Abstract
We describe conditions for rolling-circle amplification (RCA) of individual DNA molecules 5–7 kb in size by >109-fold, using φ29 DNA polymerase. The principal difficulty with amplification of small amounts of template by RCA using φ29 DNA polymerase is “background” DNA synthesis that usually occurs when template is omitted, or at low template concentrations. Reducing the reaction volume while keeping the amount of template fixed increases the template concentration, resulting in a suppression of background synthesis. Cell-free cloning of single circular molecules by using φ29 DNA polymerase was achieved by carrying out the amplification reactions in very small volumes, typically 600 nl. This procedure allows cell-free cloning of individual synthetic DNA molecules that cannot be cloned in Escherichia coli, for example synthetic phage genomes carrying lethal mutations. It also allows cell-free cloning of genomic DNA isolated from bacteria. This DNA can be sequenced directly from the φ29 DNA polymerase reaction without further amplification. In contrast to PCR amplification, RCA using φ29 DNA polymerase does not produce mutant jackpots, and the high processivity of the enzyme eliminates stuttering at homopolymer tracts. Cell-free cloning has many potential applications to both natural and synthetic DNA. These include environmental DNA samples that have proven difficult to clone and synthetic genes encoding toxic products. The method may also speed genome sequencing by eliminating the need for biological cloning.
Keywords: DNA sequencing, rolling-circle amplification, synthetic DNA
Cloning foreign DNA sequences into vectors that can replicate in Escherichia coli or other host cells is arguably the defining technique of modern molecular genetics. However, this method has limitations, because certain sequences are difficult to propagate by using available vector/host systems. Some sequences are toxic to the host cell, whereas others are prone to replication errors. Cloning is also time-consuming. The isolation of a clone from a library and its preparation as a sequencing template typically takes longer than 2 days.
For these reasons, a cell-free enzymatic method for cloning and amplifying DNA sequences has long been appealing. The “polony” method for in situ PCR amplification of individual molecules (1) was designed to replace biological cloning by in vitro amplification with the aim of speeding large sequencing projects. However, PCR has certain characteristics that make it undesirable as the basis for a general cell-free cloning method. These include mutant “jackpots” that result when mutations arise during the early cycles of a PCR reaction and a lack of processivity leading to stuttering at sequences of low complexity, especially homopolymer tracts.
We describe here the use of φ29 DNA polymerase to clone individual molecules. This enzyme was attractive for cell-free cloning because it copies DNA with high fidelity (2), has a proofreading activity (3), and is highly processive (4). Our procedure is based on the rolling-circle amplification (RCA) reaction carried out isothermally by φ29 DNA polymerase at 30°C (5). Fig. 1 diagrams the stages of this reaction and is based on the work of Lasken and colleagues (5). Strands averaging 70 kb in length are copied by the enzyme (4). Therefore, products from small circular templates contain many tandem repeats of the starting molecule. These strands in turn serve as templates for new synthesis so that the reaction undergoes an exponential phase in which the rate of initiation of new strands is proportional to the quantity of DNA already formed. In this reaction, synthesis is primed by random hexanucleotides rendered resistant to the exonuclease activity of the polymerase by the incorporation of phosphothioate linkages. Background synthesis is commonly observed when φ29 polymerase is primed with such random hexamers, if the reaction is carried out in the absence of added template. This background synthesis, which is believed to be templated by the random hexamer primers (for example, see ref. 6), has made the amplification of very small amounts of DNA difficult. We reasoned that reducing the volume of a φ29 polymerase reaction, while keeping the amount of template constant, would increase the template concentration and suppress background synthesis. This approach led to success in cell-free cloning of individual DNA molecules by using submicroliter reaction volumes.
Fig. 1.
RCA by φ29 DNA polymerase, after Dean et al. (5). Arrowheads represent random hexamer primers. This drawing is not to scale; circular templates are typically 5 kb, and product strands average 70 kb in length.
Materials and Methods
DNA Preparations. φX174 am3cs70 single-stranded virion DNA and M13mp18 single-stranded virion DNA were obtained from NEB (Beverly, MA). The DNA referred to as synφX in the text was the ligated circular double-stranded synthetic φX174 DNA described by Smith et al. (7). The Mycoplasma genitalium genomic DNA library was prepared from DNA isolated from strain G37, obtained from the American Type Culture Collection in 1996. DNA sheared to 1.5–2.2 kb was cloned into a medium copy pBR322 derivative as described in ref. 8.
Enzymes. φ29 DNA polymerase was purchased from NEB in pure form at a concentration of 10,000 units/ml. Inorganic pyrophosphatase from United States Biochemical was supplied at 40 units/ml. Restriction enzymes were from NEB.
φ29 Polymerase Amplification Reactions. Reactions contain 37 mM Tris·HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 5 mM (NH4)2SO4, 1 mM each of the four dNTPs, 1 mM DTT, 1× BSA (NEB), 0.2% Tween 20, 1 unit/ml yeast pyrophosphatase, 540 units/ml φ29 DNA polymerase, and 50 μM exonuclease-resistant hexamer mix (Fidelity Systems, Gaithersburg, MD). All components except the enzymes and hexamers were made up as a 2× concentrated buffer (2xG-buffer). Immediately before setting up reactions, enzymes and H2O were added to 2xG-buffer, this was incubated for 10 min at room temperature, and the hexamers were added so that the concentration of all components was now 1.5× the final reaction conditions (this is φ29 Mix). Reactions were initiated by adding one volume of template DNA in TE buffer [10 mM Tris/1 mM EDTA (pH 8)] to two volumes of φ29 Mix. Reactions were carried out at 30°C. For our standard 600-nl reaction, 400 nl of φ29 Mix was added to a 0.5-ml PCR tube by using a 2-μl Pipetman (Gilson, Middleton, WI) with ART 10 Reach tips (Molecular BioProducts, San Diego). Then, 200 nl of template dilution was added and the reaction was mixed by pipetting up and down ≈20 times. Reactions were overlaid with 10 μl of bio-technology grade mineral oil (Bio-Rad) to prevent evaporation, then centrifuged briefly and checked visually to make sure the aqueous phase formed a small sphere at the bottom of the tube. Reactions were incubated in a thermocycler at 30°C for 6 h, then held at 4°C until analysis.
PCR was carried out in Advantage 2 buffer, using the Advantage HF 2 PCR kit (no. K1914-1, Clontech).
Restriction Analysis of Amplification Reactions. To digest the entire reaction, 20 μl of buffer 2 plus BSA (1×, NEB) containing 10 units of PstI was added to the reaction under mineral oil, and the tube was mixed briefly and incubated at 37°C for 30–60 min. The aqueous phase was removed and loaded onto an E-Gel (Invitrogen). Unless stated otherwise, 0.8% E-Gels were electrophoresed for 40 min at 60 V. DNA size markers were HyperLadder I (Bioline, Randolph, MA) with bands at 10 kb, 8 kb, 6 kb, 5 kb, 4 kb, 3 kb, 2.5 kb, 2 kb, 1.5 kb, 1 kb, 800 bp, 600 bp, 400 bp, and 200 bp. To analyze φ29 polymerase reactions by restriction digestion and also by DNA sequencing, the reactions were diluted with 10 μl of TE and then split for analysis.
DNA Sequencing. PCR and φ29 polymerase reaction products were treated with shrimp alkaline phosphatase (SAP) and E. coli exonuclease I (Exo I) to remove primers and dNTPs before use as sequencing templates. SAP-Exo I master mix contained 0.5 μl of SAP (Boehringer Mannheim, 1 unit/μl), 0.1 μl of Exo I (NEB, 20 units/μl), 0.5 ml of 10× SAP buffer, and 8.9 μl of H2O per reaction. To sequence from a PCR reaction, 8 μl of the reaction was added to 10 μl of SAP-Exo I master mix, and the solution was incubated at 37°C for 60 min, incubated at 72°C for 15 min, and held at 4°C until use in sequencing. To sequence directly from a φ29 polymerase reaction the 600-nl reaction was diluted with 10 μl of TE. Four microliters of the reaction was added to 5 μl of SAP-Exo I master mix, and the solution was incubated as for the PCR samples above. In both cases, 4 μl of the treated templates were sequenced in 8-μl reactions by using BigDye terminator chemistry (v3.1, Applied Biosystems) with 35 extension cycles of 4 min at 60°C. This high temperature prevents priming by any residual hexamers not degraded by Exo I treatment of φ29 polymerase reactions. Reaction products were isopropanol-precipitated, rinsed with ethanol, dissolved in 20 μl of H2O, and analyzed by using an Applied Biosystems capillary sequencer (Model 3100) equipped with 50-cm capillaries.
Results
As the amount of template DNA in a φ29 polymerase RCA reaction is decreased, the amount of background synthesis increases. If the template is circular φX174 single-stranded DNA (Fig. 2A), then PstI cleaves authentic amplification product at the unique site to produce 5.4-kb linear DNA. The background product, however, does not in general contain PstI sites and migrates as large DNA, making it easy to distinguish from the RCA product of φX174 (Fig. 2 A). As the reaction volume is decreased, keeping the amount of template DNA constant, background is dramatically decreased with a marked improvement in signal-to-noise ratio (Fig. 2B). Using 50 molecules of single-stranded M13 DNA (7.2 kb) as template, a PstI digest of a 3-μl φ29 polymerase reaction displayed a distinct band (>10 ng) of M13 linear. When template was omitted from the reaction, a similar amount of DNA was produced by background synthesis, but this DNA was not cleaved by PstI and migrated more slowly than the largest marker DNA (10 kb). Larger reactions (15 and 30 μl) produced so much background synthesis that no M13 product was visible.
Fig. 2.
Factors affecting background synthesis by φ29 polymerase. (A) Background increases with decreasing template concentration. Three-microliter φ29 polymerase reactions were primed by the indicated numbers of φX single-stranded DNA molecules, calculated on the basis of dilution factors from a stock of known concentration. Reactions were digested with PstI and analyzed as described above. (B) Background synthesis is suppressed by reducing φ29 polymerase reaction volume. φ29 polymerase reactions of 30, 15, and 3 μl were assembled, each containing either 50 M13 single-stranded DNA molecules or no template DNA. The reaction products were digested with PstI, and one-half of each reaction was analyzed by gel electrophoresis as described above. The products of background synthesis, not cleaved by PstI, migrate as large DNA, slower than the largest marker DNA (10 kb).
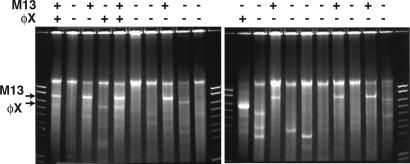
Using 600-nl φ29 polymerase reactions, we have amplified single molecules more than 109-fold to give 10 ng or more of product DNA, allowing easy visualization by ethidium bromide staining after gel electrophoresis. Fig. 3 displays gel electrophoresis of 20 duplicate 600-nl φ29 polymerase reactions digested with PstI. The template for each reaction was 200 nl of the same limiting dilution of a mixture of φX174 (5.4 kb) and M13 (7.2 kb) single-stranded DNAs. Each of these molecules contains a unique PstI site. Some lanes appear to contain neither φX nor M13 (11 of 20), some φX only (2 of 20), some M13 only (5 of 20), and some both (2 of 20). Cloning of φX174 and M13 from a mixture has been confirmed by PCR and real-time PCR analysis of reactions like those shown in Fig. 3, demonstrating that some reactions are positive for each of the molecules but completely negative for the other (data not shown). Some lanes in Fig. 3 display bands that are not the correct size for either φX174 or M13. This phenomenon is often seen in reactions to which no template is added, and we believe it results from the chance occurrence of PstI sites in the products of background synthesis. The template aliquots used in the reactions of Fig. 3 were estimated to contain an average of four φX174 molecules and two M13 molecules based on UV absorbance of the starting DNA samples before dilution. The fraction of molecules in the template dilution that participate in the amplification reaction varies somewhat from one dilution series to another but is fairly constant from day to day for the same DNA dilutions. When the DNA concentration is very low variability may result, in part, from adherence of molecules to tubes, pipettes, or particulate material.
Fig. 3.
Cell-free cloning of φX174 and M13 from a mixture. Twenty duplicate 600-nl φ29 polymerase reactions were made by using the same limiting dilution of a mixture of φX174 and M13 DNAs (see text). The reaction products were digested with PstI and analyzed as described above. The + and – symbols above the gel indicate the presence or absence of bands with the molecular weights of linear φX174 (5.4 kb) or M13 (7.2 kb) DNAs. The products of background synthesis, not cleaved by PstI, migrate as large DNA, slower than the largest marker DNA (10 kb).
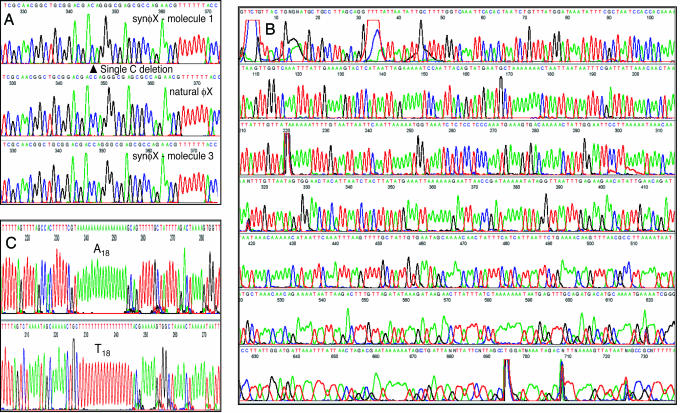
We also succeeded in cell-free cloning of double-stranded circular DNA. The synthetic φX174 genome preparation that we described previously (7) was heated to 95°C and then quick-cooled to 0°C, to promote priming by random hexamers. About 1 in 104 molecules in this preparation is an infectious phage genome, leading to an estimate of ≈10 lethal mutations per molecule (7). Fig. 4A shows a portion of the sequencing chromatograms from two molecules cloned by φ29 polymerase amplification. For comparison, we also sequenced a commercial preparation of natural φX174 DNA. We sequenced ≈1 kb from seven different molecules. Each had a distinct set of sequence differences from the natural DNA preparation. As an example, Fig. 4A displays a deletion of a single nucleotide (C) in molecule 1. Molecule 3 and the natural DNA both have the wild-type sequence at that position, as do the other five molecules sequenced (data not shown). The sequence reads for the cell-free φX174 clones are accurate to beyond 700 bp (see the supporting information, which is published on the PNAS web site).
Fig. 4.
DNA sequencing of φ29 polymerase clones. (A) Sequencing of cell-free clones of synthetic φX174 molecules. Sequencing was performed after PCR amplification of single-molecule φ29 polymerase reactions. The same region is compared for a synthetic φX clone with a single base deletion (molecule 1), one with the wild-type sequence in this region (molecule 3), and natural φX DNA. Signal strengths were as follows: molecule 1, A = 602, C = 607, G = 512, and T = 459; molecule 3, A = 324, C = 331, G = 292, and T = 300; natural φX, A = 1,242, C = 1,430, G = 1,199, and T = 1,154. (B) Direct sequencing of φ29 polymerase reaction products without PCR amplification. This figure shows the result of sequencing 18% of the product from a 600-nl reaction as described in Materials and Methods. Signal strengths were A = 48, C = 25, G = 29, and T = 52. (C) Sequence reads through an A18 tract and the complementary T18 tract on a molecule amplified by φ29 polymerase. Sequencing was performed on 18% of a 600-nl φ29 polymerase reaction, without PCR amplification. Primers, designed on the basis of the M. genitalium genome sequence, were 5′-GTTAAAGGGCGACTAATAG-3′ for the A18 sequence and 5′-AACTTAATACTTTGGTCAG-3′ for the T18 sequence. Signal strengths were A = 202, C = 104, G = 120, and T = 226 for the A18 sequence and A = 107, C = 57, G = 65, and T = 121 for the T18 sequence.
We were also able to clone segments of bacterial genomic DNA in 600-nl φ29 polymerase reactions. We started with a conventional genomic library of M. genitalium DNA sheared to ≈2 kb and ligated into a medium copy number pBR322 derivative (8). It should be noted that this M. genitalium DNA library was never propagated in E. coli. For the purposes of the present experiment, circularization of the M. genitalium DNA fragments is essential, but properties of the vector are irrelevant. The library DNA was diluted to give φ29 polymerase reactions primed by individual molecules. Fig. 5 Upper shows the analysis of such reactions cleaved with PstI. Because the vector is 3.5 kb in size, we expect the clones to have a total size of ≈5.5 kb. Most positive lanes in Fig. 5 have a band in this size range, but the sizes vary somewhat, as would be expected because the cloned DNA was not perfectly homogeneous in size. The inserts were amplified by PCR using the M13 forward and reverse sequencing primer sites that flank the cloning sites in the vector (Fig. 5 Lower). Some lanes contain two bands of slightly different sizes, indicating that two molecules were amplified in those reactions. PCR products that appeared on the gel to be pure species were sequenced from both ends by using the M13 sequencing primers. Six of seven cloned molecules gave clearly readable sequence that matched the M. genitalium genome sequence perfectly throughout the readable range (out to ≈650 bp or greater; see the supporting information). One of the seven reactions appeared to contain a mixture of two sequences. The products of 600-nl φ29 polymerase reactions can also be sequenced directly, without PCR amplification. The chromatogram for such a sequence is shown in Fig. 4B, which displays a sequence that matches the M. genitalium genome accurately to beyond 750 bp (see the supporting information).
Fig. 5.
Cell-free cloning of M. genitalium genomic DNA. A library consisting of 1.5- to 2.5-kb fragments of M. genitalium DNA ligated to a pBR322 derivative was diluted 100-fold in TE from an initial DNA concentration of ≈0.3 ng/μl, heated at 95°C for 2 min, and quenched on ice. The DNA was diluted an additional 1,000-fold (Upper Left) or 2,000-fold (Upper Right) and used as template in two sets of 600-nl φ29 polymerase reactions (eight duplicate reactions per set), as described in Materials and Methods. One-half of each reaction was cleaved with PstI and analyzed by gel electrophoresis (Upper). Inserts were amplified by PCR from the remaining portion of each reaction, using M13 forward and reverse sequencing primers that flank the cloning site in the vector, and analyzed on the lower gels.
The high processivity of φ29 polymerase allows accurate amplification of homopolymer tracts from single molecules. DNA from an M. genitalium clone containing an A18/T18 tract was diluted and used to prime single-molecule φ29 polymerase reactions. The DNA from these reactions was sequenced directly and portions of the chromatograms are shown in Fig. 4C. One strand shows a run of 18 A residues, and the complementary strand shows 18 T residues, as expected. The sequence is clearly readable beyond the homopolymer run on both strands and matches the M. genitalium genome sequence accurately for the readable length of the run (>700 bp; see the supporting information).
Discussion
Much recent work has focused on the use of φ29 polymerase to amplify small amounts of DNA. Sequencing templates can be produced by amplification of plasmids from single bacterial colonies using RCA (9). Also, useful amounts of genomic DNA can be amplified from small numbers of cells by using the enzyme in the multiple displacement amplification (MDA) reaction (10). Cell-free cloning of small circular DNA involves even smaller amounts of template (<10–17 g for a 5-kb DNA molecule), and background synthesis, which occurs in the absence of added template when φ29 polymerase is primed by random hexamers, becomes a serious problem. Using submicroliter reaction volumes, we have amplified single circular DNA molecules to give amounts of DNA that can easily be visualized by gel electrophoresis and can be sequenced. The use of small reactions increases the template concentration and suppresses the amount of background synthesis. Reactants do not appears to be limiting in these reactions, which contain enough of the four dNTPs to produce ≈1 μg of DNA per μl of reaction volume. It appears to be a fortunate coincidence that reduction of commonplace reaction volumes by only about one order of magnitude allows cell-free cloning, by using reaction volumes near the lower limit for conventional pipettes. In this article, we describe the use of 600-nl reactions that can be prepared by using conventional manual pipetting devices. However, preliminary results indicate that it is advantageous to reduce volumes even further (unpublished data). Robotic nanoliter pipetting devices [for example, Parallab 350 (Brooks Automation, Chelmsford, MA] may yield improvements in the method and make its large-scale application more practical.
The only known requirement for a DNA to be cloned by φ29 polymerase is that it is circular, and of such a size that RCA can produce multiple tandem copies of the molecule. In principle, no vector is needed, because a molecule could be circularized in a blunt-end ligation reaction. Alternatively, special φ29 cloning vectors can be designed to facilitate circularization of the target DNA and subsequent use of the products for sequencing or other purposes (unpublished results). Such φ29 cloning vectors can be quite small (a few hundred base pairs or less) because they do not need to support replication in bacteria. The largest molecule we have cloned with φ29 polymerase is M13 (7.2 kb), but we have not tested larger molecules, and it seems likely that circular DNA 10 kb or larger might be cloned by this method. It also seems likely that the use of very small reaction volumes would be useful in the amplification of genomic DNA from single cells by the MDA reaction.
One application of the method described here is the cell-free cloning and sequencing of single synthetic DNA molecules without propagation in bacteria. This method can be used for molecules that are unclonable in E. coli. For example, synthetic φX174 genomes containing frameshifts in essential genes can be cloned by using φ29 polymerase (Fig. 4A). These genomes cannot be propagated as phages or, in general, propagated in E. coli by cloning in plasmid vectors either, because several of the φX174 gene products are toxic to E. coli. We therefore expect that cell-free cloning will be valuable for dealing with synthetic DNA that is difficult to clone by conventional methods.
In applications of cell-free cloning to synthetic DNA it is important that the amplified DNA has exactly the sequence of the starting molecule. We demonstrated that molecules cloned by φ29 polymerase give accurate DNA sequencing results. However, sequencing determines the consensus sequence and does not detect random mutations that occur during amplification. It is therefore of interest to try to estimate the mutation frequency in the φ29 product amplified from a single molecule. However, this analysis is not straightforward because RCA is not a simple doubling process. Consequently, the usual equation, which relates polymerase error rate, mutation frequency, and number of doublings, does not apply. Because many copies are made directly from the original input molecule, jackpots of mutants should not be a problem because they are with mutations arising during the first cycles of a PCR reaction. The φ29 polymerase appears to have an accuracy comparable to other polymerases with a 3′ proofreading exonuclease (2), which are generally in the range of 10–6 to 10–8 (11). Calculations based on published estimates of the φ29 polymerase error rate (2, 9, 11), and reasonable models of the reaction, indicate that the majority of molecules resulting from 109-fold amplification of a 5-kb circular DNA by φ29 polymerase should have exactly the same sequence as the parental molecule (see the supporting information).
Cell-free cloning may also be applied to facilitate DNA sequencing projects. It should be useful in situations where cloning in E. coli is difficult or impossible. For example, some environmental DNA samples are difficult to clone in E. coli (for example, see ref. 12). Cloning using φ29 DNA polymerase should be useful in the sequence analysis of such samples. In conventional genome sequencing projects the method could substantially reduce the time required for preparation of sequencing templates and also reduce problems with unclonable sequences. Mate-paired sequence reads from the ends of φ29 polymerase cell-free clones should also facilitate the assembly of data generated by a new generation of sequencers such as the Genome Sequencer 20 (454 Life Sciences/Roche Applied Science) (13, 14). Such devices have extremely high throughput and do not rely on E. coli cloning but currently produce only short read lengths of lower quality than conventional methods. Combining such “454 data” with φ29 polymerase cell-free clone data could potentially result in an extremely rapid and accurate genome sequencing method.
Supplementary Material
Acknowledgments
We thank members of the Synthetic Biology group for helpful discussions, John Glass for help with real-time PCR, and Mahir Maruf for preparation of plasmid DNA. This work was supported by grants from the U.S. Department of Energy and the J. Craig Venter Science Foundation.
Conflict of interest statement: No conflicts declared.
Abbreviation: RCA, rolling-circle amplification.
References
- 1.Mitra, R. D. & Church, G. M. (1999) Nucleic Acids Res. 27, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteban, J. A., Salas, M. & Blanco, L. (1993) J. Biol. Chem. 268, 2719–2726. [PubMed] [Google Scholar]
- 3.Garmendia, C., Bernad, A., Esteban, J. A., Blanco, L. & Salas, M. (1992) J. Biol. Chem. 267, 2594–2599. [PubMed] [Google Scholar]
- 4.Blanco, L., Bernad, A., Lazaro, J. M., Martin, G., Garmendia, C. & Salas, M. (1989) J. Biol. Chem. 264, 8935–8940. [PubMed] [Google Scholar]
- 5.Dean, F. B., Nelson, J. R., Giesler, T. L. & Lasken, R. S. (2001) Genome Res. 11, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holbrook, J. F., Stabley, D. & Sol-Church, K. (2005) J. Biomol. Tech. 16, 125–133. [PMC free article] [PubMed] [Google Scholar]
- 7.Smith, H. O., Hutchison, C. A., III, Pfannkoch, C. & Venter, J. C. (2003) Proc. Natl. Acad. Sci. USA 100, 15440–15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 9.Nelson, J. R., Cai, Y. C., Giesler, T. L., Farchaus, J. W., Sundaram, S. T., Ortiz-Rivera, M., Hosta, L. P., Hewitt, P. L., Mamone, J. A., Palaniappan, C. & Fuller, C. W. (2002) BioTechniques 32, Suppl., 44–47. [PubMed] [Google Scholar]
- 10.Sato, M., Ohtsuka, M. & Ohmi, Y. (2005) Biomol. Eng. 22, 129–132. [DOI] [PubMed] [Google Scholar]
- 11.Kunkel, T. A. (2004) J. Biol. Chem. 279, 16895–16898. [DOI] [PubMed] [Google Scholar]
- 12.Breitbart, M., Salamon, P., Andresen, B., Mahaffy, J. M., Segall, A. M., Mead, D., Azam, F. & Rohwer, F. (2002) Proc. Natl. Acad. Sci. USA 99, 14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., Berka, J., Braverman, M. S., Chen, Y. J., Chen, Z., et al. (2005) Nature 437, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers, Y. H. & Venter, J. C. (2005) Nature 437, 326–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.