Abstract
Major surface protein 2 (MSP-2), identified as a protection-inducing immunogen against Anaplasma marginale challenge, is an immunodominant outer membrane protein with orthologues in all examined Anaplasma species. Although immunization with live Anaplasma centrale has long been used to induce protection against acute disease upon challenge with virulent A. marginale, its MSP-2 structure and whether MSP-2 variants are generated during persistence of the vaccine strain was unknown. In this study, we showed that the A. centrale vaccine strain persisted for a minimum of 4 years postvaccination and generated sequential MSP-2 variants. Comparison of amino acid sequences encoded by A. centrale msp-2 transcripts from the initial postimmunization period and from sequential time points during persistence of the vaccine strain revealed a central hypervariable domain flanked by conserved amino and carboxy-terminal regions. This structure corresponded to that shown in A. marginale MSP-2, where the central hypervariable region encodes variant B-cell epitopes in the extracellular domain and the flanking transmembrane domains are rich in CD4+-T-cell epitopes. Importantly, at least four CD4+-T-cell epitopes are conserved between the two species, a finding consistent with A. marginale challenge triggering a recall response of CD4+ T cells induced by A. centrale vaccination. The genomic arrangement is conserved between A. centrale and A. marginale with multiple msp-2 pseudogenes and a single operon-linked expression site for the full-length msp-2. This conservation of both genomic structure for generating MSP-2 variants and the CD4+-T-cell epitopes between these two genetically distinct Anaplasma species indicates that they present a similar repertoire of MSP-2 epitopes to the immune system and that this similarity may be responsible for all or part of the A. centrale vaccine efficacy.
The identification of Anaplasma marginale as a specific infectious pathogen of cattle in 1908 was the first description of what are now recognized as ehrlichiae (28, 29). Remarkably, within 4 years of this initial description, Sir Arnold Theiler described the use of a related organism, Anaplasma centrale, as a live vaccine to minimize the severity of A. marginale infection (30). In experiments carried out from 1909 to 1911, Theiler described the need to establish an A. centrale infection and allow recovery from this mild form of the disease prior to field exposure to ticks carrying A. marginale (30). Furthermore, he established that A. centrale vaccination did not prevent A. marginale infection upon challenge but rather resulted in lower levels of A. marginale rickettsemia and a reduction in severity of clinical signs compared to unvaccinated controls. Based on these findings, A. centrale live vaccines were used to protect cattle imported from England to South Africa in 1909 and have since remained in use in tropical and subtropical regions (15, 24, 25, 33).
Current mechanistic understanding of immunity to A. marginale has been derived both from experimental vaccine trials, which identified major surface protein 2 (MSP-2) as a protection-inducing immunogen (22), and from the study of infection-induced immunity. After recovery from acute A. marginale infection, during which rickettsemia levels exceed 109organisms per ml, cattle remain persistently infected but are immune to high-level rickettsemia and disease if subsequently challenged with homologous or heterologous strains (15, 23). Persistent infection is asymptomatic and characterized by cyclic waves of rickettsemia (12), with each cycle reflecting the emergence and replication of A. marginale organisms expressing an antigenically variant MSP-2 (8, 9, 20). Analysis of A. marginale MSP-2 antigenic variants has revealed highly conserved amino and carboxy-terminal regions with the central one-third of the protein being hypervariable (7–9). This central region represents the primary surface domain and encodes the immunodominant but hypervariable B-cell epitopes (7–9). In contrast, the conserved flanking regions are rich in T-cell epitopes and induce strong recall CD4+-T-cell responses against MSP-2 regardless of the variation in the central surface domain (3, 5). This conserved anamnestic T-cell response is postulated to provide rapid T-cell help for primary B-cell responses to the hypervariable surface epitopes and control the emergent variants prior to reaching the high rickettsemia levels responsible for clinical disease (3, 20). This role for the conserved T-cell epitopes is supported by the rapid generation of variant-specific immunoglobulin G2 antibody after emergence of a new MSP-2 variant and the control of rickettsemia during persistent A. marginale infection to levels 100 to 1,000 times below the threshold for clinical disease (4, 8, 9).
Does Theiler’s A. centrale vaccine work by mimicking natural persistent infection? If this is correct, A. centrale should persist in vaccinates and generate MSP-2 variants that mimic the antigenic structure of those expressed by A. marginale. Specifically, this would comprise a diverse set of MSP-2 central extracellular domains and highly conserved flanking regions that contain CD4+-T-cell epitopes shared between A. centrale and A. marginale. In this study, we report testing these hypotheses with A. centrale-immunized cattle.
MATERIALS AND METHODS
A. centrale vaccine and cattle immunization.
The A. centrale vaccine strain was originally obtained from South Africa in 1952 and has since been used as the standard vaccine in Israel (24). The procedures on vaccine quality control, determination of dose, thawing, and administration have been described in detail (24). Briefly, all calves were screened for A. marginale infection prior to vaccination by using serology, microscopic examination of Giemsa-stained blood smears, and PCR (24, 32). The 3- to 4-month-old calves were then inoculated intravenously with 1.0 ml of vaccine containing erythrocytes infected with 108 live A. centrale.The study population included three groups maintained under tick-free conditions: (i) two Holstein calves (no. 337 and 410) examined during acute rickettsemia after A. centrale vaccination, (ii) two Holstein calves (no. 405 and 429) during acute rickettsemia after A. centrale vaccination and sequentially for 12 months after vaccination, and (iii) eight Holstein dairy cows (no. 376, 378, 468, 477, 478, 573, 578, and 773) examined at a single time point 30 to 49 months postvaccination. Blood was obtained from individual vaccinates at various time points postinoculation, and the persistence of the vaccine strain was determined by subinoculation of 50 ml of blood into seronegative, susceptible calves that were monitored by microscopic detection of A. centrale-infected erythrocytes (13, 15) or by using A. centrale-specific PCR.
A. centrale PCR.
A one-stage nested PCR for A. centrale was developed based on the mpb-58 sequence and was shown to be capable of specific discrimination between A. centrale and A. marginale (V. Shkap, T. Molad, L. Fish, and E. Pipano, Abstr. 2nd Int. Vet. Vaccines Diagn. Conf., abstr. P52, 2000). The one-step nested PCR has a detection limit of ca. 500 A. centrale-infected erythrocytes per ml of blood, corresponding to a level of 0.00001% infected erythrocytes in the peripheral blood. Briefly, the external forward and reverse primers were, respectively, 5′-TGCACCGTCCTGCCGTGACC and 5′-AACCCACGCGGGCAGCTTGA. The internal forward and reverse primers were 5′-TCCAGTAACAAGCAGTTC and 5′-TGAACCTACGGTACACAT, respectively. The two rounds of amplification were separated by limiting the amount of the external primers. The reaction was performed in a final reaction volume of 25 μl containing a 0.004 μM concentration of each external primer, a 0.5 μM concentration of each internal primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1× PCR buffer (10 mM Tris-HCl, pH 8.4; 50 mM KCl; 0.08% Nonidet P-40), 2.5 mM MgCl2, and 0.75 U of Taq DNA polymerase. Cycling conditions were as follows: 1 cycle of preheating at 95°C for 3 min, 14 cycles of 94°C for 50 s, 60°C for 50 s, and 72°C for 50 s; 26 cycles of 94°C for 30 s, 49°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. Amplicons were analyzed by electrophoresis in 1.5% agarose gels and staining with 0.015% ethidium bromide.
Cloning and sequencing of full-length msp-2 cDNA.
The msp-2 transcripts were identified from A. centrale acute rickettsemia or from subsequent peaks of persistent rickettsemic cycles within individual vaccinates. Cycles were identified by using the quantitative msp-5 PCR as previously described in detail (8). At the peak of each rickettsemic cycle, total RNA was extracted from whole blood and reverse transcribed by using random hexamers, as described previously (8, 9). Full-length msp-2 cDNA clones were generated by PCR with primers derived from the 5′ and 3′ ends of msp-2. The forward primer was 5′-TTGGATCCATGAGTGCTGTAAGTAATAGGAAGC and the reverse primer was 5′-TCTCTCGAGCTAGAAGGCAAACCTAACACCCA. These primers were derived from the conserved 5′ and 3′ ends of the open reading frame of the existing full-length genomic clones, DF5 msp-2 and 11.2 msp-2 (7, 21). To amplify only the 600-bp central hypervariable region of msp-2, two primers flanking this region were used: 5′-GTGGATCCGGAGGAGCAAGGGTTGAAGTA and 5′-AGCTCGAGTTACCACCGATACC-AGCACAA (8). These sequences are conserved among msp-2 genes and transcripts of different A. marginale and A. ovis strains (3, 8, 9, 21, 26) and represent nucleotide positions 376 to 396 and positions 967 to 945, respectively, as numbered in the 11.2 msp-2 sequence (21). PCR products were ligated into pCR2.1 by using the TA Cloning Kit (Invitrogen). Competent Escherichia coli XL1-Blue was transformed with the ligated vector and plated with 5 mM IPTG (isopropyl-1-β-d-thiogalactopyranoside) and BCIP (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue-white screening. Presence of msp-2 inserts in plasmids from transformed colonies was confirmed by restriction digests or PCR, as previously described (8, 9). Plasmid DNA was extracted from each clone and sequenced in both directions.
Sequence analysis.
For alignment and presentation of amino acid sequences, CLUSTAL W (31) and the Boxshade Server (http://www.ch.embnet.org/software/BOX_form.html) were used. The GenBank accession numbers for the A. centrale msp-2 transcripts are AY040556 to AY040563.
Southern blot detection of genomic A. centrale msp-2 and the msp-2 expression site.
DNA was extracted from 109 A. centrale and from the South Idaho strain of A. marginale and digested by using FspI as previously described for A. marginale (1). After electrophoresis on a 0.7% agarose gel and transfer to a nylon membranes by standard techniques, the membrane was hybridized sequentially with each of three probes: (i) full-length orf4 from the expression site, (ii) msp-2 (bp 375 to 965), and (iii) the 5′ terminus of msp-2 (bp 2 to 335). The probes were generated by PCR by using the following corresponding primer sets: (i) for orf4, 5′-TTGCACACACTACCCTCTTC and 5′-GCCGCTACATAGAGCTCACCC; (ii) for msp-2, 5′-TGGAGGAGCAAGGGTTGAAGT and 5′-TTACCACCGATACCAGCACAA; and (iii) for the 5′ terminus msp-2, 5′-TGAGTGCTGTAAGTAATAGG and 5′-CTATCCTTGAAGCTAATCTTGGGTTC. Labeling of the probes with digoxigenin, hybridization with DIG Easy Hyb (Roche, Inc.) overnight at 42°C, washing (twice with 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-0.1% sodium dodecyl sulfate at room temperature and twice in the same buffer at 60°C), and detection by enhanced chemiluminescence was done as previously described (1, 2). The filter was stripped between hybridizations with each of the probes by using two washes in 0.2 M NaOH-0.1% sodium dodecyl sulfate at 37°C for 15 min.
Peptide stimulation of MSP-2 specific CD4+-T-cell lines.
The derivation and characterization of the short-term T-cell lines used in this study have been described in detail elsewhere (3). Briefly, these T-cell lines were derived from three MSP-2-immunized calves with different class II haplotypes (calf no. 59 [DRB3 22/7, DQA 9B/2, and DQB 9B/2], calf no. 60 [DRB3 22/23, DQA 9B/7D, and DQB 9B/7A], and calf no. 61 [DRB3 8/8, DQA 12/12, and DQB 12/12]). After stimulation with A. marginale for 1 week and depletion of γ/δ and CD8+ T cells by treatment with monoclonal antibody and complement (3), the cell lines were 94 to 99% CD4+ T cells. Proliferation assays were conducted in replicate wells of 96-well plates by using the T-cell lines (3 × 104 cells per well) in complete RPMI 1640 medium containing antigen and, as antigen-presenting cells, irradiated autologous peripheral blood mononuclear cells (2 × 105 cells per well), as previously described (3–5). The tested peptide antigens were derived from MSP-2 sequences that stimulated CD4+-T-cell lines from each of the three calves of different major histocompatibility complex (MHC) class II haplotypes (3) or else represented the polymorphic sequence identified in A. centrale during the present study. These sequences are shown in Fig. 5. The peptides were tested at concentrations from 0.4 to 10.0 μg/ml. As positive control antigens, A. marginale lysate, purified native MSP-2, and the A. marginale peptide P12 (positions 312 to 341) were tested at the same concentrations. As negative control antigens, lysates of uninfected erythrocytes or an A. marginale 30-mer peptide (P1) that does not contain a T-cell epitope (3) was tested at the same concentrations. Cells were radiolabeled during the last 18 h of a 3- to 4-day stimulation period by using 0.25 μCi of [3H]thymidine, collected by using an automated cell harvester, and counted with a liquid scintillation counter. The results are presented as the mean counts per minute of replicate cultures ± 1 standard deviation.
FIG. 5.
Structure of MSP-2 and location of CD4+-T-cell epitopes. The central hypervariable region (cross-hatched) is flanked by conserved N- and C-terminal regions (solid). The positions of the CD4+-T-cell epitopes recognized by cattle with at least three different MHC class II haplotypes (3) are indicated by solid lines, and the amino acid positions (numbering based on 11.2 MSP-2 [21]). The sequences of the peptides representing the epitopes are provided for both A. marginale and A. centrale. The tested peptides include the 312-to-341 P12 peptide and 18-, 19-, and 20-mer peptides representing the different sequences of A. marginale and A. centrale. The amino acid substitution is indicated by an underlined boldface letter.
RESULTS
Persistence of the A. centrale vaccine strain.
A. centrale was detected in blood obtained from each of the 10 Holstein dairy cows vaccinated 5 to 49 months earlier (Table 1). Blood from each of the eight vaccinates tested transmitted A. centrale upon subinoculation into individual susceptible, seronegative calves (Table 1). Each of the recipient calves developed microscopically detectable A. centrale rickettsemia within 40 days of inoculation. The two vaccinates that were tested only using the A. centrale mpb58 PCR (no. 405 and 429) were positive and were used to evaluate expression of msp-2 transcripts during persistence as described in detail below. Two additional vaccinates (no. 337 and 410) were examined only during the acute infection after A. centrale vaccination, as described below, and were not further examined for persistence.
TABLE 1.
Detection of A. centrale vaccine strain persistence
| A. centrale vaccinate (animal no.) | Time postvaccination (mo) | Detection of A. centrale |
|---|---|---|
| 405 | 5 | Nested mpb58 PCR |
| 429 | 11 | Nested mpb58 PCR |
| 773 | 30 | Subinoculation into calf 369 |
| 578 | 35 | Subinoculation into calf 336 |
| 573 | 37 | Subinoculation into calf 334 |
| 376 | 41 | Subinoculation into calf 359 |
| 378 | 41 | Subinoculation into calf 355 |
| 468 | 48 | Subinoculation into calf 351 |
| 477 | 49 | Subinoculation into calf 350 |
| 478 | 49 | Subinoculation into calf 362 |
Sequence and structure of MSP-2 expressed after A. centrale vaccination.
The MSP-2 expressed by A. centrale in the acute infection after vaccination were identified in the blood of vaccinated animals 337 and 410. The predicted A. centrale MSP-2 sequence, derived from a full-length msp-2 transcript obtained by reverse transcription-PCR from the blood of vaccinated animal 337, was compared to A. marginale 11.2 MSP-2 (the initial A. marginale msp-2 sequenced [21]). The overall primary (Fig. 1A) and secondary (Fig. 1B) structure of the encoded MSP-2 is common between the A. centrale vaccine strain and A. marginale. While overall there is 77% identity between the vaccine strain MSP-2 and A. marginale, alignment revealed that this conservation was primarily in the amino- and carboxy-terminal domains (Fig. 1A). There is 83% identity in the region from amino acids 1 to 189 between A. centrale and A. marginale and 92% identity in the region from amino acid 272 to the carboxy terminus (numbering based on 11.2 MSP-2 [21]). The predicted membrane-spanning structure of these amino- and carboxy-terminal regions is highly conserved between A. marginale and A. centrale (Fig. 1B). In contrast, the central extracellular domain was variable between A. centrale and A. marginale 11.2, with only 44% identity in the region between amino acids 190 and 271. This variation is generated by deletions and substitutions in A. centrale MSP-2 relative to A. marginale MSP-2 (Fig. 1A) and does alter the predicted structure (Fig. 1B).
FIG. 1.
Comparison of the full-length A. marginale 11.2 MSP-2 (21) with A. centrale MSP-2 obtained postimmunization from vaccinate 337. (A) Amino acid sequence alignment. Areas of amino acid substitutions, insertions, and deletions are indicated by a white background, and areas of amino acid identity have a black background. (B) Secondary structure prediction. The predicted membrane orientation calculated by using TMPred (http://www.ch.embnet.org/software/TMPRED_form.html) is plotted on the y axis versus the amino acid position on the x axis for A. marginale 11.2 MSP-2 (upper panel) and A. centrale vaccinate 337 MSP-2 (lower panel). The solid line (i > 0) is the calculation performed from the N to the C terminus; the dashed line (0 > i) is the calculation performed from the C to the N terminus.
Examination of a second A. centrale vaccinate, animal 410, in the acute infection after vaccination revealed two distinct full-length msp-2 transcripts. One of these, 410A, encoded a MSP-2 very similar to that in animal 337 but the second, 410B, encoded a variant MSP-2 (Fig. 2). Alignment of the amino acid sequences encoded by these two transcripts with the sequence from vaccinate 337 revealed absolute conservation of the amino-terminal domain, a single amino acid substitution in the carboxy-terminal domain, and a highly variable region between amino acid 181 and approximately amino acid 265 (Fig. 2). This region corresponds to the A. marginale MSP-2 hypervariable extracellular domain (7, 8, 26), and the changes among the A. centrale MSP-2 variants reflected insertions, deletions, and substitutions (Fig. 2).
FIG. 2.
Alignment of the full-length A. centrale MSP-2 sequence obtained postimmunization from vaccinate 337 with the two MSP-2 sequences obtained postimmunization from vaccinate 410. The amino acid identity and substitutions, insertions, and deletions are indicated as in Fig. 1A.
Expression of msp-2 variants in A. centrale vaccinates.
To address whether variation in the central extracellular domain is generated during persistence of the vaccine strain, the central 600-bp portion of msp-2 transcripts was sequenced from vaccinated animal 429 during acute infection following vaccination (1 May 2000) and at the peaks of the first two cycles of persistent rickettsemia (6 June 2000 and 31 August 2000). Consistent with results from A. marginale infection in which multiple variants emerge during persistence, a total of 37 msp-2 transcripts were identified. Alignment of the encoded MSP-2 amino acid sequences revealed a predominant region of variation extending from position 197 to position 270. The identity in the hypervariable region ranged from 75 to 95% when the peak 1 and peak 2 MSP-2 types were compared. Alignment of the two most different encoded MSP-2 types from peaks 1 and 2 illustrates the substitutions, insertions, and deletions that typify the hypervariable regions (Fig. 3). To verify that variation was consistently generated in the A. centrale vaccinates, msp-2 transcripts were sequenced from three peaks of rickettsemic cycles in vaccinated animal no. 405 (peak 1, 5 May 2000; peak 2, 8 June 2000; peak 4, 8 October 2000 [peak 3 was not examined]). A total of 28 different msp-2 transcripts were identified. These revealed the same pattern of central extracellular domain variation, typified by 72% identity between the most divergent MSP-2 types of peaks 1 and 2 and 73% identity between MSP-2 types of peaks 2 and 4 (accession no. AY040561 to AY040563 [data not shown]).
FIG. 3.
Alignment of the A. centrale MSP-2 hypervariable regions between clones from two sequential persistent rickettsemic peaks. Peak 1-1 and peak 2-1 each represent a single MSP-2 type from, respectively, the first or second persistent rickettsemic cycle of vaccinate 429. Sequences are aligned starting from amino acid position 126 and extending to position 326 for peak 1-1 and to position 333 for peak 2-1. The amino acid identity and substitutions, insertions, and deletions are indicated as in Fig. 1A.
Genomic structure of A. centrale msp-2.
Genomic DNA was extracted from the vaccine strain and digested with FspI to excise the entire expression site locus (1). This locus is composed of an promoter and an operon of four genes, orf4, orf3, orf2, and msp-2 (1). There are no FspI sites within this locus in either A. centrale or A. marginale. Probing the digested DNA with the orf4 probe identified the operon expression locus in A. centrale and, as previously reported (1, 2), in A. marginale (Fig. 4). The same blot was stripped and rehybridized by using a msp-2 probe (bp 375 to 695) that includes the highly conserved region from bp 375 to 550 present in all described msp-2 genes, including the pseudogenes that lack a complete 5′ end, as well as the expression site complete msp-2 gene (1). This second hybridization revealed multiple fragments in both A. centrale and A. marginale, a pattern previously reported for A. marginale DNA cleaved with FspI or additional restriction enzymes that cleave outside of the msp-2 coding sequence (1, 21). In contrast to the multiple msp-2 sequences detected by the bp-375-to-695 probe, stripping and then rehybridizing with a probe representing the 5′ terminus of msp-2 (bp 2 to 335) identified only the single band corresponding to the operon expression site (Fig. 4). This demonstrates the presence of multiple msp-2 pseudogenes, lacking the 5′ end of full-length msp-2, in A. centrale as well as in A. marginale. There was only a single msp-2 gene complete with a 5′ end, and this sole full-length gene was in the operon expression site defined by orf4 (Fig. 4).
FIG. 4.
Genomic structure of msp-2. Genomic DNA from either the A. centrale vaccine strain (AC) or A. marginale (AM) was digested with FspI and, by using a Southern blot, hybridized sequentially with probes representing orf4, msp-2 (bp 375 to 695), and msp-2 5′ end (bp 2 to 335). The blot was stripped between hybridizations. The expression site (ES) defined by orf4 is indicated in the right margin. The molecular size markers (M) are indicated in the left margin.
Conservation of CD4+-T-cell epitopes between A. centrale and A. marginale MSP-2.
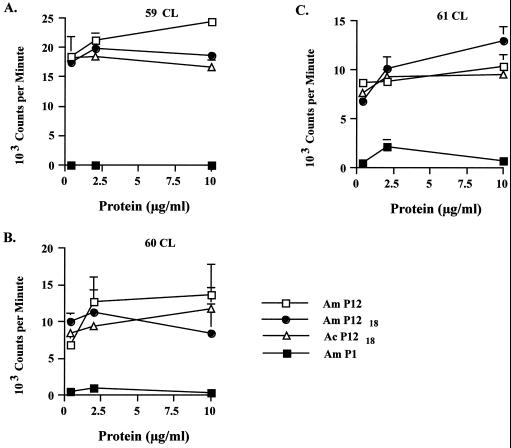
Notable features of the MSP-2 structure in A. centrale and A. marginale include the hypervariable extracellular domain and the flanking amino- and carboxy-terminal membrane domains (Fig. 1). Although the amino- and carboxy-terminal domains are, in general, conserved between A. marginale and A. centrale MSP-2 (Fig. 1A), the occurrence of amino acid substitutions in A. centrale relative to A. marginale may affect the CD4+-T-cell epitopes identified in these regions. Mapping of the CD4+-T-cell epitopes within the amino- and carboxy-terminal domains of A. marginale MSP-2 identified a minimum of four epitopes that were recognized by cattle with three different MHC class II haplotypes (3). Comparison of the sequences between A. marginale and the vaccine strain of A. centrale (Fig. 5) revealed that three of these epitopes were completely conserved and the fourth (designated P12 [3]), represented by amino acids 312 to 341, had a single amino acid substitution in A. centrale (V→I). This 312-to-341 region contains a dominant CD4+-T-cell epitope in A. marginale MSP-2 that induces both strong proliferation and gamma interferon production (3). Stimulation of MSP-2-specific CD4+-T-cell lines with the parent P12 (312 to 341), the A. marginale P1218 (324 to 341 peptide) represented in A. marginale, or the A. centrale P1218 peptide with the V→I substitution found in A. centrale revealed similar proliferation (Fig. 6). Notably, T-cell lines from each of three cattle (animals 59, 60, and 61) with different MHC class II haplotypes responded to the peptide derived from either A. marginale or A. centrale (Fig. 6). There were no significant differences in the proliferative responses to the P12, A. marginale P1218, and A. centrale P1218 peptides by any of the T-cell lines. In addition, each T-cell line responded similarly to A. marginale P1219 (323 to 341 peptide), A. marginale P1220 (322 to 341 peptide), A. centrale P1219 peptide, and A. centrale P1220 (data not shown), indicating that the V→I substitution found in A. centrale had no effect on the epitope at either terminal or subterminal positions. All cell lines responded to A. marginale lysate (Fig. 6) and native MSP-2 (data not shown) but not to either a control peptide P1 (Fig. 6) or uninfected erythrocyte lysate (data not shown).
FIG. 6.
Conservation of MSP-2 CD4+-T-cell epitopes between A. marginale and the A. centrale vaccine strain. MSP-2-specific cell lines from each of three calves (A, animal 59; B, animal 60; and C, animal 61) were stimulated with a 30-mer peptide representing A. marginale MSP-2 amino acids 312 to 341 (Am P12) previously shown to contain a CD4+-T-cell epitopes (3), an 18-mer peptide representing A. marginale MSP-2 amino acids 324 to 341 (Am P1218), an 18-mer peptide representing the same region but with a single amino acid substitution in A. centrale MSP-2 (Ac P1218), or a 30-mer peptide representing an A. marginale sequence that does not contain a T-cell epitope (P1 [3]). All peptides were tested over a concentration range of 0.4 to 10 μg per ml. The results are presented as mean counts per minute of replicate cultures ± 1 standard deviation.
DISCUSSION
A. centraleis genetically distinct from A. marginale based upon analysis of the 16S rRNA gene sequences of both organisms (11). The question of whether A. centrale is a subspecies of A. marginale, as originally suggested by Theiler (30), or a separate species is unresolved (6, 11). Nonetheless, there are clear differences in other genes, antigens, and virulence between the two organisms (14, 16, 19, 27). The presence of a closely related MSP-2 ortholog in A. centrale is unsurprising given that both Anaplasma ovis and Anaplasma phagocytophila, the agent of human granulocytic ehrlichiosis, also contain a MSP-2 ortholog (3, 10, 17, 18, 35). The overall structure of these MSP-2 orthologs is retained in each of these Anaplasma species—conservation in the amino- and carboxy-terminal thirds of the protein with a highly variable domain in the central extracellular domain (3, 10, 17, 18, 35).Originally shown in A. marginale persistently infected cattle, variation is generated in this central hypervariable MSP-2 domain, while the amino- and carboxy-terminal regions are essentially invariant (3, 8, 9, 26). The MSP-2 ortholog of A. phagocytophila demonstrates a similar pattern of variation in the central domain when strains are compared (10, 16, 35). In the present work, we have shown that the A. centrale vaccine strain also generates sequential variation in the central domain, whereas the flanking regions are highly conserved. Comparison of the encoded MSP-2 sequences obtained during acute and persistent infection after A. centrale vaccination revealed substitutions, deletions, and insertions in the central hypervariable domain. This pattern of variation and the degree of variation, as little as 72% identity between sequential time points, is similar to the variation identified in the A. marginale central extracellular domain and has been shown to result in B-cell epitope variation (9, 10, 26). Thus, persistence of the A. centrale vaccine strain would be expected to induce a population of B-cell responses against MSP-2 variants, including those that may cross-react with A. marginale variants. This is consistent with prior data demonstrating that cross-reactive antibody from persistent infection, resulting from infection with either the A. centrale vaccine strain or A. marginale, is primarily directed at MSP-2 (19, 34).
The amino- and carboxy-terminal thirds of MSP-2 are not identical between A. centrale and A. marginale, although they are conserved among MSP-2 variants within each species. In A. marginale this conservation is reflected by the ability of CD4+-T-cell lines and clones against these regions to recognize all strains and is consistent with the priming and continual restimulation of CD4+ T cells to provide B-cell help for production of variant-specific antibody during persistent infection (3, 5). CD4+-T-cell epitopes mapped in A. marginale MSP-2 include four that are recognized by T cells of cattle with three different MHC class II haplotypes (3). Three of these epitopes are within sequences that are identical between A. centrale and A. marginale (amino acids 141 to 170, 292 to 321, and 332 to 361), while the variation in the fourth (amino acids 312 to 341) was represented by a single amino acid substitution at position 324 that did not alter the epitope. This epitope conservation suggests that A. marginale challenge would stimulate recall responses in A. centrale-vaccinated cattle. In contrast, the polymorphic region from amino acid positions 19 to 41 in the amino terminus, in which 12 of 23 residues differ between A. centrale and A marginale (Fig. 1A), does not appear to contain a CD4+-T-cell epitope recognized by any of the MHC class II alleles represented in the calves (3). There are some CD4+-T-cell epitopes in the amino- and carboxy-terminal thirds that map to sequences that are polymorphic between A. centrale and A. marginale, e.g., the 30-mer peptide representing amino acid positions 372 to 401. This peptide, recognized by CD4+-T-cell lines from some but not all of the MHC class II haplotypes (3), has eight substitutions in A. centrale relative to A. marginale (Fig. 1A). Whether multiple conserved CD4+-T-cell epitopes would be sufficient to trigger protective recall responses even though other CD4+-T-cell epitopes are polymorphic is an intriguing question but has not yet been addressed.
A. marginale MSP-2 variants are generated by the recombination of pseudogenes, each composed of the hypervariable domain flanked by segments of the conserved 5′ and 3′ regions, into a single expression site (1, 2). The expression site itself is an operon composed of a single promoter followed by four open reading frames: orf4, orf3, orf2, and finally msp-2 (1, 2). Consistent with a conserved pattern of MSP-2 variation, A. centrale maintains this genomic structure with multiple msp-2 pseudogenes but with only a single full-length msp-2 gene that maps to the orf4-bearing expression site. Amplification and sequencing of this msp-2 expression site locus in A. centrale (data not shown) identifies the presence of the promoter, orf4, orf3, and orf2 as predicted.
The efficacy of A. centrale vaccination is dependent upon actual infection of the recipient and cannot be reproduced by inoculation with killed whole organisms. This is consistent with the need to generate sequential MSP-2 variants during infection and to induce both a population of variant-specific B cells and CD4+ T cells to conserved epitopes capable of controlling the high-level rickettsemia upon A. marginale challenge. The efficacy of this vaccine regimen, now in use for more than 90 years, suggests that strategies to mimic these induced immune responses are a key to development of an effective, non-blood-based vaccine.
Acknowledgments
This work was supported by U.S.-Israel Binational Agricultural Research and Development grant US-2799-96C and NIH grants R01AI44005 and R0145580.
We thank Kim Kegerreis and Max Tarchanov for excellent technical assistance, Y. Krigel and B. Leibovitz for assistance with animals, and Terry McElwain for critical review of the manuscript.
Editor: E. I. Tuomanen
REFERENCES
- 1.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of the ehrlichia Anaplasma marginale by expression of MSP-2 sequence mosaics. Infect. Immun. 68:6133–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, W. C., T. C. McGuire, D. Zhu, H. A. Lewin, J. Sosnow, and G. H. Palmer. 2001. Highly conserved regions of the immunodominant major surface protein 2 (MSP2) of the ehrlichial pathogen Anaplasma marginale are rich in naturally derived CD4+ T lymphocyte epitopes that elicit strong recall responses. J. Immunol. 166:1114–1125. [DOI] [PubMed] [Google Scholar]
- 4.Brown, W. C., V. Shkap, D. Zhu, T. C. McGuire, W. Tuo, T. F. McElwain, and G. H. Palmer. 1998. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 66:5406–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, W. C., D. Zhu, V. Shkap, T. C. McGuire, E. F. Blouin, K. M. Kocan, and G. H. Palmer. 1998. The repertoire of Anaplasma marginale antigens recognized by CD4+ T lymphocyte clones from protectively immunized cattle is diverse and includes major surface protein 2 (MSP-2) and MSP-3. Infect. Immun. 66:5414–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, F. Jongejan, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia, and Ehrlichia with Neorickettsia, descriptions of five new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145–2165. [DOI] [PubMed] [Google Scholar]
- 7.Eid, G., D. M. French, A. M. Lundgren, A. F. Barbet, T. F. McElwain, and G. H. Palmer. 1996. Expression of major surface protein 2 antigenic variants during acute Anaplasma marginale rickettsemia. Infect. Immun. 64:836–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French, D. M., W. C. Brown, and G. H. Palmer. 1999. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect. Immun. 67:5834–5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French, D. M., T. F. McElwain, T. C. McGuire, and G. H. Palmer. 1998. Expression of Anaplasma marginale major surface protein 2 variants during persistent cyclic rickettsemia. Infect. Immun. 66:1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ijdo, J. W., W. Sun, Y. Zhang, L. A. Magnarelli, and E. Fikrig. 1998. Cloning of the gene encoding the 44-kilodalton antigen of the agent of human granulocytic ehrlichiosis and characterization of the humoral response. Infect. Immun. 66:3264–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inokuma, H., Y. Terada, T. Kamio, D. Raoult, and P. Brouqui. 2001. Analysis of the 16S rRNA gene sequence of Anaplasma centrale and its phylogenetic relatedness to other ehrlichiae. Clin. Diagn. Lab. Immunol. 8:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieser, S. T., I. S. Eriks, and G. H. Palmer. 1990. Cyclic rickettsemia during persistent Anaplasma marginale infection of cattle. Infect. Immun. 58:1117–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krigel, Y., E. Pipano, and V. Shkap. 1992. Duration of carrier state following vaccination with live Anaplasma centrale. Trop. Anim. Health Prod. 24:209–210. [DOI] [PubMed] [Google Scholar]
- 14.Kuttler, K. 1967. A study of the immunological relationship of Anaplasma marginale and Anaplasma centrale. Res. Vet. Sci. 8:467–471. [PubMed] [Google Scholar]
- 15.Losos, G. J. 1986. Anaplasmosis, p.743–795. In G. J. Losos (ed.), Infectious tropical diseases of domestic animals. Longman House, Essex, United Kingdom.
- 16.Molloy, J. B., R. E. Bock, J. M. Templeton, A. G. Bruyeres, P. M. Bowles, G. W. Blight, and W. K. Jorgensen. 2001. Identification of antigenic differences that discriminate between cattle vaccinated with Anaplasma centrale and cattle naturally infected with Anaplasma marginale. Int. J. Parasitol. 31:179–186. [DOI] [PubMed] [Google Scholar]
- 17.Murphy, C. I., J. R. Storey, J. Recchia, L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, J. S. Bakken, R. T. Coughlin, and G. A. Beltz. 1998. Major antigenic proteins of the agent of human granulocytic ehrlichiosis are encoded by members of a multigene family. Infect. Immun. 66:3711–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer, G. H., J. R. Abbott, D. M. French, and T. F. McElwain. 1998. Persistence of Anaplasma ovis infection and conservation of the msp-2 and msp-3 multigene families within the genus Anaplasma. Infect. Immun. 66:6035–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer, G. H., A. F. Barbet, A. J. Musoke, J. Katende, F. R. Rurangirwa, V. Shkap, E. Pipano, W. C. Davis, and T. C. McGuire. 1988. Recognition of conserved surface protein epitopes on Anaplasma centrale and Anaplasma marginale isolates from Israel, Kenya and the United States. Int. J. Parasitol. 18:33–38. [DOI] [PubMed] [Google Scholar]
- 20.Palmer, G. H., W. C. Brown, and F. R. Rurangirwa. 2000. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. 2:167–176. [DOI] [PubMed] [Google Scholar]
- 21.Palmer, G. H., G. Eid, A. F. Barbet, T. C. McGuire, and T. F. McElwain. 1994. The immunoprotective Anaplasma marginale major surface protein 2 is encoded by a polymorphic multigene family. Infect. Immun. 62:3808–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer, G. H., S. M. Oberle, A. F. Barbet, W. C. Davis, W. L. Goff, and T. C. McGuire. 1988. Immunization with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect. Immun. 56:1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer, G. H., F. R. Rurangirwa, K. M. Kocan, and W. C. Brown. 1999. Molecular basis for vaccine development against the ehrlichial pathogen Anaplasma marginale. Parasitol. Today 15:281–286. [DOI] [PubMed] [Google Scholar]
- 24.Pipano, E., Y. Krigel, M. Frank, A. Marcovics, and E. Mayer. 1986. Frozen Anaplasma centrale vaccine against anaplasmosis in cattle. Br. Vet. J. 142:553–556. [DOI] [PubMed] [Google Scholar]
- 25.Potgeiter, F. T. 1979. Epizootiology and control of anaplasmosis in South Africa J. South Afr. Vet. Assoc. 50:367–372. [PubMed] [Google Scholar]
- 26.Rurangirwa, F. R., D. Stiller, D. M. French, and G. H. Palmer. 1999. Restriction of major surface protein 2 (MSP2) variants during tick transmission of the ehrlichia Anaplasma marginale. Proc. Natl. Acad. Sci. USA 96:3171–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shkap, V., E. Pipano, T. C. McGuire, and G. H. Palmer. 1991. Identification of immunodominant polypeptides common between Anaplasma centrale and A. marginale. Vet. Immunol. Immunopathol. 29:31–40. [DOI] [PubMed] [Google Scholar]
- 28.Theiler, A. 1910. Report of the Government Veterinary Bacteriologist 1908–1909, p.1–29. Department of Agriculture, Union of South Africa, Johannesburg, South Africa.
- 29.Theiler, A. 1911. Further investigations into anaplasmosis of South African cattle, p.7–46. First Report of the Director of Veterinary Research, Union of South Africa, Johannesburg, South Africa.
- 30.Theiler, A. 1912. Gallsickness of imported cattle and the protective inoculation against this disease. Agric. J. Union South Africa 3:7–46. [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torioni de Echaide, S., D. P. Knowles, T. C. McGuire, G. H. Palmer, C. E. Suarez, and T. F. McElwain. 1998. Detection of cattle naturally infected with Anaplasma marginale in a region of endemicity by nested PCR and a competitive enzyme-linked immunosorbent assay using recombinant major surface protein 5. J. Clin. Microbiol. 36:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turton, J. A., T. C. Katsande, M. B. Matingo, W. K. Jorgensen, U. Ushewokunze-Obatolu, and R. J. Dalgliesh. 1998. Observations on the use of Anaplasma centrale for immunization of cattle against anaplasmosis in Zimbabwe. Onderstepoort J. Vet. Res. 65:81–86. [PubMed] [Google Scholar]
- 34.Zakimi, S., N. Tsuji, and K. Fujisaki. 1994. Protein analysis of Anaplasma marginale and Anaplasma centrale by two-dimensional polyacrylamide gel electrophoresis. J. Vet. Med. Sci. 56:1025–1027. [DOI] [PubMed] [Google Scholar]
- 35.Zhi, N., N. Ohashi, Y. Rikihisa, H. W. Horowitz, G. P. Wormser, and K. Hechemy. 1998. Cloning and expression of the 44-kilodalton major outer membrane protein gene of the human granulocytic ehrlichiosis agent and application of the recombinant protein to serodiagnosis. J. Clin. Microbiol. 36:1666–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]