Abstract
Infectivity of Anaplasma spp. develops when infected ticks feed on a mammalian host (transmission feed). Specific Anaplasma marginale major surface protein 2 (MSP2) variants are selected for within the tick and are expressed within the salivary glands. The aims of this study were to determine when and where MSP2 variant selection occurs in the tick, how MSP2 expression is regulated in salivary glands of transmission-feeding ticks, and whether the number of A. marginale organisms per salivary gland is significantly increased during transmission feeding. The South Idaho strain of A. marginale was used, as MSP2 expression is restricted to two variants, SGV1 and SGV2, in Dermacentor andersoni. Using Western blot, real-time PCR, and DNA sequencing analyses it was shown that restriction and expression of MSP2 occurs early in the midgut within the first 48 h of the blood meal, when ticks acquire infection. A. marginale is present in the tick salivary glands before transmission feeding is initiated, but the msp2 mRNA and MSP2 protein levels per A. marginale organism increase only minimally and transiently in salivary glands of transmission-feeding ticks compared to that of unfed ticks. A. marginale numbers per tick increase gradually in salivary glands of both transmission-fed and unfed ticks. It is concluded that MSP2 variant selection is an early event in the tick and that MSP2 variants SGV1 and SGV2 are expressed both in the midgut and salivary glands. While MSP2 may be required for infectivity, there is no strict temporal correlation between MSP2 expression and the development of infectivity.
Ixodid ticks transmit important pathogens of animals and humans, including bacteria of the genera Anaplasma, Borrelia, Ehrlichia, and Rickettsia. In contrast to other arthropod vectors of microbial pathogens, contact between ixodid ticks and mammalian hosts is long, lasting several days, and requires adaptation of tick organs to modify the interface with the host and form the feeding lesion (32). Tick-transmitted pathogens are taken into the tick midgut together with the blood meal (acquisition feed). Invasion of midgut cells is followed by pathogen replication and colonization of most tick tissues, including the salivary gland, presumably via the hemocoel (5, 20, 26). Pathogens replicate during the subsequent transmission feed in the tick salivary gland and midgut, and their infectivity develops and increases while the tick feeds (10, 18). This development and increase in infectivity may reflect replication leading to a higher pathogen titer or may be due to unique or enhanced expression of specific pathogen molecules. For example, Borrelia hermsii expresses serotype 33 of the variable major protein only in ticks, and expression of outer surface protein C (OspC) of Borrelia burgdorferi is transcriptionally upregulated in transmission-feeding ticks (8, 25, 29). OspC has been implicated in facilitating migration of B. burgdorferi from the tick midgut to the tick salivary gland (13).
Key determinants of rickettsial pathogen transmission by ixodid ticks are unknown. Anaplasma phagocytophila and A. marginale (order Rickettsiales, family Anaplasmataceae) are not transmitted when homogenates of infected unfed ticks are inoculated into sheep and cattle, respectively. Infection is only transmitted after the ticks are allowed to feed. Furthermore, infectivity increases, as reflected by decreased prepatent periods, as the transmission feeding period is extended (14, 17, 22). The only rickettsial protein known to be differentially expressed in the bloodstream of the mammalian host and the salivary gland of ticks is the major surface protein 2 (MSP2) of A. marginale. A. marginale MSP2 variants are sequentially generated during persistent infection in cattle by recombination of pseudogenes into an operon-linked expression site (1, 3). As a result, multiple MSP2 variants can be identified in the bloodstream at any given time point (11). However, during development within the tick, new MSP2 variants are selected for and are expressed (7, 28). What selects for or against specific MSP2 variants in the tick and how this selection is regulated are unknown.
In the research presented here we use the South Idaho strain of A. marginale, which expresses a restricted set of two MSP2 variants, SGV1 and SGV2, in the salivary gland of Dermacentor andersoni ticks, to address three questions. First, in which tissue of the tick and at what time point during tick infection does the switch to the tick-specific MSP2 variants SGV1 and SGV2 occur? Second, are MSP2 variants SGV1 and SGV2 transcribed from a second expression site, distinct from the operon? And lastly, is enhanced MSP2 expression or an increase in the number of A. marginale organisms in the tick salivary gland during transmission feeding associated with the development of increased infectivity?
MATERIALS AND METHODS
Tick infection and feeding.
The South Idaho strain of A. marginale and laboratory-reared adult male ticks of the Reynolds Creek stock of D. andersoni were used (10, 27). Nine male Holstein calves (designated 809, 814, 839, 847, 854, 855, 857, 873, and 882) were confirmed to be free of A. marginale by competitive inhibition-enzyme-linked immunosorbent assay for MSP5 (16). Four of the five calves used for acquisition feeds (809, 839, 854, and 873) were inoculated intravenously with approximately 109 infected erythrocytes of the South Idaho strain of A. marginale (27). The fifth calf (847) was infected by tick transmission of the South Idaho strain of A. marginale by using D. andersoni that had been infected by feeding on calf 839 in order to obtain ticks infected under natural conditions. For acquisition feeding, approximately 400 uninfected D. andersoni ticks were placed under a tightly sealed skin patch over the flank or on the back of each calf and allowed to feed for 7 to 8 days during the period of peak rickettsemia (≥109 infected red blood cells/ml). This was done to maximize the infection rate of ticks (10). Ticks were removed and incubated at 26°C and 90 to 98% humidity for 7 days to allow clearance of the blood meal from midguts. Ticks were allocated to two groups and either were transmission fed on uninfected calves (814, 847, 855, 857, and 882) or, as an unfed control, remained at 26°C and 90 to 98% humidity. Ticks acquisition fed on calves 809, 839, and 847 were collected during the transmission feed on calves 814, 847, and 855, respectively, at 6, 18, 42, and 72 h and, in case of calf 839, also on day 8 of the acquisition feed and days 3 and 7 of the holding period. Samples from these ticks were used to quantify msp5 DNA and msp2 transcript and MSP2 protein levels (see below). In feeding experiments using calves 854 and 873 for acquisition feeding and calves 857 and 882 for transmission feeding, ticks were collected during the acquisition feed (day 0, i.e., before attachment, and days 1 through 7), holding period (days 3 and 7), and transmission feed (day 3). Samples from these ticks were used for sequence analysis (see below).
Tick and blood samples.
Blood was collected in EDTA from all calves at the beginning, day 3, and end of the tick acquisition feed. Ticks, collected at the time points described above, were dissected, and salivary glands and midguts were isolated. Salivary gland pairs and midguts were pooled to obtain DNA, RNA, and protein from 10 ticks each per time point. For genomic DNA, tick salivary glands and midguts and cell pellets from blood samples were collected in cell lysis buffer and digested with proteinase K overnight at 37°C followed by incubation at 65°C for 2 h, and DNA was isolated with the Puregene DNA isolation kit (Gentra Systems). Total RNA was extracted from tick salivary glands and midguts using TRIzol (BRL) after sample collection in RNAlater (Ambion). DNA and RNA pellets were reconstituted in 50 μ l of nuclease-free water. To obtain protein samples, tick salivary glands and midguts and erythrocyte pellets were collected and sonicated in proteinase inhibition buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 5 mM iodoacetamide, 0.1 mM Na-p-tosyl-l-lysine chloromethyl ketone, and 1 mM phenylmethylsulfonyl fluoride) and mixed with sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis buffer (16).
Identifications of msp2 variants from blood and tick stages.
MSP2 variants are expressed as part of an operon (Fig. 1A) in blood stages of at least three A. marginale strains and in tick stages of the Oklahoma strain (1, 2). The msp2 gene present in the operon expression site is the only full-length copy of the gene in blood stages (3). DNA isolated from ticks and blood was amplified with primers specific for full-length msp2 (3) and spanning the hypervariable region of MSP2 (Fig. 1A), MSP2:184-201 (5′ TTC GGC AGC ATC AAG GAC 3′) and MSP2:891-874 (5′ CAT TAC AGA AGT AGA CCC 3′). To specifically amplify operon-linked msp2, primer ORF2:261-282 (5′ GGG GAA AAG ACG CTT GGT AGG 3′), located at the 3′ end of orf2, was used as the forward primer in combination with MSP2:891-874 as the reverse primer (Fig. 1A). Cycling parameters were melting at 95°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 45 s, for 35 cycles. Amplification products were detected by electrophoresis in 1.5% agarose gels containing ethidium bromide. PCR products were gel purified with the Promega PCR purification kit, blunt ended with Pfu polymerase (Stratagene), and cloned with the Zero-blunt TOPO PCR cloning kit for sequencing (Invitrogen). Sequences were obtained from 90 clones from tick salivary glands and midguts and 20 clones from blood samples with the Big Dye kit and an ABI PRISM automated sequencer (PE-Applied Biosystems) and were compiled by using the VECTOR NTI software package (InforMAX).
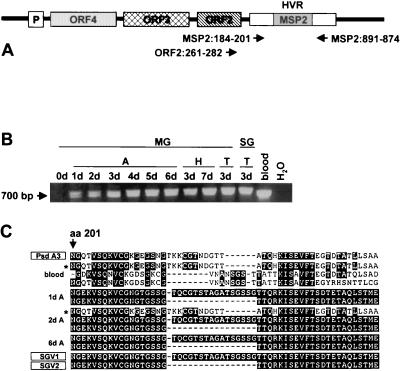
FIG. 1.
MSP2 variants in the blood and tick organs during acquisition and transmission feeding of D. andersoni. (A) Representation of the msp2 operon indicating the expression site structure and primer pair positions. HVR indicates the location of the hypervariable region of msp2 (11); P indicates the location of the operon promoter. Numbering of nucleotides is based on ORF2 and the pCKR11.2 msp2 sequences (24). (B) PCR analysis for full-length msp2 in ticks. Ticks were acquisition fed (A) on calf 873, held at 26°C (H), and transmission fed (T) on calf 882. Midguts (MG) were collected before ticks were attached (0d), on days 1 through 6 during the acquisition feeding (1d through 6d), days 3 and 7 (3d and 7d) of the holding period, and day 3 (3d) of the transmission feeding. Salivary glands (SG) were obtained on day 3 of the transmission feeding. Blood (blood) was collected from calf 873 on day three during the acquisition feeding. The reactions were carried out with primer pair MSP2:184-201 and MSP2:891-874. A reaction mixture without template served as negative control (H2O). Amplicons were identified by agarose gel electrophoresis and ethidium bromide staining. The position of the 700-bp fragment is indicated in the left margin. (C) Amino acid sequence alignment of MSP2 expressed in the blood of calf 873 during acquisition feeding of the ticks and in midgut samples from days 1 (1d A), 2 (2d A), and 6 (6d A) of the acquisition feeding. Only one sequence of each type obtained by cloning at each time point is shown. Sequences are compared to the published sequences of the tick-specific MSP2 variants SGV1 and SGV2 (27) and to the sequence of pseudogene A3 (Psd A3) (3). The first depicted amino acid corresponds to amino acid 201 (aa 201) of full-length MSP2 (24). The asterisk designates corresponding clones obtained from the tick midgut and blood sample.
Real-time PCR for msp5 DNA and msp2, msp5, and orf4 RNA.
To quantify organism numbers, real-time PCR for the single-copy gene msp5 (33) was performed. To control for RNA quality when performing quantitation of msp2 transcripts, msp5 and orf4 transcript levels were determined. msp5 was used because it is expressed at very low levels. orf4 was used because it is located at the 5′ end of the polycistronic RNA transcribed from the msp2 operon and because endonucleolytic degradation of prokaryotic mRNA proceeds 5′ to 3′ (21). The cDNA copy numbers determined by real-time PCR for orf4 were within a range of 1.5 times less than that of cDNA copies amplified with msp2-specific primers, suggesting partial degradation of transcripts. Quantification of msp5 DNA and msp2, msp5, and orf4 transcripts was done in triplicate on samples obtained from the transmission-fed ticks and the unfed control ticks. Just prior to use in reverse transcription, RNA samples were treated twice with RNase-free DNase (Ambion) followed by heat inactivation at 65°C and addition of RNase inhibitor (Boehringer Mannheim). Reverse transcription was carried out with the first-strand synthesis kit (Boehringer Mannheim) and random hexamers. Primers utilized in real-time PCRs were designed with the Primer Express software package (Perkin Elmer). Primers used for amplification of msp5 were MSP5:529-549 (5′ CTC ACA GGC GAA GAA GCA GAC 3′) and MSP5:667-649 (5′ GCC CGA CAT ACC TGC CTT T 3′); primers used for amplification of msp2 were MSP2:114-132 (5′ AGG CGG CGA GGG TCT ATT T 3′) and MSP2:260-241 (5′ GCA TCT CGC TTG TAC GGG AA 3′); and primers used for amplification of orf4 were OFR4:312-331 (5′ TGC AGC TCA GCG CTT ATA GC 3′) and ORF4:447-428 (5′ CCA ATG TGA TAC TCG GCC GT 3′). Real-time PCR was performed with the ABI PRISM 5700 Sequence Detection System using MicroAmp Optical 96-Well reaction plates and SYBR Green PCR Master Mix (all from Perkin Elmer). Primer optimization reactions were done using all possible combinations of the concentrations 50, 300, and 500 nM for the forward and reverse primers of a pair at three different MgCl concentrations (2, 5, and 8 mM). Cycling parameters for all runs were as follows: hold 1, 50°C, 2 min (AmpErase UNG incubation), and hold-2, 95°C, 10 min (AmpliTaq Gold activation) followed by 55 cycles of melting at 95°C, 15 s; annealing at 60°C, 15 s; and extension at 72°C, 15 s. This was followed by gradual melting of the amplification products in one-degree increments from 60°C to 90°C to generate a melting curve for evaluation of product specificity. Sensitivity and efficiency of the PCR assays were determined using serial, 10-fold dilutions of a pBS vector containing full-length msp2, a TOPOpBAD vector containing full-length msp5, and a TOPO-TA vector containing full-length orf4, the short intergenic sequence and the 5′ end of orf3 (1, 4, 24). Efficiency of real-time PCRs was calculated from standard curves obtained by amplification of 103, 2 × 103, 5 × 103, 104, and 2 × 104 copies of the individual plasmid vectors (15). Specificity was confirmed by a single, narrow peak in melting curve analyses and a single band at the expected size in 1.5% agarose gel analyses of the amplification products. Efficiency of reverse transcription was determined by using an msp2 RNA competitor. The competitor template was constructed by using an upstream primer with three regions: a central target gene binding region with the sequence 5′ GTG TTG ATG GCT CTG GCT GC 3′ (nucleotides 40 to 59) flanked upstream by the core T7 promoter and a GC clamp and downstream by a second target gene binding site, 5′ AGC ATT CGG CAG CAT CAA GGA 3′ (nucleotides 181 to 200). The use of two target binding sites introduced a 121-bp deletion in the 5′ region of the competitor and allowed for the differentiation of the amplification products of the (shorter) competitor and target. Primer MSP2:526-548 (5′ CTC TTA GTC ATC TTA CCT AAA GC 3′) was used as the downstream primer to amplify the 388-bp competitor template. Transcription into the RNA competitor was done with the reverse transcription-PCR competitor construction kit (Ambion, Inc.) and the upstream primer MSP2:40-59 (5′ GTG TTG ATG GCT CTG GCT GC 3′). The RNA competitor was purified, quantified, and stored at −20°C until used. Efficiency of reverse transcription was determined to be ∼1% by using an msp2 RNA competitor in known quantities followed by quantification of cDNA by real-time PCR. Generally, less than 20% of mRNA molecules are converted to cDNA (Ambion technical notes). Quantities given for cDNA copies are therefore corrected 100 times to a more closely reflected msp2 transcript level. The msp2 cDNA copies per A. marginale organism were calculated by dividing the number of msp2 cDNA copies per tick by the number of msp5 DNA copies per tick.
Western blot analysis for MSP2 and MSP5.
Electrophoresis of protein samples from blood, midguts, and salivary glands was carried out on SDS-containing 12% polyacrylamide slab gels with a 3.9% stacking gel for 7 to 11 h at 20 to 50 mA. Following transfer to nitrocellulose, membranes were blocked in phosphate-buffered saline-0.1% Tween 20 containing 5% milk. MSP2 was detected by using polyclonal rabbit serum R883 generated against native purified MSP2, and MSP5 was detected by using the monoclonal antibody ANAF16C1 (16, 24). Peroxidase-labeled goat anti-rabbit and anti-mouse antibodies (Cappel) were added, and bound antibody was detected by using enhanced chemiluminescence (Amersham International). Salivary glands and midguts from uninfected ticks and uninfected erythrocytes were handled identically and served as negative controls.
Statistics.
DNA copies and cDNA levels were compared between groups and time points using a two-way analysis of variance after ranking the data by using the NCSS 2000 software package. A P value of <0.05 was defined as significant.
RESULTS
Operon-linked msp2 variants in blood and tick stages.
Multiple MSP2 variants are expressed from the operon-linked expression site during acute rickettsemia and in each of the rickettsemic cycles that characterize persistent infection (1, 3, 11). Analysis of the blood of calf 873 at the time of D. andersoni acquisition feeding revealed the expected presence of multiple MSP2 variants (Fig. 1B and C). PCR using primers specific for full-length msp2 followed by cloning and sequencing of PCR products showed that none of the 10 clones from the blood of calf 873 were SGV1 or SGV2. In contrast, sequencing of amplicons from midguts of ticks acquisition fed on calf 873 for 1, 2, and 6 days identified both SGV1 and SGV2 (Fig. 1C). The MSP2 population in the tick midgut at early time points was not limited to SGV1 and SGV2. On day 2 a MSP2 variant previously detected in the blood was present among the sequenced clones (Fig. 1C; sequence is designated with an asterisk). Clone 1 was identical to a previously described MSP2 variant, designated A3, that was shown to be expressed in blood during acute infection with the South Idaho strain of A. marginale (3). The experiment was repeated using calf 854, and the same pattern of restriction from multiple MSP2 variants in the blood to SGV1 and SGV2 in the tick midgut was observed (data not shown).
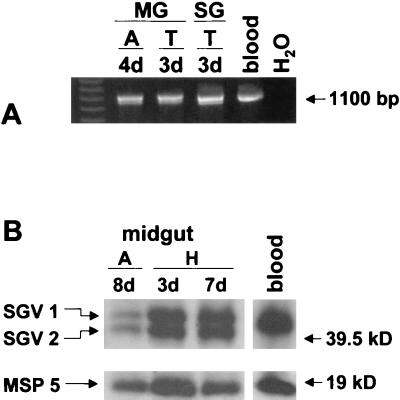
To determine if MSP2 variants SGV1 and SGV2 are present in the operon-linked expression site, PCR was performed using a primer pair spanning from the 3′ end of orf2 past the hypervariable region of msp2 (Fig. 1A). Amplification resulted in fragments of the expected size (Fig. 2A) (27), and the identity of SGV1 and SGV2 was confirmed by sequence analysis in tick midgut and salivary gland samples obtained during the acquisition feeding on calf 873 and transmission feeding on calf 882. The experiment was repeated using ticks acquisition fed on calf 854 and transmission fed on calf 857. Operon-linked SGV1 and SGV2 was identified in all but 1 of 45 clones obtained from the tick midgut samples and from all 45 clones derived from salivary gland samples. Of the 45 clones derived from midgut samples, 27 (60%) were SGV1 and 17 (37.8%) were SGV2. The remaining clone was A3, as described above. Of the 45 clones derived from salivary glands, 33 were SGV1 (73.3%) and 12 were SGV2 (26.7%). To confirm expression, MSP2 protein was detected by using Western blots of midguts from ticks infected by feeding on calf 873. Two distinct MSP2 proteins were detected during the acquisition feeding and in the holding period when the ticks were incubated (Fig. 2B). These two proteins have the predicted molecular size for SGV1 and SGV2, with SGV2 being approximately 2 kDa smaller due to a 16-amino-acid deletion (27). The experiment was repeated using midgut and blood samples from the acquisition feeding on calf 854, and the same results were obtained (data not shown).
FIG. 2.
Expression of MSP2 variants SGV1 and SGV2 from the operon site. (A) PCR analysis for operon-linked msp2 performed on midguts (MG) and salivary glands (SG) obtained at day 4 of the acquisition feeding (A) on calf 873 and day 3 of the transmission feeding (T) on calf 882. Blood was collected from calf 873 on day 3 during the acquisition feeding (blood). Reactions were carried out using the primer pair ORF2:261-282 and MSP2:891-874. A reaction mixture without template served as negative control (H2O). Amplicons were electrophoresed using agarose gels and were stained with ethidium bromide. The position of the 1,100-bp fragment is indicated in the right margin. Amplicons from tick salivary glands and midguts were cloned and sequenced and confirmed to be SGV1 and SGV2 (data not shown). (B) Western blot analysis of midgut samples obtained from ticks acquisition fed for 8 days on calf 839 (A) and held for 3 or 7 days at 26°C (H). MSP2 was detected with a polyclonal serum, R883, against purified native MSP2 (24). MSP5 was used to control for equal loading of organism numbers, and MSP5 protein was detected by using monoclonal antibody ANAF16C1 (16). The positions of SGV1, SGV2, and MSP5 are indicated in the left margin, and molecular size markers are indicated in the right margin.
Quantitation of msp2 transcript and protein expression.
A quantitative real-time PCR assay was developed to measure msp2 transcript levels in tick salivary glands and midguts. Sensitivity of the real-time PCR assay was 2.3 copies (10 attograms) of plasmid DNA. Efficiency of msp2 real-time PCR was consistently between 0.91 and 0.98 (15). Ticks were acquisition fed on calf 839 and transmission fed on calf 847, and salivary gland samples were collected at 0, 6, 18, 42, and 72 h of the transmission feeding. The msp2 cDNA levels per salivary gland pair were determined by using a standard curve generated with serial plasmid dilutions (Fig. 3). msp2 cDNA levels increased from 105 copies per salivary gland pair during the acquisition feed to ≥108 copies per salivary gland pair during transmission feeding. The increase in salivary glands of transmission-fed ticks was paralleled by increases in unfed ticks (Table 1). Only at 18 h are msp2 cDNA copy numbers clearly higher in salivary glands of transmission-fed ticks than in unfed controls. When msp2 cDNA copies per A. marginale are analyzed, levels remain relatively constant throughout the duration of the transmission feeding and the difference between fed and unfed ticks at 18 h is nearly eliminated (data not shown). The higher expression of msp2 transcripts in transmission-fed ticks at 18 h correlates with increased MSP2 protein expression in this group (Fig. 4). The MSP2 to MSP5 ratio obtained by densitometry for salivary glands samples was 11.6 for fed ticks and 9.6 for unfed ticks, a 1.2-fold difference. At no other time point is MSP2 protein expression in transmission-fed ticks different from that in unfed ticks. The apparent increase in MSP2 protein levels in salivary glands of transmission-fed ticks at 42 h reflects higher antigen load, as inferred from higher levels of MSP5 protein in the same lane compared to MSP5 levels at the same time point in unfed ticks (Fig. 4) and the same densitometric ratio of MSP2 to MSP5 for both lanes (data not shown). The msp2 cDNA copy numbers and MSP2 protein quantities are depicted for ticks obtained during the transmission feed on calf 847 and are representative for data obtained in experiments done in triplicate on samples from three transmission feeds using calves 814, 847, and 855. A transient increase in msp2 transcript and MSP2 protein levels at 18 h was detected in all feeding experiments when data for transmission-fed ticks were compared to that for unfed ticks.
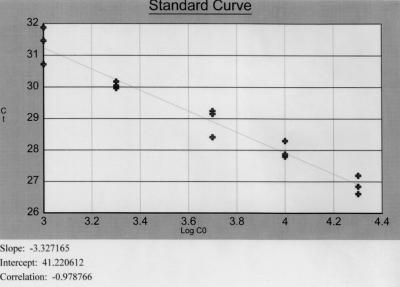
FIG. 3.
Quantitative real-time PCR for msp2 DNA. Standard curve generated with serial dilutions of a pBS vector containing full-length msp2 (24). The log calculated copy number of plasmid DNA (Log CO; x axis) is plotted against the corresponding threshold cycle (Ct; y axis). The slope, intercept, and correlation are indicated. Efficiency was calculated to be 0.998 (15).
TABLE 1.
Quantification of A. marginale and msp2 transcript levels in D. andersoni salivary glands
| Time point | Tick groupc | No. of msp5 DNA copies per salivary gland paira
|
No. of msp2 cDNA copies per salivary gland pairab
|
||
|---|---|---|---|---|---|
| Median | Range | Median | Range | ||
| Acquisition | |||||
| 8 days | 2.43 × 105 | 1.24 × 105–9.52 × 105 | 2.43 × 105 | 1.81 × 105–5.82 × 105 | |
| Holding | |||||
| 3 days | 1.39 × 105 | 5.80 × 104–1.87 × 105 | 4.06 × 107 | 9.39 × 106–8.27 × 107 | |
| 7 days | 5.84 × 105 | 2.93 × 105–1.04 × 106 | 8.27 × 107 | 3.57 × 107–1.34 × 108 | |
| Transmission | |||||
| 6 h | Fed | 3.82 × 105 | 7.66 × 104–6.90 × 105 | 2.16 × 108 | 6.70 × 107–4.10 × 108 |
| 26°C | 4.97 × 105 | 1.46 × 105–1.94 × 106 | 2.14 × 108 | 1.51 × 108–3.15 × 108 | |
| 18 h | Fed | 4.64 × 105 | 4.50 × 105–1.65 × 106 | 6.10 × 108 | 4.92 × 108–8.11 × 108 |
| 26°C | 6.82 × 105 | 5.10 × 105–2.40 × 106 | 1.09 × 108 | 7.23 × 107–1.97 × 108 | |
| 42 h | Fed | 7.59 × 105 | 6.43 × 105–8.52 × 105 | 1.51 × 108 | 8.16 × 107–3.13 × 108 |
| 26°C | 1.35 × 106 | 8.30 × 105–1.57 × 106 | 4.02 × 108 | 9.24 × 107–1.11 × 109 | |
| 72 h | Fed | 1.70 × 105 | 1.01 × 105–7.90 × 105 | 9.81 × 108 | 5.05 × 107–1.07 × 109 |
| 26°C | 3.06 × 105 | 1.02 × 105–3.88 × 105 | 5.00 × 108 | 1.66 × 108–1.81 × 109 | |
The values are the medians and ranges of three replicates from each pool of 10 salivary gland pairs.
Quantities are corrected 100 times to more closely reflect msp2 transcript levels based on ∼1% efficiency of reverse transcription.
Ticks were either transmission fed on calf 847 (Fed) or kept at 26°C and were assayed at the indicated times.
FIG. 4.
Expression of A. marginale MSP2 protein during transmission feeding. Ticks were either transmission fed on calf 847 (F) or held at 26°C (26). Expression of MSP2 was examined at 6, 18, 42, and 72 h by using Western blots. MSP2 and MSP5 were detected as described for Fig. 2B. The positions of SGV1, SGV2, and MSP5 are indicated in the left margin, and molecular size markers are indicated in the right margin.
Quantitation of A. marginale organisms per tick.
To determine the number of A. marginale per salivary gland pair at the time of transmission, quantitative real-time PCR for the single-copy gene msp5 (33) was performed. The PCR assay was validated as described for msp2 and had a sensitivity of 21 copies (100 attograms) of plasmid DNA, and efficiency ranged from 0.95 to 0.99 (15). Ticks were acquisition fed on calf 839 and transmission fed on calf 847, and salivary gland samples were collected at 0, 6, 18, 42, and 72 h of the transmission feed. The number of bacteria present in salivary glands of transmission-fed ticks was compared to quantities present in control ticks incubated at 26°C at each time point (Table 1). The number of msp5 DNA copies increased and then terminally declined in the salivary glands of transmission-fed ticks and unfed ticks. Within each group of ticks the difference in msp5 DNA copies between time points 42 h and 72 h are statistically significant. Differences in msp5 copies between fed and incubated ticks are not statistically significant at any of the examined time points. Experiments were done in triplicate and also repeated with samples from ticks acquisition fed on calves 809 and 847 and transmission fed on calves 814 and 855. A parallel increase and terminal decline in msp5 DNA over time in salivary glands of transmission-fed and unfed ticks was detected in all feeding experiments.
Presence of A. marginale in salivary glands of acquisition-feeding ticks.
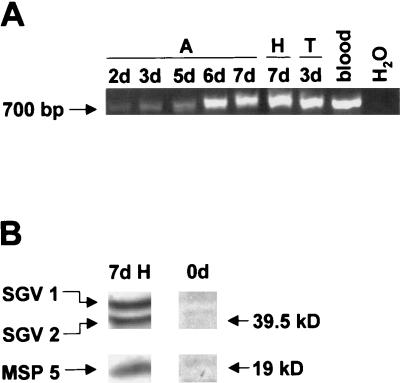
To determine at which time point during the acquisition feed A. marginale colonizes the salivary gland, PCR with primers specific for full-length msp2 was performed on samples obtained from ticks fed on calf 873. Amplicons of the expected size were detected in salivary glands from day 2 of the acquisition feed (Fig. 5A). To confirm expression of full-length msp2 in the salivary gland of ticks before the second attachment and transmission feed, Western blots were performed on salivary glands obtained at the end of the holding period following the acquisition feed on calf 873. Two distinct MSP2 proteins of the predicted molecular size for SGV1 and SGV2 were detected (Fig. 5B).
FIG. 5.
Presence of A. marginale in the salivary gland of acquisition-feeding ticks. (A) Full-length msp2 was detected by PCR in salivary glands obtained on days 2, 3, 5, 6, and 7 of the acquisition feed (A) on calf 873, day 7 of the holding period (H), and day 3 of the transmission feed (T) on calf 882. Blood was collected from calf 873 on day 3 during the acquisition feeding (blood). The reactions were carried out with primer pair MSP2:184-201 and MSP2:891-874. A reaction mixture without template served as negative control (H2O). The position of the 700-bp fragment is indicated in the left margin. (B) Expression of MSP2 in salivary glands was examined before attachment of the ticks (0d) and day 7 (7d) of the holding period (H) after the acquisition feed on calf 873 by using Western blots. MSP2 and MSP5 were detected as described for Fig. 2B. The positions of SGV1, SGV2, and MSP5 are indicated in the left margin, and molecular size markers are indicated in the right margin.
DISCUSSION
The expression of MSP2 in the blood is limited to a single, operon-linked expression site (1, 3). MSP2 A3, detected in the blood at the time of tick acquisition feeding (Fig. 1B), is expressed in acute infection with the South Idaho strain of A. marginale and represents recombination of pseudogene A3 into the operon expression site (3). The expression of MSP2 A3 in the tick midgut at day 2 of the acquisition feed indicates the presence of A. marginale organisms in the blood meal that have not yet switched to tick-associated MSP2 variants. Whether these bacteria reside in the midgut lumen or have already entered midgut epithelial cells has not been determined. If the switch to tick variants occurs in the midgut lumen, a role for specific MSP2 proteins in midgut cell invasion by A. marginale is conceivable and could be explored in view of the observed differences in the transmissibility of various A. marginale strains (31, 34). A similar role has been proposed for a second A. marginale outer membrane protein, MSP1a (6). Attachment of B. burgdorferi within Ixodes scapularis, mediated by the outer membrane protein OspA, which is also differentially expressed between tick and blood stages, provides precedent for a role of outer membrane proteins in tick midgut invasion (23).
The tick-associated MSP2 variants SGV1 and SGV2 are detectable in the midgut within the first 48 h of the acquisition feed. Notably, SGV1 and SGV2 are also in the operon expression site at this time point. Barbet et al. have recently shown transcription of msp2 from the operon expression site in infected ticks using the Oklahoma strain of A. marginale, which does not undergo restriction to tick-specific MSP2 variants (2). We had hypothesized that SGV1 and SGV2 were expressed from a second, non-operon-linked site under control of a separate promoter(s). The change in expression could occur rapidly by switching from the operon promoter to the SGV promoter(s). The data presented here refute that hypothesis. The switch from A3 and other MSP2 variants to SGV1 and SGV2 appears to reflect the same mechanism of recombination into a single expression site previously shown for generation of A. marginale MSP2 variants in the blood (3). This is also consistent with the Southern blot analysis of the South Idaho strain showing only a single 5′ msp2 end in the genome, located within the expression site (3).
Subsequent to the switch from blood- to tick-specific variants, only SGV1 and SGV2 are found in the midgut and, by day 5 of the acquisition feed, in the salivary gland. Migration of A. marginale into the salivary gland prior to the transmission feed had previously been reported by Rurangirwa et al. (27) and has also been demonstrated in I. scapularis nymphs infected as larvae with the agent of the human granulocytic ehrlichiosis (5). Our data confirm this early invasion of A. marginale into the salivary gland. It contradicts findings by others, primarily based on histological examination, that migration to salivary glands was a late event, occurring only upon transmission feeding (12, 19). The lower detection threshold of histology compared to that of PCR most likely explains the prior failure to detect early salivary gland invasion.
Interestingly, SGV1 and SGV2 appear to be equally represented in the expression site and are also expressed approximately equally as protein. This is consistent with previous data from salivary glands of transmission-fed ticks in which approximately equal levels of SGV1 and SGV2 transcripts were identified (27). How is the expression of a relatively constant ratio of SGV1 and SGV2 achieved by using a single genomic expression site? The most likely explanation is the presence of two closely balanced bacterial populations, each expressing one of the tick-specific variants. The predicted amino acid sequences of SGV1 and SGV2 are identical except for a 16-amino-acid deletion in the hypervariable region of SGV2 (27). This suggests that identical regions in the two proteins could mediate a common function and that bacterial populations expressing either SGV1 or SGV2 are equally selected for within the tick. This is consistent with the presence of coexisting A. marginale populations in the blood that express different MSP2 variants as demonstrated by immunofluorescence using MSP2 variant-specific antibody (11). Alternatively, a constant switch between the two variants within individual bacteria could occur. To date, there is no data supporting this alternative mechanism.
We tested two hypotheses addressing how A. marginale infectivity increases in the salivary gland during transmission feeding. The first hypothesis was that expression of tick-specific variants of A. marginale MSP2 is increased in stimulated salivary glands of transmission-feeding ticks. This hypothesis was derived from studies of B. burgdorferi showing a switch from expression of OspA to OspC in the transmission-feeding tick vector and a correlation with increased spirochetal infectivity (8, 25, 30). Enhanced msp2 transcript and MSP2 protein expression occurred in A. marginale within salivary glands obtained from transmission-feeding D. andersoni ticks compared to that from unfed ticks, but only at 18 h. Based on the parallel increase in transcript and protein abundance at this time point, increased MSP2 expression could be transcriptionally regulated as B. burgdorferi OspC is (9) or, less likely, could be regulated by changes in mRNA half-life. However, the difference in A. marginale was transient and not statistically significant for msp2 transcript levels. Consequently, the hypothesis that expression of tick-specific variants of A. marginale MSP2 is increased in stimulated salivary glands of transmission feeding was rejected. While MSP2 may be required for infectivity, there is no strict correlation between the levels of MSP2 expression and development of infectivity.
The second hypothesis addressing enhanced infectivity was that bacterial numbers increase in the salivary gland during the transmission feed of ticks and are higher in fed than in unfed ticks. Dot blot analyses of whole-tick homogenates of transmission-fed D. andersoni ticks indicated an increase in the number of A. marginale organisms per tick and an increase in the percentage of infected ticks over time when transmission feeding was mimicked by incubation of ticks at 37°C (18). Examination of histological preparations of salivary glands suggested that the number of colonized salivary gland acini was higher in ticks incubated at 37°C than in ticks incubated at 26°C (18). Using quantitative real-time PCR for the single-copy gene msp5 (33), the number of A. marginale per salivary gland pair was determined in transmission feeding ticks and was compared to numbers in salivary glands of unfed ticks incubated at 26°C. A. marginale numbers increased from approximately 4 × 105 to approximately 7 × 105 and then terminally dropped to levels below those detected after the first 6 h of the transmission feeding in both groups of ticks. The detected peak levels of 105 organisms per salivary gland pair is in agreement with previous semiquantitative estimates based on Southern blot analysis using an msp1β probe (18). Interestingly, no statistically significant difference in organism numbers was detected between fed and unfed ticks. This may reflect that replication of A. marginale is time dependent rather than stimulated by physiologic changes of the salivary gland induced by the feeding process (32). The number of A. marginale in salivary glands during the transmission feed alone cannot explain increased infectivity upon tick feeding, and the hypothesis was therefore rejected, albeit with a caveat. We cannot exclude the fact that the number of A. marginale detected in tick salivary glands may be a function of bacterial replication in salivary gland cells in combination with loss of bacteria with the saliva. It is conceivable that A. marginale replicates faster in salivary glands stimulated by the feeding process and that loss of bacteria due to salivation reduces the number to levels reached in unstimulated salivary glands of ticks incubated at 26°C. Quantitation of A. marginale in the saliva of transmission-feeding ticks would be required to explore this hypothesis; however, this hypothesis is difficult to test, as in vitro membrane feeding of Dermacentor spp. does not mimic natural transmission feeding.
The mechanisms by which infectivity of Anaplasma increases in the salivary gland of transmission feeding ticks (14, 17) remain unknown. Using the South Idaho strain of A. marginale, which has the advantage of restricted MSP2 variant expression in the tick, we have rejected two hypotheses: (i) expression of tick-specific variants of A. marginale MSP2 is increased in stimulated salivary glands of transmission feeding ticks, and (ii) bacterial numbers increase in the salivary gland during the transmission feed of ticks and are higher in fed than in unfed ticks. Based on studies with Borrelia showing that a switch of outer membrane proteins correlates with increased spirochetal infectivity (8, 25, 30), we retain the hypothesis that differential expression of outer membrane proteins of A. marginale in fed and unfed ticks is a key factor in increased infectivity observed with transmission-feeding ticks. A more global approach to identify outer membrane proteins (other than MSP2) that are specifically expressed during transmission feeding is needed to address this hypothesis and can be pursued by using microarray analysis and genome sequence data (3) of A. marginale.
Acknowledgments
We are deeply indebted to Tom Schmittgen and Brian Zakraysek at the Department of Veterinary and Comparative Anatomy, Pharmacology, and Physiology for access to the ABI PRISM 5700 Sequence Detection System, technical assistance, and helpful discussion. The technical assistance of Ralph Horn, Beverly Hunter, and Carla Robertson is greatly appreciated, as is the administrative support of Don Knowles.
This work was supported by National Institutes of Health grant AI 44005, U.S. Department of Agriculture grant USDA-ARS-CRIS 5348-32000-012-00D, and by a postdoctoral fellowship from the Deutsche Akademie der Naturforscher Leopoldina.
Editor: R. N. Moore
REFERENCES
- 1.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect. Immun. 68: 6133–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbet, A. F., J. Yi, A. Lundgren, B. R. McEwen, E. F. Blouin, and K. M. Kocan. 2001. Antigenic variation of Anaplasma marginale: major surface protein 2 diversity during cyclic transmission between ticks and cattle. Infect. Immun. 69: 3057–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98: 4130–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camacho-Nuez, M., M. de Lourdes, C. E. Suarez, T. C. McGuire, W. C. Brown, and G. H. Palmer. 2000. Expression of polymorphic msp1β genes during acute Anaplasma marginale rickettsemia. Infect. Immun. 68: 1946–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, S., K. Deponte, N. L. Marcantonio, J. W. Ijdo, E. Hodzic, P. Katavolos, S. W. Barthold, S. R. Telford, F. S. Kantor, and E. Fikrig. 1998. Granulocytic ehrlichiosis in tick-immune guinea pigs. Infect. Immun. 66: 1803–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, and K. M. Kocan. 2001. Differential adhesion of major surface proteins 1a and 1b of the ehrlichial cattle pathogen Anaplasma marginale to bovine erythrocytes and tick cells. Int. J. Parasitol 31: 145–153 [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente, J., and K. M. Kocan. 2001. Expression of Anaplasma marginale major surface protein 2 variants in persistently infected ticks. Infect. Immun. 69: 5151–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Silva, A. M., S. R. Telford, L. R. Brunet, S. W. Barthold, and E. Fikrig. 1996. Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J. Exp. Med. 183: 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Silva, A. M., N. S. Zeidner, Y. Zhang, M. C. Dolan, J. Piesman, and E. Fikrig. 1999. Influence of outer surface protein A antibody on Borrelia burgdorferi within feeding ticks. Infect. Immun. 67: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriks, I. S., D. Stiller, and G. H. Palmer. 1993. Impact of persistent Anaplasma marginale rickettsemia on tick infection and transmission. J. Clin. Microbiol 31: 2091–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French, D. M., W. C. Brown, and G. H. Palmer. 1999. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect. Immun. 67: 5834–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge, N. L., K. M. Kocan, E. F. Blouin, and G. L. Murphy. 1996. Developmental studies of Anaplasma marginale (Rickettsiales:Anaplasmataceae) in male Dermacentor andersoni (Acari:Ixodidae) infected as adults by using nonradioactive in situ hybridization and microscopy. J. Med. Entomol 33: 911–920 [DOI] [PubMed] [Google Scholar]
- 13.Gilmore, R. D. J., and J. Piesman. 2000. Inhibition of Borrelia burgdorferi migration from the midgut to the salivary glands following feeding by ticks on OspC-immunized mice. Infect. Immun. 68: 411–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodzic, E., D. Fish, C. M. Maretzki, A. M. De Silva, S. Feng, and S. W. Barthold. 1998. Acquisition and transmission of the agent of human granulocytic ehrlichiosis by Ixodes scapularis ticks. J. Clin. Microbiol. 36: 3574–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20: 291–299 [DOI] [PubMed] [Google Scholar]
- 16.Knowles, D., D. E. Torioni, G. Palmer, T. McGuire, D. Stiller, and T. McElwain. 1996. Antibody against an Anaplasma marginale MSP5 epitope common to tick and erythrocyte stages identifies persistently infected cattle. J. Clin. Microbiol. 34: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocan, K. M., S. J. Barron, S. A. Ewing, and J. A. Hair. 1985. Transmission of Anaplasma marginale by adult Dermacentor andersoni during feeding on calves. Am. J. Vet. Res. 46: 1565–1567 [PubMed] [Google Scholar]
- 18.Kocan, K. M., W. L. Goff, D. Stiller, W. Edwards, S. A. Ewing, P. L. Claypool, T. C. McGuire, J. A. Hair, and S. J. Barron. 1993. Development of Anaplasma marginale in salivary glands of male Dermacentor andersoni. Am. J. Vet. Res. 54: 107–112 [PubMed] [Google Scholar]
- 19.Kocan, K. M., D. Stiller, W. L. Goff, P. L. Claypool, W. Edwards, S. A. Ewing, T. C. McGuire, J. A. Hair, and S. J. Barron. 1992. Development of Anaplasma marginale in male Dermacentor andersoni transferred from parasitemic to susceptible cattle. Am. J. Vet. Res. 53: 499–507 [PubMed] [Google Scholar]
- 20.Kocan, K. M., K. D. Teel, and J. A. Hair. 1980. Demonstration of Anaplasma marginale Theiler in ticks by tick transmission, animal inoculation, and fluorescent antibody studies. Am. J. Vet. Res. 41: 183–186 [PubMed] [Google Scholar]
- 21.Lewin, B. 1997. Genes VI, 6th ed. Oxford University Press, Oxford, United Kingdom
- 22.MacLeod, J. 1936. Studies in tick-born fever in sheep. II. Experiments on transmission and distribution of the disease. Parasitology 28: 320–329 [Google Scholar]
- 23.Pal, U., A. M. De Silva, R. R. Montgomery, D. Fish, J. Anguita, J. F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Investig. 106: 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer, G. H., G. Eid, A. F. Barbet, T. C. McGuire, and T. F. McElwain. 1994. The immunoprotective Anaplasma marginale major surface protein 2 is encoded by a polymorphic multigene family. Infect. Immun. 62: 3808–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piesman, J. 1993. Dynamics of Borrelia burgdorferi transmission by nymphal Ixodes dammini ticks. J. Infect. Dis. 167: 1082–1085 [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro, J. M., T. N. Mather, J. Piesman, and A. Spielman. 1987. Dissemination and salivary delivery of Lyme disease spirochetes in vector ticks (Acari:Ixodidae). J. Med. Entomol 24: 201–205 [DOI] [PubMed] [Google Scholar]
- 27.Rurangirwa, F. R., D. Stiller, D. M. French, and G. H. Palmer. 1999. Restriction of major surface protein 2 (MSP2) variants during tick transmission of the ehrlichia Anaplasma marginale. Proc. Natl. Acad. Sci. USA 96: 3171–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rurangirwa, F. R., D. Stiller, and G. H. Palmer. 2000. Strain diversity in major surface protein 2 expression during tick transmission of Anaplasma marginale. Infect. Immun. 68: 3023–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwan, T. G., and B. J. Hinnebusch. 1998. Bloodstream- versus tick-associated variants of a relapsing fever bacterium. Science 280: 1938–1940 [DOI] [PubMed] [Google Scholar]
- 30.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete Borrelia burgdorferi during the chain of infection in ticks and mice. J. Clin. Microbiol. 38: 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, R. D., M. G. Levy, M. S. Kuhlenschmidt, J. H. Adams, D. L. Rzechula, T. A. Hardt, and K. M. Kocan. 1986. Isolate of Anaplasma marginale not transmitted by ticks. Am. J. Vet. Res. 47: 127–129 [PubMed] [Google Scholar]
- 32.Sonenshine, D. E. 1991. Biology of ticks, vol. 1. Oxford University Press, Inc., New York, N.Y
- 33.Visser, E. S., T. C. McGuire, G. H. Palmer, W. C. Davis, V. Shkap, E. Pipano, and D. P. J. Knowles. 1992. The Anaplasma marginale msp5 gene encodes a 19-kilodalton protein conserved in all recognized Anaplasma species. Infect. Immun. 60: 5139–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickwire, K. B., K. M. Kocan, S. J. Barron, S. A. Ewing, R. D. Smith, and J. A. Hair. 1987. Infectivity of three Anaplasma marginale isolates for Dermacentor andersoni. Am. J. Vet. Res. 48: 96–99. [PubMed] [Google Scholar]