closer to those of other basidiomycetes with respect to the main regulators involved in sexual development than previously suspected. Our findings reveal that both transcriptional regulators and a pheromone and pheromone receptor system are present in the C. neoformans MAT locus, but the arrangement of the locus is distinct compared to those of other model basidiomycetes in which the two regulators are unlinked.
In addition to these regulatory genes, ∼15 other genes were identified in the different mating-type alleles (Table 2). Some encode components of the pheromone response pathway that regulates mating, fruiting, or virulence of C. neoformans (14, 15, 35, 45-47, 49, 63, 69, 71, 73). Interestingly, a similar unusual cluster of genes that may be involved in pheromone signaling was recently reported in another opportunistic human fungal pathogen, Pneumocystis carinii (65). Genome sequencing revealed a locus that shares similarities with the mating-type locus of C. neoformans and contains genes encoding components of a putative pheromone response pathway (65). Whether this region represents a true mating-type locus of P. carinii is not known, and no sexual cycle has been described for this pathogenic fungus. The finding that the basidiomycete C. neoformans and the ascomycete P. carinii share similarly arranged mating-type loci raises the question of whether the MAT locus plays a role in the virulence of P. carinii, as has already been established for the MATα locus of C. neoformans.
Our studies reveal that three different types of mating-type loci exist in fungi. The first comprises the classical MAT loci of ascomycetes, in which mating type is determined by specialized transcription factors encoded by a single, compact locus. The second comprises the tetrapolar mating systems of the basidiomycetes, in which mating type is determined by two distinct, unlinked loci encoding transcriptional regulators and pheromone and pheromone receptor systems. The third is the novel mating-type locus in C. neoformans and a related region in P. carinii, in which mating-specific transcription factors, a pheromone and pheromone receptor system, and elements of the pheromone-activated MAP kinase cascade are part of a single, contiguous multigene locus. Because C. neoformans is a basidiomycete and P. carinii is an ascomycete, this class of MAT locus either evolved prior to the divergence of the two major fungal phyla or resulted from convergent evolution.
Unlike most model basidiomycetes but similar to C. neoformans, U. hordei has a bipolar mating system and recombination in the mating-type region is suppressed. Two opposite mating-type and pathogenicity alleles, MAT-1 and MAT-2, have been identified and have been found to span 500- and 460-kb regions, respectively (44). MAT-1 and MAT-2 include one locus encoding mating-type-specific transcription factors and a second locus containing tightly linked pheromone and receptor genes. Both loci reside on the same chromosome and are separated by 450 to 500 kb of intervening DNA in which recombination is suppressed. These findings explain at a molecular level how a tetrapolar mating system can be converted into a bipolar system by linking of the commonly found mating-type loci on a single chromosome and the involvement of mechanisms that suppress recombination across the intervening sequences (2, 44).
An interesting question is how recombination is suppressed across the MAT loci of C. neoformans and U. hordei. The sequence of the interval between the two loci of U. hordei is being determined and contains many repetitive sequences and transposable elements that may contribute to the suppression of recombination (J. Kronstad, personal communication). Our analysis of the C. neoformans mating-type alleles reveals two factors that may also play a role. First, mating-type-specific alleles of several genes vary from 5 to ∼50% in sequence (46), and some genes are unique to one or the other allele. Second, gene positions in the mating-type alleles are extensively rearranged (including inversions). For example, the RPO41α and RPO41a genes are almost identical (97%) but are oriented in opposite directions in serotype D. Thus, crossover events between these two alleles would result in one acentric and one dicentric chromosome, both of which would be unstable. These sequence and structural differences likely prevent proper alignment of this chromosomal region during meiosis and thereby suppress recombination.
Interestingly, for the ascomycete Neurospora tetrasperma, genetic and cytological studies have shown that during meiosis the chromosomes containing the mating-type locus are unpaired over a large interval that includes the MAT locus, and recombination is suppressed in this region. In addition, specific sites flanking this region trigger recombination events that may function to ensure proper chromosome segregation during meiosis (26, 48). These observations suggest that the chromosomes containing the fungal MAT loci share features with mammalian sex chromosomes.
The mating-type-determining region of C. neoformans shares features with both the self-incompatibility locus that governs pollen recognition in species of the crucifer plant Brassica (50) and the mating-type locus of the green alga Chlamydomonas reinhardtii. For example, the multiallelic S locus in Brassica spp. is composed of divergent and rearranged sequences linking the SRK and SCR genes involved in pollen-stigmata interactions (5). In Chlamydomonas, the mating-type locus is located in a region of ∼830 kb in which recombination is suppressed (23). In addition, a 190-kb core region thought to contain the mating-type determining factors is highly rearranged via several translocations, inversions, duplications, and deletions (21-23). These chromosomal aberrations are thought to be responsible for suppressing recombination in the core region and the flanking 640 kb of genomic DNA. The mating-type region in C. reinhardtii is located close to one end of linkage group VI (23). This is similar to C. neoformans because analysis from the ongoing genome project reveals that the MAT locus resides ∼170 kb from one telomere of this 1.8-Mb chromosome. Whether chromosomal location has any impact on the function of the mating-type loci in these organisms is not known, but it is interesting that the HML and HMR silent mating-type cassettes in S. cerevisiae are also located near the ends of yeast chromosome III.
Sex determination in higher eukaryotes is often accompanied by the presence of dimorphic sex chromosomes. An interesting model that explains the evolution of sex chromosomes is based on the initial requirement for genetic differences in multiple loci for the definition of sexual identity. Since the generation of self-fertile or sterile progeny is unfavorable, mechanisms had to evolve to ensure tight linkage between the genes involved (11, 12), possibly including the evolution of nonhomologous genes and chromosomal rearrangements. Once established, these mechanisms suppress the exchange of genetic material in these regions, and genetic divergence between genomic regions results in the evolution of a “diallelic” sex chromosome system. It has been proposed that animal sex chromosomes evolved from autosomes that were initially homologous except for a small sex-determining region (30). Following suppression of recombination in this region, subsequent divergence between the two “autosomes” resulted in the evolution of the sex chromosomes responsible for the hetero- (XY) and homogametic (XX) sexes in mammals (13, 43, 57, 58). The pseudoautosomal region on the mammalian Y chromosome may reflect its ancestral autosomal origin.
The ∼1.8-Mb mating-type chromosome in C. neoformans shares features with mammalian sex chromosomes. Although the sex-determining region comprises only ∼7% of this fungal chromosome, recombination is suppressed in the mating-type region but does occur in more distal regions of the chromosome. While recombination is suppressed between most of the sex chromosomes of mammals, recombination does occur in the pseudoautosomal region and is thought to be essential for proper chromosome segregation. Similar to mammalian sex chromosomes, the MAT locus of C. neoformans is characterized by nonhomologous genes and extensive rearrangements. In addition, the lack of genetic exchange favors the accumulation of repetitive sequences and transposable elements within the sex-determining region, which favors intrachromosomal rearrangements and drives divergence. The MAT loci in the green alga C. reinhardtii and the fungi N. tetrasperma, P. carinii, and U. hordei all share similar features with sex chromosomes. Since sex is thought to have originally evolved in lower eukaryotes, such as yeasts and algae, it is intriguing that the sex-determining systems of several unicellular eukaryotes share features resembling an early step in the evolutionary pathway to the dimorphic sex chromosomes of multicellular eukaryotes.
Acknowledgments
We thank Christina Hull, Robin Wharton, and John Perfect for advice and comments and Jim Kronstad for providing high-density BAC filter arrays and BAC data.
This study was supported by R01 grant AI50113 and P01 grant AI44975 (NIAID) to the Duke mycology research unit. Joseph Heitman is a Burroughs Welcome Scholar in molecular pathogenic mycology and an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakkeren, G., and J. W. Kronstad. 1994. Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc. Natl. Acad. Sci. USA 91:7085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beach, D. L., J. Thibodeaux, P. Maddox, E. Yeh, and K. Bloom. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10:1497-1506. [DOI] [PubMed] [Google Scholar]
- 4.Bölker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 5.Boyes, D. C., M. E. Nasrallah, J. Vrebalov, and J. B. Nasrallah. 1997. The self-incompatibility (S) haplotypes of Brassica contain highly divergent and rearranged sequences of ancient origin. Plant Cell 9:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
- 7.Casselton, L. A., and N. S. Olesnicky. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 2001. The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. Proc. Nat. Acad. Sci. USA 98:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. C., B. L. Wickes, G. F. Miller, L. A. Penoyer, and K. J. Kwon-Chung. 2000. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J. Exp. Med. 191:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlesworth, B. 1991. The evolution of sex chromosomes. Science 251:1030-1033. [DOI] [PubMed] [Google Scholar]
- 12.Charlesworth, B. 1994. Evolutionary genetics. The nature and origin of mating types. Curr. Biol. 4:739-741. [DOI] [PubMed] [Google Scholar]
- 13.Charlesworth, B. 1978. Model for evolution of Y chromosomes and dosage compensation. Proc. Natl. Acad. Sci. USA 75:5618-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung, S., M. Karos, Y. C. Chang, J. Lukszo, B. L. Wickes, and K. J. Kwon-Chung. 2002. Molecular analysis of CPRα, a MATα-specific pheromone receptor gene of Cryptococcus neoformans. Eukaryot. Cell 1:432-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke, D. L., G. L. Woodlee, C. M. McClelland, T. S. Seymour, and B. L. Wickes. 2001. The Cryptococcus neoformans STE11α gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol. Microbiol. 40:200-213. [DOI] [PubMed] [Google Scholar]
- 16.Coppin, E., R. Debuchy, S. Arnaise, and M. Picard. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61:411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dee, J. 1982. Genetics of Physarum polycephalum, p. 211-251. In H. C. Aldrich and J. W. Daniel (ed.), Cell biology of Physarum and Didymium, vol. 1. Academic Press, New York, N.Y.
- 18.Dzelzkalns, V. A., J. B. Nasrallah, and M. E. Nasrallah. 1992. Cell-cell communication in plants: self-incompatibility in flower development. Dev. Biol. 153:70-82. [DOI] [PubMed] [Google Scholar]
- 19.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 20.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 21.Ferris, P. J., E. V. Armbrust, and U. W. Goodenough. 2002. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160:181-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferris, P. J., and U. W. Goodenough. 1997. Mating type in chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferris, P. J., and U. W. Goodenough. 1994. The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76:1135-1145. [DOI] [PubMed] [Google Scholar]
- 24.Forche, A., J. Xu, R. Vilgalys, and T. G. Mitchell. 2000. Development and characterization of a genetic linkage map of Cryptococcus neoformans var. neoformans using amplified fragment length polymorphisms and other markers. Fungal Genet. Biol. 31:189-203. [DOI] [PubMed] [Google Scholar]
- 25.Fox, D. S., M. C. Cruz, R. A. L. Sia, H. Ke, G. M. Cox, M. E. Cardenas, and J. Heitman. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol. Microbiol. 39:835-849. [DOI] [PubMed] [Google Scholar]
- 26.Gallegos, A., D. J. Jacobson, N. B. Raju, M. P. Skupski, and D. O. Natvig. 2000. Suppressed recombination and a pairing anomaly on the mating-type chromosome of Neurospora tetrasperma. Genetics 154:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillissen, B., J. Borgemann, C. Sandmann, B. Schroeer, M. Bolker, and R. Kahmann. 1992. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68:647-657. [DOI] [PubMed] [Google Scholar]
- 28.Goodwin, T. J. D., and R. T. M. Poulter. 2001. The diversity of retrotransposons in the yeast Cryptococcus neoformans. Yeast 18:865-880. [DOI] [PubMed] [Google Scholar]
- 29.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 30.Graves, J. A., and J. W. Foster. 1994. Evolution of mammalian sex chromosomes and sex-determining genes. Int. Rev. Cytol. 154:191-259. [DOI] [PubMed] [Google Scholar]
- 31.Heitman, J., B. Allen, J. A. Alspaugh, and K. J. Kwon-Chung. 1999. On the origins of the congenic MATα and MATa strains of pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28:1-5. [DOI] [PubMed] [Google Scholar]
- 32.Herskowitz, I. 1989. A regulatory hierarchy for cell specialization in yeast. Nature 342:749-757. [DOI] [PubMed] [Google Scholar]
- 33.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, A. D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5:552-558. [DOI] [PubMed] [Google Scholar]
- 35.Karos, M., Y. C. Chang, C. M. McClelland, D. L. Clarke, J. Fu, B. L. Wickes, and K. J. Kwon-Chung. 2000. Mapping of the Cryptococcus neoformans MATα locus: presence of mating type-specific mitogen-activated protein kinase cascade homologs. J. Bacteriol. 182:6222-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karpova, T. S., S. L. Reck-Peterson, N. B. Elkind, M. S. Mooseker, P. J. Novick, and J. A. Cooper. 2000. Role of actin and Myo2p in polarized secretion and growth of Saccharomyces cerevisiae. Mol. Biol. Cell 11:1727-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kothe, E. 2001. Mating-type genes for basidiomycete strain improvement in mushroom farming. Appl. Microbiol. Biotechnol. 56:602-612. [DOI] [PubMed] [Google Scholar]
- 38.Kothe, E. 1996. Tetrapolar fungal mating types: sexes by the thousands. FEMS Microbiol. Rev. 18:65-87. [DOI] [PubMed] [Google Scholar]
- 39.Kronstad, J. W., and S. A. Leong. 1990. The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev. 4:1384-1395. [DOI] [PubMed] [Google Scholar]
- 40.Kronstad, J. W., and C. Staben. 1997. Mating type in filamentous fungi. Annu. Rev. Genet. 31:245-276. [DOI] [PubMed] [Google Scholar]
- 41.Kües, U., W. V. J. Richardson, A. M. Tymon, E. S. Mutasa, B. Gottgens, S. Gaubatz, A. Gregoriades, and L. A. Casselton. 1992. The combination of dissimilar alleles of the Aα and Aβ gene complexes, whose proteins contain homeo domain motifs, determines sexual development in the mushroom Coprinus cinereus. Genes Dev. 6:568-577. [DOI] [PubMed] [Google Scholar]
- 42.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lahn, B. T., and D. C. Page. 1999. Four evolutionary strata on the human X chromosome. Science 286:964-967. [DOI] [PubMed] [Google Scholar]
- 44.Lee, N., G. Bakkeren, K. Wong, J. E. Sherwood, and J. W. Kronstad. 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA 96:15026-15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lengeler, K. B., P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 2000. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc. Natl. Acad. Sci. USA 97:14455-14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClelland, C. M., J. Fu, G. L. Woodlee, T. S. Seymour, and B. L. Wickes. 2002. Isolation and characterization of the Cryptococcus neoformans MATa pheromone gene. Genetics 160:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merino, S. T., M. A. Nelson, D. J. Jacobson, and D. O. Natvig. 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore, T. D. E., and J. C. Edman. 1993. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nasrallah, J. B. 2002. Recognition and rejection of self in plant reproduction. Science 296:305-308. [DOI] [PubMed] [Google Scholar]
- 51.Oefner, P. J., S. P. Hunicke-Smith, L. Chiang, F. Dietrich, J. Mulligan, and R. W. Davis. 1996. Efficient random subcloning of DNA sheared in a recirculating point-sink flow system. Nucleic Acids Res. 24:3879-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Shea, S. F., P. T. Chaure, J. R. Halsall, N. S. Olesnicky, A. Leibbrandt, I. F. Connerton, and L. A. Casselton. 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pardo, E. H., S. F. O'Shea, and L. A. Casselton. 1996. Multiple versions of the A mating type locus of Coprinus cinereus are generated by three paralogous pairs of multiallelic homeobox genes. Genetics 144:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pöggeler, S. 2001. Mating-type genes for classical strain improvements of ascomycetes. Appl. Microbiol. Biotechnol. 56:589-601. [DOI] [PubMed] [Google Scholar]
- 55.Quadbeck-Seeger, C., G. Wanner, S. Huber, R. Kahmann, and J. Kamper. 2000. A protein with similarity to the human retinoblastoma binding protein 2 acts specifically as a repressor for genes regulated by the b mating type locus in Ustilago maydis. Mol. Microbiol. 38:154-166. [DOI] [PubMed] [Google Scholar]
- 56.Raper, J. R. 1966. Genetic sexuality of higher fungi. Ronald Press, New York, N.Y.
- 57.Rice, W. R. 1994. Degeneration of a nonrecombining chromosome. Science 263:230-232. [DOI] [PubMed] [Google Scholar]
- 58.Rice, W. R. 1987. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 116:161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rudge, S. A., A. J. Morris, and J. Engebrecht. 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rudge, S. A., T. R. Pettitt, C. Zhou, M. J. Wakelam, and J. A. Engebrecht. 2001. SPO14 separation-of-function mutations define unique roles for phospholipase D in secretion and cellular differentiation in Saccharomyces cerevisiae. Genetics 158:1431-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schein, J. E., K. L. Tangen, R. Chiu, H. Shin, K. B. Lengeler, W. K. MacDonald, I. Bosdet, J. Heitman, S. J. M. Jones, M. A. Marra, and J. W. Kronstad. 2002. Physical maps for genome analysis of serotype A and D strains of the fungal pathogen Cryptococcus neoformans. Genome Res. 12:1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schafer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 63.Shen, W.-C., R. C. Davidson, G. M. Cox, and J. Heitman. 2002. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot. Cell 1:366-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh, G., and A. M. Ashby. 1998. Cloning of the mating type loci from Pyrenopeziza brassicae reveals the presence of a novel mating type gene within a discomycete MAT 1-2 locus encoding a putative metallothionein-like protein. Mol. Microbiol. 30:799-806. [DOI] [PubMed] [Google Scholar]
- 65.Smulian, A. G., T. Sesterhenn, R. Tanaka, and M. T. Cushion. 2001. The ste3 pheromone receptor gene of Pneumocystis carinii is surrounded by a cluster of signal transduction genes. Genetics 157:991-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Specht, C. A., M. M. Stankis, L. Giasson, C. P. Novotny, and R. C. Ullrich. 1992. Functional analysis of the homeodomain-related proteins of the Aα locus of Schizophyllum commune. Proc. Natl. Acad. Sci. USA 89:7174-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stankis, M. M., C. A. Specht, H. Yang, L. Giasson, R. C. Ullrich, and C. P. Novotny. 1992. The Aα mating locus of Schizophyllum commune encodes two dissimilar multiallelic homeodomain proteins. Proc. Natl. Acad. Sci. USA 89:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaillancourt, L. J., M. Raudaskoski, C. A. Specht, and C. A. Raper. 1997. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics 146:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, P., C. B. Nichols, K. B. Lengeler, M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2002. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot. Cell 1:257-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wendland, J., L. J. Vaillancourt, J. Hegner, K. B. Lengeler, K. J. Laddison, C. A. Raper, and E. Kothe. 1995. The mating-type locus Bα1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 14:5271-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wickes, B. L., U. Edman, and J. C. Edman. 1997. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol. Microbiol. 26:951-960. [DOI] [PubMed] [Google Scholar]
- 72.Yin, H., D. Pruyne, T. C. Huffaker, and A. Bretscher. 2000. Myosin V orientates the mitotic spindle in yeast. Nature 406:1013-1015. [DOI] [PubMed] [Google Scholar]
- 73.Yue, C., L. M. Cavallo, J. A. Alspaugh, P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 1999. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 153:1601-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]