Abstract
Abiotic stresses cause extensive losses to agricultural production worldwide. Acclimation of plants to abiotic conditions such as drought, salinity, or heat is mediated by a complex network of transcription factors and other regulatory genes that control multiple defense enzymes, proteins, and pathways. Associated with the activity of different transcription factors are transcriptional coactivators that enhance their binding to the basal transcription machinery. Although the importance of stress-response transcription factors was demonstrated in transgenic plants, little is known about the function of transcriptional coactivators associated with abiotic stresses. Here, we report that constitutive expression of the stress-response transcriptional coactivator multiprotein bridging factor 1c (MBF1c) in Arabidopsis (Arabidopsis thaliana) enhances the tolerance of transgenic plants to bacterial infection, heat, and osmotic stress. Moreover, the enhanced tolerance of transgenic plants to osmotic and heat stress was maintained even when these two stresses were combined. The expression of MBF1c in transgenic plants augmented the accumulation of a number of defense transcripts in response to heat stress. Transcriptome profiling and inhibitor studies suggest that MBF1c expression enhances the tolerance of transgenic plants to heat and osmotic stress by partially activating, or perturbing, the ethylene-response signal transduction pathway. Present findings suggest that MBF1 proteins could be used to enhance the tolerance of plants to different abiotic stresses.
Abiotic stress conditions cause extensive losses to agricultural production worldwide (Boyer, 1982; Bray et al., 2000; Hoerling and Kumar, 2003; Rosegrant and Cline, 2003; Peters et al., 2004). Key to the tolerance of plants to abiotic stresses is a complex network of transcription factors and other regulatory genes that control multiple defense enzymes, proteins, and pathways (Bray et al., 2000; Cushman and Bohnert, 2000). Although the important role of many stress-response transcription factors was demonstrated in transgenic plants subjected to abiotic stresses (e.g. Kasuga et al., 1999; Mishra et al., 2002; Maruyama et al., 2004; Vogel et al., 2005), little is known about the function of other components of the plant transcriptional machinery during stress.
Transcriptional coactivators play a crucial role in eukaryotic gene expression by communicating between transcription factors and/or other regulatory components and the basal transcription machinery. They are divided into two classes: transcriptional coactivators that recruit or possess enzymatic activities that modify chromatin structure (e.g. acetylation of histone) and transcriptional coactivators that recruit the general transcriptional machinery to a promoter where a transcription factor(s) is bound (Näär et al., 2001). Multiprotein bridging factor 1 (MBF1) is a highly conserved transcriptional coactivator involved in the regulation of diverse processes such as endothelial cell differentiation, hormone-regulated lipid metabolism, central nervous system development, and His metabolism (Takemaru et al., 1997, 1998; Brendel et al., 2002; Liu et al., 2003). MBF1 proteins from different organisms interact with transcription factors such as c-Jun, GCN4, and ATF1, or with different nuclear receptors, and link them with the TATA-binding protein (Takemaru et al., 1997, 1998; Brendel et al., 2002; Busk et al., 2003; Liu et al., 2003). The flowering plant Arabidopsis (Arabidopsis thaliana) contains three different genes encoding MBF1. Functional assays demonstrate that all three Arabidopsis genes can complement MBF1 deficiency in yeast (Tsuda et al., 2004). MBF1a (At2g42680) and MBF1b (At3g58680) are developmentally regulated (Tsuda and Yamazaki, 2004). In contrast, the steady-state level of transcripts encoding MBF1c (At3g24500) is specifically elevated in Arabidopsis in response to pathogen infection, salinity, drought, heat, hydrogen peroxide, and application of the plant hormones abscisic acid or salicylic acid (Rizhsky et al., 2004b; Tsuda and Yamazaki, 2004; a search of 1,800 ATH1 chips at https://www.genevestigator.ethz.ch/). The level of transcripts encoding MBF1c or its orthologs is also elevated in response to a combination of drought and heat in Arabidopsis, tobacco (Nicotiana tabacum), and the desert legume Retama raetam (Pnueli et al., 2002; Rizhsky et al., 2002, 2004b). However, the relative contribution of MBF1c to biotic and abiotic stress tolerance is unknown.
Here, we report that constitutive expression of the transcriptional coactivator MBF1c in Arabidopsis enhances the tolerance of transgenic plants to bacterial infection, salinity, heat, and osmotic stress, and that the enhanced tolerance of transgenic plants to heat and osmotic stress is maintained even when these two stresses are combined. We further show that MBF1 expression enhances the tolerance of transgenic plants to heat and osmotic stress by partially activating, or perturbing, the ethylene-response signal transduction pathway. MBF1 proteins could, therefore, be used to enhance the tolerance of plants to different abiotic stresses.
RESULTS
Production and Characterization of Transgenic Arabidopsis Plants Expressing MBF1c
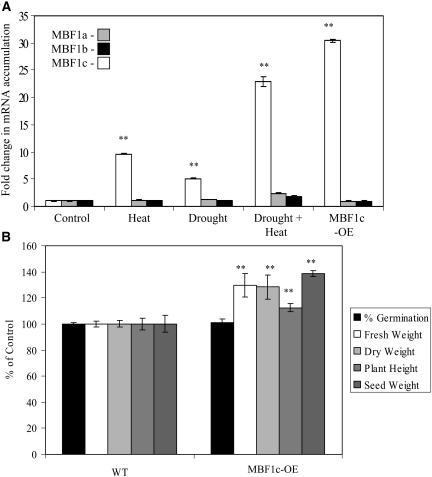
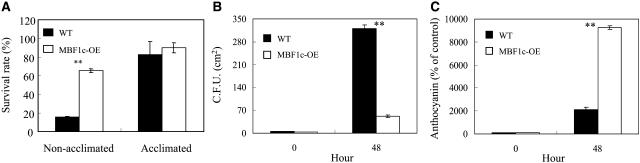
To test the function of MBF1c in Arabidopsis, we generated transgenic plants that constitutively express MBF1c under the control of the 35S cauliflower mosaic virus promoter and subjected them to biotic or abiotic stresses. Transcripts encoding MBF1c accumulated in transgenic plants grown under controlled conditions to levels that were comparable to or higher than those detected in wild-type plants subjected to heat stress, drought, or a combination of heat stress and drought (Fig. 1A; Supplemental Fig. 1; Rizhsky et al., 2004b). In contrast, the level of transcripts encoding MBF1a or MBF1b did not significantly accumulate in wild-type plants subjected to heat stress, drought, or a combination of heat stress and drought (Fig. 1A). The expression of MBF1a or MBF1b was also not altered in transgenic plants expressing MBF1c (MBF1c-OE; Fig. 1A). Transgenic plants expressing MBF1c appeared similar in their growth and development to wild-type plants. However, as shown in Figure 1B, transgenic plants expressing MBF1c were 20% larger than control plants and produced more seeds. In addition, compared to wild-type plants, transgenic plants expressing MBF1c bolted 2 to 3 d earlier. As shown in Figure 2A, the basal thermotolerance of 4- to 5-d-old MBF1c-expressing seedlings was higher than that of wild-type seedlings of similar age and size (measured as survival rate following a 2-h 45°C heat stress). As shown in Figure 2B, 2-week-old MBF1c-expressing plants were more resistant than wild-type plants to bacterial growth (measured as suppressed in planta bacterial population size following inoculation of plants with 50 colony forming units [cfu] cm−2). In addition, when grown under high-light conditions (1,000 μmol m−2 s−1 for 48 h), 2-week-old MBF1c-expressing plants accumulated a higher level of anthocyanin than wild type (Fig. 2C). In contrast, the tolerance of 4- to 5-d-old transgenic seedlings expressing MBF1c to cold stress was similar to that of wild-type seedlings (data not shown).
Figure 1.
Characterization of transgenic plants expressing MBF1c (MBF1c-OE). A, Relative expression of MBF1 transcripts (MBF1a, At2g42680; MBF1b, At3g58680; MBF1c, At3g24500) in transgenic plants expressing MBF1c (MBF1c-OE) or wild-type plants subjected to heat stress, drought, or a combination of heat stress and drought (after Rizhsky et al. [2004b]). B, Growth and productivity of wild-type and MBF1c-expressing (MBF1c-OE) transgenic plants. Plants were grown at 21°C, 14-h light cycle, 100 μmol m−2 s−1, and a relative humidity of 70%. Production of transgenic plants and RNA blots were performed as described in “Materials and Methods.” **, Student's t test significant at P < 0.01.
Figure 2.
Enhanced tolerance of transgenic plants expressing MBF1c (MBF1c-OE) to heat stress and bacterial growth. A, Survival rates of wild-type and transgenic seedlings in response to heat stress (45°C for 2 h), showing enhanced basal thermotolerance of MBF1c-expressing plants. B, In planta bacterial population measurements showing enhanced resistance of MBF1c-expressing plants to Pseudomonas syringae inoculation. Bacteria (50 cfu cm−2 prepared in water) was infiltrated into leaves with a syringe. Forty-eight hours after inoculation, bacteria was extracted from leaves, plated on agar plates, and scored for cfu cm−2. C, Augmented accumulation of anthocyanins in MBF1c-expressing plants in response to light stress (1,000 μmol m−2 s−1 for 48 h). Stress assays and pathogen infection were performed as described in “Materials and Methods.” **, Student's t test significant at P < 0.01.
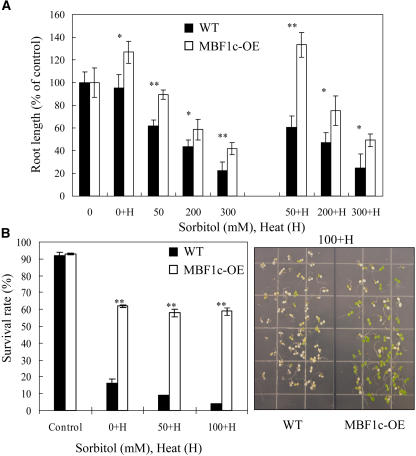
To study the tolerance of MBF1c-expressing plants to osmotic or salinity stress, we subjected 4- to 5-d-old wild-type and MBF1c-expressing seedlings to these stresses on agar plates and measured their root growth. In addition, to examine whether the tolerance of transgenic plants to these stresses is maintained when they are combined with heat stress, we subjected 4- to 5-d-old seedlings to a combination of heat and osmotic stress and to a combination of heat and salinity stress. As shown in Figure 3A, MBF1c-expressing plants were more tolerant than wild-type plants to heat or osmotic stress. Furthermore, the tolerance of transgenic plants to heat or osmotic stress was maintained even when these two stresses were combined (Fig. 3A). MBF1c-expressing plants were more tolerant than wild-type plants to a low level of salinity stress (i.e. 50 mm; data not shown). In contrast to the results obtained with heat and osmotic stress combination (Fig. 3A), the tolerance of MBF1c-expressing plants to salinity stress was not maintained when salinity stress was combined with heat stress (data not shown). As shown in Figure 3, B and C, the tolerance of transgenic plants expressing MBF1c to a combination of osmotic and heat stress was also evident from survival assays in which transgenic plants were compared to wild-type plants in survival assays similar to those shown in Figure 2A. In contrast, the survival rate of transgenic seedlings expressing MBF1c to a combination of salinity and heat stress was similar to that of wild type (data not shown).
Figure 3.
Enhanced tolerance of transgenic seedlings expressing MBF1c (MBF1c-OE) to heat stress, osmotic stress, or a combination of osmotic and heat stress. A, Root growth of wild-type and transgenic seedlings subjected to heat stress (38°C, 48 h), osmotic stress (sorbitol, 50, 200, and 300 mm), or their combination. B, Survival rate measurements of MBF1c-expressing seedlings subjected to heat stress (45°C for 2 h) or heat stress combined with osmotic stress (sorbitol, 50, 100 mm). C, A photograph of wild-type and transgenic seedlings subjected to heat stress (45°C for 2 h) combined with osmotic stress (sorbitol, 100 mm). Stress assays were performed as described in “Materials and Methods.” **, Student's t test significant at P < 0.01; *, Student's t test significant at P < 0.05.
Transcriptional Profiling of MBF1c-Expressing Plants
To test whether constitutive expression of MBF1c in transgenic plants results in the accumulation of different stress-response transcripts under controlled conditions, similar to the effect of constitutively expressing a defense-response transcription factor (e.g. Maruyama et al., 2004; Vogel et al., 2005), we performed transcriptional profiling of wild-type and transgenic plants. Table I summarizes all transcripts with a known or putative function elevated in MBF1c-expressing plants under controlled conditions. As shown in Table I, the constitutive expression of MBF1c resulted in the accumulation of transcripts encoding a number of stress-response transcription factors and signal transduction genes. These include WRKY and CBF-like transcription factors, MAPK3/11, and calcium-binding proteins. Ethylene was shown to play an important role in the defense response of plants against heat stress (Larkindale et al., 2005). In this respect it was interesting to find that the steady-state level of a number of transcripts involved in ethylene signaling was elevated in MBF1c-expressing plants. These include transcripts encoding ethylene-response-binding factors and the rate-limiting ethylene biosynthesis enzyme 1-aminocyclopropane-1-carboxylic acid (ACC) synthase. Although a relatively large number of transcripts enhanced by ethylene treatment of wild-type plants (e.g. De Paepe et al., 2004) did not accumulate in transgenic plants expressing MBF1c, compared to the transcriptome of wild-type plants subjected to different abiotic stresses, ethylene-response transcripts were overrepresented in the transcriptome of transgenic plants expressing MBF1c (Table I; Supplemental Fig. 2).
Table I.
Transcripts elevated in MBF1c-expressing plants under controlled conditions
Transcripts with a putative or known function significantly elevated in transgenic plants expressing MBF1c compared to wild-type plants (cutoff >1.5 log2). ATH1 and AGI locus identification numbers are given on left. Transcript annotation and fold change in log2 are given on right. Plant growth under controlled conditions, transcriptome profiling, and data analysis were performed as described in “Materials and Methods.” A complete list of all transcripts significantly elevated or suppressed in MBF1c-expressing plants (cutoff >1.5 log2) is included in Supplemental Tables I and II, respectively. Transcripts indicated by a “*” were also found to be elevated in wild-type plants in response to heat stress or a combination of drought and heat stress (Rizhsky et al., 2004b).
| Array Element | Locus Identifier | Fold (log2) | sd | Annotation |
|---|---|---|---|---|
| Transgene | ||||
| 258133_at | AT3G24500 | 4.70 | 0.40 | MBF1c* |
| Transcription factors and signal transduction transcripts | ||||
| 248400_at | AT5G52020 | 3.50 | 1.35 | APETALA2 (AP2) domain transcription factor |
| 258947_at | AT3G01830 | 3.07 | 0.12 | Calmodulin-like |
| 249197_at | AT5G42380 | 2.93 | 0.06 | Similar to calmodulin |
| 255937_at | AT1G12610 | 2.90 | 0.10 | Similar to transcriptional activator CBF1 |
| 261984_at | AT1G33760 | 2.87 | 0.15 | AP2 domain-containing transcription factor |
| 262360_at | AT1G73080 | 2.60 | 1.14 | Leu-rich repeat transmembrane protein kinase |
| 259879_at | AT1G76650 | 2.60 | 0.26 | Calmodulin |
| 258682_at | AT3G08720 | 2.47 | 0.45 | Ser/Thr protein kinase (PK19) |
| 264147_at | AT1G02205 | 2.20 | 0.17 | Receptor-like protein glossy1 (gl1) |
| 255511_at | AT4G02075 | 2.17 | 0.31 | Zinc finger (C3HC4-type RING finger) |
| 257919_at | AT3G23250 | 2.17 | 0.16 | Myb family transcription factor (MYB15) |
| 249862_at | AT5G22920 | 2.13 | 0.12 | Zinc finger (C3HC4-type RING finger) |
| 261892_at | AT1G80840 | 1.97 | 0.21 | WRKY40 |
| 251745_at | AT3G55980 | 1.93 | 0.12 | Zinc-finger transcription factor (PEI1) |
| 247137_at | AT5G66210 | 1.90 | 0.17 | Calcium-dependent protein kinase |
| 267028_at | AT2G38470 | 1.87 | 0.15 | WRKY33 |
| 253915_at | AT4G27280 | 1.87 | 0.06 | Calcium-binding EF hand protein |
| 253485_at | AT4G31800 | 1.87 | 0.15 | WRKY18 |
| 248389_at | AT5G51990 | 1.83 | 0.31 | DRE-binding protein/CRT/DRE-binding factor |
| 252193_at | AT3G50060 | 1.80 | 0.10 | R2R3-MYB transcription factor |
| 250099_at | AT5G17300 | 1.77 | 0.29 | Myb family transcription factor |
| 257022_at | AT3G19580 | 1.77 | 0.12 | Zinc-finger (C2H2 type) protein 2 |
| 246028_at | AT5G21170 | 1.77 | 0.06 | 5′-AMP-activated protein kinase |
| 265737_at | AT2G01180 | 1.73 | 0.21 | Phosphatidic acid phosphatase family protein |
| 259428_at | AT1G01560 | 1.70 | 0.26 | Mitogen-activated protein kinase 11 |
| 260856_at | AT1G21910 | 1.70 | 0.26 | AP2 domain-containing transcription factor |
| 245247_at | AT4G17230 | 1.70 | 0.20 | Scarecrow-like transcription factor 13 |
| 251259_at | AT3G62260 | 1.70 | 0.17 | Protein phosphatase 2C |
| 258188_at | AT3G17790 | 1.70 | 0.10 | Acid phosphatase type 5 |
| 255568_at | AT4G01250 | 1.67 | 0.15 | WRKY family transcription factor |
| 267460_at | AT2G33810 | 1.67 | 0.12 | Squamosa-promoter binding protein like 3 |
| 248606_at | AT5G49450 | 1.60 | 0.10 | bZIP transcription factor |
| 246777_at | AT5G27420 | 1.60 | 0.10 | RING-H2 zinc finger protein |
| 267083_at | AT2G41100 | 1.57 | 0.12 | Touch-responsive, calmodulin-related protein 3 |
| 263783_at | AT2G46400 | 1.57 | 0.06 | WRKY46 |
| 247426_at | AT5G62570 | 1.53 | 0.15 | Calmodulin-binding protein |
| 252592_at | AT3G45640 | 1.50 | 0.17 | Mitogen-activated protein kinase 3 |
| 251636_at | AT3G57530 | 1.50 | 0.10 | Calcium-dependent protein kinase |
| Ethylene-associated transcripts | ||||
| 253259_at | AT4G34410 | 2.93 | 0.15 | Ethylene-responsive element binding |
| 257918_at | AT3G23230 | 2.70 | 0.12 | Ethylene-responsive element binding protein 4 |
| 266821_at | AT2G44840 | 2.13 | 0.15 | Ethylene-response element binding protein |
| 245250_at | AT4G17490 | 2.07 | 0.12 | Ethylene-responsive element binding factor-like protein 6 |
| 261470_at | AT1G28370 | 1.90 | 0.10 | Ethylene-responsive element binding factor |
| 248448_at | AT5G51190 | 1.83 | 0.12 | Ethylene-responsive element binding factor |
| 254926_at | AT4G11280 | 1.70 | 0.06 | ACC synthase 6 |
| 248799_at | AT5G47230 | 1.50 | 0.17 | Ethylene-responsive element binding factor 5 |
| Pathogen and stress-associated transcripts | ||||
| 253161_at | AT4G35770 | 3.60 | 0.35 | Senescence-associated, sen1 |
| 245757_at | AT1G35140 | 3.13 | 0.15 | Phosphate-responsive protein phi-1 |
| 252368_at | AT3G48520 | 2.70 | 0.10 | Cytochrome P450* |
| 264153_at | AT1G65390 | 2.40 | 0.20 | Disease resistance protein RPS4 |
| 261037_at | AT1G17420 | 2.30 | 0.10 | Lipoxygenase |
| 260399_at | AT1G72520 | 2.27 | 0.21 | Lipoxygenase |
| 255064_at | AT4G08950 | 2.27 | 0.06 | Phosphate-responsive protein |
| 266072_at | AT2G18700 | 2.23 | 0.06 | Trehalose-6-P synthase |
| 248964_at | AT5G45340 | 2.20 | 0.10 | Cytochrome P450 |
| 256763_at | AT3G16860 | 2.17 | 0.15 | Phytochelatin synthetase related |
| 249645_at | AT5G36910 | 2.13 | 0.15 | Thionin Thi2.2 |
| 246099_at | AT5G20230 | 2.07 | 0.12 | Blue copper-binding protein |
| 251804_at | AT3G55430 | 2.00 | 0.10 | β-1,3-Glucanase PR-2 |
| 246114_at | AT5G20250 | 1.93 | 0.06 | Raffinose synthase |
| 264213_at | AT1G65390 | 1.90 | 0.26 | Disease resistance protein (TIR class) |
| 265648_at | AT2G27500 | 1.87 | 0.15 | β-1,3-Glucanase PR-2 |
| 263019_at | AT1G23870 | 1.83 | 0.72 | Glycosyl transferase/trehalose-phosphatase |
| 247280_at | AT5G64260 | 1.83 | 0.12 | Phosphate-responsive protein phi-1 |
| 265111_at | AT1G62510 | 1.80 | 0.10 | Protease inhibitor/seed storage/lipid transfer* |
| 261901_at | AT1G80920 | 1.80 | 0.00 | DNAJ N-terminal domain protein |
| 256526_at | AT1G66090 | 1.77 | 0.06 | Disease resistance protein, RPP1-WsA |
| 247279_at | AT5G64310 | 1.70 | 0.10 | Arabinogalactan protein 1 |
| 261443_at | AT1G28480 | 1.67 | 0.12 | Glutaredoxin |
| 262382_at | AT1G72920 | 1.63 | 0.15 | Disease resistance protein (TIR-NBS class) |
| 249746_at | AT5G24590 | 1.60 | 0.26 | Turnip crinkle virus-interacting protein |
| 245668_at | AT1G28330 | 1.60 | 0.10 | Dormancy-associated protein, putative |
| 259443_at | AT1G02360 | 1.57 | 0.25 | Chitinase PR-3 |
| 249264_s_at | AT5G41740 | 1.57 | 0.21 | Disease resistance protein (TIR-NBS-LRR class) |
| 264339_at | AT1G70290 | 1.57 | 0.15 | Trehalose-6-P synthase |
| Metabolism, development, and cell structure | ||||
| 248622_at | AT5G49360 | 3.60 | 0.10 | Glycosyl hydrolase family 3 protein |
| 247866_at | AT5G57550 | 2.23 | 0.15 | Xyloglucan:xyloglucosyl transferase |
| 262456_at | AT1G11260 | 2.13 | 0.06 | Glucose transporter (STP1) |
| 256300_at | AT1G69490 | 2.10 | 0.26 | No apical meristem family protein |
| 256772_at | AT3G13750 | 2.07 | 0.31 | β-Galactosidase, putative/lactase |
| 265680_at | AT2G32150 | 1.83 | 0.21 | Haloacid dehalogenase-like hydrolase |
| 251774_at | AT3G55830 | 1.80 | 0.10 | Glycosyltransferase family protein 47 |
| 259445_at | AT1G02400 | 1.80 | 0.30 | Gibberellin 2-oxidase, putative |
| 265511_at | AT2G05540 | 1.77 | 0.06 | Gly-rich protein |
| 249742_at | AT5G24490 | 1.67 | 0.06 | 30S ribosomal protein |
| 260914_at | AT1G02640 | 1.67 | 0.21 | Glycosyl hydrolase family 3 protein |
| 263443_at | AT2G28630 | 1.63 | 0.06 | β-Ketoacyl-CoA synthase family protein |
MBF1c expression resulted in the accumulation of transcripts encoding pathogenesis-related (PR) proteins, such as chitinase (PR-3) and glucanase (PR-2). Expression of PR-2 and PR-3 was associated with enhanced tolerance of plants to pathogens (Mittler et al., 1995), and might explain the enhanced tolerance of MBF1c plants to bacterial growth (Fig. 2B). With the exception of transcripts encoding a protein with a DNAJ domain, transcripts encoding classical heat shock proteins (HSPs) or drought-response late embryogenesis abundant proteins did not accumulate in transgenic plants under control conditions, suggesting that the enhanced tolerance of these plants to osmotic and heat stress (Figs. 2 and 3) is not associated with constitutive expression of HSPs and late embryogenesis abundant proteins. Of the transcripts elevated in transgenic plants expressing MBF1c, only two transcripts (At3g48520-cytochrome P450 and At1g62510-lipid transfer protein) were also elevated by heat stress or a combination of heat stress and drought in wild-type plants (Table I; Rizhsky et al., 2004b). A complete list of all transcripts significantly elevated or suppressed in MBF1c-expressing plants grown under controlled conditions is included in Supplemental Tables I and II, respectively. It should be noted that the results presented in Table I were obtained with young plants grown in soil and could not be directly correlated with tolerance of 4- to 5-d-old seedlings grown on agar plates (Figs. 2 and 3).
The results presented in Table I suggest that MBF1c does not act as a classical drought- or heat-response transcription factor transgene that constitutively enhances the expression of defense transcripts involved in the response of plants to drought or heat (see e.g. Kasuga et al., 1999; Mishra et al., 2002). The effects of MBF1c on plant tolerance to environmental stress (Figs. 2 and 3) might therefore be linked to its putative coactivator function in cells (Tsuda et al., 2004). Thus, MBF1c might augment the plant's response during stress by binding to different stress-response promoters and facilitating their activation.
Augmented Response of MBF1c-Expressing Plants to Heat Stress
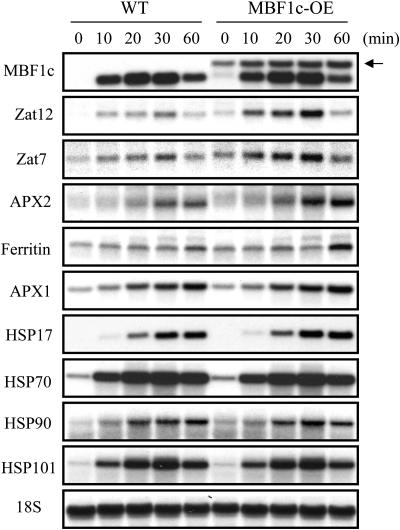
To test whether MBF1c expression in transgenic plants facilitates the accumulation of different stress-response transcripts during heat stress, possibly by acting as a transcriptional coactivator, we compared the accumulation of different stress-response transcripts during heat stress between wild-type and transgenic plants. For this purpose we specifically chose heat- or oxidative-stress-response transcripts that were not elevated in MBF1c-expressing plants in the absence of stress (i.e. absent from Table I). The choice of oxidative-stress-response transcripts was based on a recent report in which MBF1 was shown to regulate the redox response of AP-1 during oxidative stress in Drosophila (Jindra et al., 2004). As shown in Figure 4, in response to heat stress, transcripts encoding the defense proteins ascorbate peroxidase 2 (APX2), ferritin, and the zinc-finger proteins Zat7 and Zat12 accumulated in MBF1c-expressing plants to a higher level than in wild-type plants. In contrast, transcripts encoding APX1 and different HSPs accumulated to a similar level in wild-type and transgenic plants. Constitutive expression of ferritin, Zat12, or Zat7 was shown to enhance the tolerance of transgenic plants to biotic and abiotic stresses (Deak et al., 1999; Rizhsky et al., 2004a). It is possible that the augmented accumulation of these transcripts during stress in transgenic plants (Fig. 4) contributes to the enhanced tolerance of transgenic plants to abiotic stresses (Figs. 2 and 3).
Figure 4.
Augmented response of MBF1c-expressing plants (MBF1c-OE) to heat stress. A time-course RNA gel-blot analysis of 2-week-old wild-type and MBF1c-expressing plants subjected to heat stress (38°C, 10, 20, 30, and 60 min), showing the augmented accumulation of transcripts encoding Zat12, Zat7, APX2, and ferritin in transgenic plants. Time-course experiments were repeated three times with similar results. Representative RNA blots are shown. RNA blots and stress assays were performed as described in “Materials and Methods.” Plants were grown at 21°C, 14-h light cycle, 100 μmol m−2 s−1, and a relative humidity of 70% and subjected to heat stress as described above. Arrow on right side of top section indicates the transgenic transcript of MBF1c. **, Student's t test significant at P < 0.01.
Sugars such as Suc or trehalose play a key role in plant tolerance to drought and heat stress and might have a protective or stabilizing role that could enhance stress tolerance (Bray et al., 2000; Cushman and Bohnert, 2000; Garg et al., 2002; Kaplan et al., 2004; Rizhsky et al., 2004b). Comparative profiling of sugars in transgenic and wild-type plants subjected to heat stress (Supplemental Fig. 3) revealed that the relative level of trehalose was higher in MBF1c-expressing plants compared to wild-type plants under controlled conditions. The relative level of trehalose was further enhanced in transgenic plants in response to heat stress (Supplemental Fig. 3). The higher level of treahalose and its enhanced accumulation during heat stress in transgenic plants could be linked to the accumulation of transcripts encoding trehalose-6-P synthase in transgenic MBF1c plants grown under controlled conditions (i.e. At2g18700 and At1g70290; Table I). Trehalose overaccumulation was shown to enhance the tolerance of transgenic plants to abiotic stresses (Garg et al., 2002; Penna, 2003). These findings might explain the enhanced tolerance of MBF1c plants to abiotic stresses (Figs. 2 and 3). Further studies, including direct measurements of trehalose biosynthesis and degradation in transgenic plants, are, however, required to elucidate the role of trehalose in enhancing the tolerance of MBF1c-expressing plants to abiotic stresses.
MBF1c Expression Enhances the Tolerance of Transgenic Plants to Abiotic Stresses by Partially Activating, or Perturbing, the Ethylene-Response Signal Transduction Pathway
The accumulation of transcripts associated with ethylene signaling in transgenic plants (Table I; Supplemental Fig. 2) suggests that MBF1c expression partially activates, or perturbs, the ethylene-response signal transduction pathway. To test this possibility, we examined whether etiolated seedlings of transgenic plants exhibit the classical triple response associated with ethylene perception (Guzman and Ecker, 1990). As shown in Figure 5A, etiolated seedlings of MBF1c-expressing plants, when compared to wild type, exhibited a stronger triple-response phenotype in the presence or absence of ACC. This result could suggest that MBF1c expression enhances the biosynthesis of ethylene in transgenic plants. Alternatively, MBF1c expression could enhance the sensitivity of transgenic plants to ethylene. To test whether ethylene signaling is involved in the enhanced tolerance of MBF1c-expressing plants to osmotic or heat stress, we tested the effects of the ethylene-signaling inhibitor aminoethoxyvinylglycine (AVG) or silver thiosulfate (STS) on wild-type and transgenic plants subjected to heat stress or heat stress combined with osmotic stress. As shown in Figure 5B, AVG application suppressed the tolerance of MBF1c-expressing plants to heat stress or heat stress combined with osmotic stress. Similar results were obtained with STS (data not shown). To test whether ethylene signaling is required for plant tolerance to a heat stress, osmotic stress, or a combination of heat and osmotic stress, we compared wild-type plants to ein-2 mutants, impaired in ethylene sensing (Guzman and Ecker, 1990). As shown in Figure 5C, ein-2 mutants are more sensitive than wild type to these stresses.
Figure 5.
Enhanced tolerance to abiotic stress in MBF1c-expressing plants is mediated by ethylene signaling. A, Triple-response phenotype of etiolated MBF1c-expressing seedlings in the presence or absence of ACC compared to wild type. B, Survival rate measurements showing suppression of MBF1c-induced tolerance to abiotic stress by the ethylene-signaling inhibitor AVG. C, Root growth measurements of ein2 seedlings, impaired in ethylene sensing, showing enhanced sensitivity to osmotic and heat stress compared to wild type. Stress assays, application of AVG, and ein2 analysis were performed as described in “Materials and Methods.” **, Student's t test significant at P < 0.01.
DISCUSSION
Enhancing plant tolerance to biotic or abiotic stress conditions by activating a stress-response signal transduction pathway in transgenic plants is a powerful and promising approach (Kasuga et al., 1999; Cushman and Bohnert, 2000; Kovtun et al., 2000; Umezawa et al., 2004). Here we report that constitutive expression of the eukaryotic transcriptional coactivator MBF1c in Arabidopsis enhances the tolerance of transgenic plants to bacterial infection, salinity, heat, and osmotic stress, and that the enhanced tolerance of transgenic plants to heat or osmotic stress is maintained even when these two stresses are combined. We further show that MBF1c expression enhances the tolerance of transgenic plants to heat and osmotic stress by perturbing, or partially activating, the ethylene-response signal transduction pathway. Evidence supporting this finding include the accumulation of different ethylene-response transcripts as well as transcripts encoding ACC synthase in MBF1c-expressing plants (Table I), the partial triple-response phenotype of etiolated MBF1c-expressing seedlings (Fig. 5A), and the inhibition of MBF1c-induced tolerance to stress by inhibitors of the ethylene response (Fig. 5B). In contrast to examples in which enhanced tolerance to abiotic stresses was associated with suppressed growth of transgenic plants (Kasuga et al., 1999), constitutive expression of MBF1c did not suppress plant growth (Fig. 1B). The accumulation of MBF1c-encoding transcripts in different plants in response to drought, heat stress, and a combination of drought and heat stress (Fig. 1A; Pnueli et al., 2002; Rizhsky et al., 2002, 2004b); the augmented response of transgenic plants to heat stress (Fig. 4); and the enhanced tolerance of transgenic plants to osmotic stress and heat stress (Figs. 2 and 3) suggest that MBF1 proteins could potentially be used to enhance the tolerance of different plants and crops to these stresses. The interpretation of the results obtained with osmotic and heat stress combination on agar plates (Figs. 2 and 3) should, however, take into consideration the high humidity conditions that occur during these assays. These do not reflect the conditions that occur in the field and may decrease the effects of the osmotic stress on plant acclimation to the stress combination.
Expression of MBF1c in transgenic plants resulted in the constitutive expression of several signal transduction and defense transcripts (Table I), as well as in the augmented accumulation of different stress-associated transcripts in response to heat (Fig. 4). These findings suggest that constitutive expression of MBF1c in transgenic plants alters the accumulation of specific transcripts under controlled conditions and during stress. It is possible that in transgenic plants, MBF1c links between different transcription factors and the basal transcriptional machinery to form a complex that associates with, and activates, specific promoters. The transcripts identified by our study as hyperresponsive or constitutively expressed in transgenic plants (Fig. 4; Table I), might be ideal subjects for future studies to address this possibility. The finding that only a small part of the defense response of plants against drought, heat stress, or a combination of drought and heat stress (Rizhsky et al., 2004b) is constitutively activated in MBF1c-expressing plants grown under controlled conditions (Table I) suggests that augmentation of defense responses by MBF1c during stress (Fig. 4), rather than constitutive activation of defenses (Table I), is the main mode of action of MBF1c in transgenic plants. Compared to the broad effects on gene expression and plant development reported in transgenic plants expressing a transcriptional coactivator that affects histone acetylation (Stockinger et al., 2001; Vlachonasios et al., 2003), the effects of MBF1c expression in transgenic plants appeared to be more limited (Fig. 1; Table I), suggesting that MBF1c binds to specific promoters mainly associated with stress or pathogen responses.
Developing plants with enhanced tolerance to different abiotic stresses and their combination is essential for agricultural production worldwide (Boyer, 1982; Cushman and Bohnert, 2000; Moffat, 2002). Our analyses of transgenic plants expressing MBF1c demonstrate that this transcriptional coactivator plays an important role in plant protection against different environmental stresses. In addition, at least with osmotic and heat stress, the tolerance MBF1c induces in plants toward these stresses was maintained even when they were combined. Our findings thus offer a transgenic strategy to develop plants and crops with enhanced tolerance to different abiotic stresses.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Molecular Analysis
Arabidopsis plants (Arabidopsis thaliana cv Columbia) were grown in peat pellets (Jiffy-7, Shippagan) under controlled conditions: 21°C, 14-h light cycle, 100 μmol m−2 s−1, and a relative humidity of 70% (E-30, AR-66; Percival Scientific). Plant transformation was performed with the binary vector pB001 as described by Rizhsky et al. (2004a). Transgenic plants were selected based on herbicide tolerance (bar) and screened by RNA blots. T4 homozygous lines pooled from three independent transformation events were used for this study (Supplemental Fig. 1). RNA was isolated and analyzed as described previously (Davletova et al., 2005). Sugars were isolated and analyzed by gas chromatography-mass spectrometry as described by Rizhsky et al. (2004b). For the analysis of transcript accumulation in response to heat stress, 2-week-old plants were heat stressed at 38°C, 100 μmol m−2 s−1, and sampled at 0, 10, 20, 30, and 60 min. All experiments were performed in triplicates and repeated at least three times.
DNA Chip Analysis
In three independent experiments, RNA was isolated from 17-d-old control and MBF1c-expressing plants grown under controlled conditions as described above. All experiments were sampled at the same time of day (10 am). At least 75 plants were used for each RNA sample, and RNA was isolated using Trizol. RNA samples were used to perform chip hybridization analyses (Arabidopsis ATH1 chips; Affymetrix) at the University of Iowa DNA facility (http://dna9.intmed.uiowa.edu/microarrays.htm). Conditions for RNA isolation, labeling, and hybridization are described by Davletova et al. (2005). All GeneChip arrays were processed first by robust multi-array average (RMA; Irizarry et al., 2003) using the R package affy (Gautier et al., 2004). Specifically, expression values were computed from raw CEL files by first applying the RMA model of probe-specific correction of perfect-match probes. These corrected probe values were then normalized via quantile normalization, and a median polish was applied to compute one expression measure from all probe values. Resulting RMA expression values were log2 transformed. These are standard methods for processing Affymetrix data. Please see the affy manual at www.bioconductor.org/repository/devel/vignette/affy.pdf for details. Density plots and boxplots of RMA expression value distributions of all arrays were very similar with no apparent outlying arrays (data not shown). Digestion curves describing trends in RNA degradation between the 5′ end and the 3′ end in each probeset were generated, and all six proved very similar, with a downward trend at the 5′ end (data not shown). To determine whether genes were differentially expressed, an ANOVA was performed on the RMA expression values. For an overview on the application of ANOVA to microarray data, please see Kerr et al. (2000). The model described in Davletova et al. (2005) was used for this analysis, and transcripts with adjusted P values < 0.05 were extracted for further analysis. Of these, genes with differential expression of more than 1.5 log2 were selected. The R package limma was used for ANOVA methods (www.bioconductor.org/repository/devel/vignette/affy.pdf). The experiments described in this paper were performed side-by-side with the experiments reported by Rizhsky et al. (2004b) and could therefore be compared to these experiments. Microarray data from this experiment were submitted to the Nottingham Arabidopsis Stock Centre arrays at http://affymetrix.arabidopsis.info/.
Stress Assays and Application of Ethylene Inhibitors
Bacterial (Pseudomonas syringae cv tomato) infection, light stress, and anthocyanin measurements were performed as described previously (Mittler et al., 1997, 1999; Bariola et al., 1999; Pnueli et al., 2003). To avoid complications resulting from differences in plant size and reproduction stage, all stress assays were performed with 4- to 5-d-old wild-type and transgenic seedlings, or 14-d-old wild-type and transgenic plants. At these growth stages no differences were observed between the size and developmental stage of wild-type or transgenic plants (data not shown). In addition, the stress tolerance of MBF1c-expressing plants was compared to that of wild-type plants, as well as that of empty vector plants. No differences were found between the tolerance of wild-type plants and empty vector plants (data not shown). For the analysis of abiotic stress tolerance, seeds of wild-type and MBF1c-expressing lines were surface sterilized and placed in rows on 1% agar plates (0.5× Murashige and Skoog [MS] medium), containing different concentrations of NaCl (50, 100, 150, and 200 mm) or sorbitol (50, 100, 200, and 300 mm). Each row of seeds placed on a plate was divided into two parts: wild type and MBF1c expressing. Thus, the different seeds were placed side by side on the same plate. Plates were incubated for 48 h at 4°C, placed vertically in a growth chamber (21°C–22°C, constant light, 100 μmol m−2 s−1) and scored for percentage of germination and root length 5 d following transfer to the growth chamber. Four- or 5-d-old seedlings grown on 0.5× MS agar plates in a growth chamber were also subjected to heat stress (38°C, 1, 6, 24, and 48 h) and scored for root length 1 d following recovery from heat stress. For stress combination, seeds were surface sterilized and germinated on MS plates containing sorbitol or NaCl as described above and maintained vertically in a growth chamber. Five- to 7-d-old seedlings (grown on MS plates or plates supplemented with sorbitol or NaCl) were heat stressed (38°C) for 48 h and returned to controlled growth condition. Control seedlings (grown on MS plates or MS plates supplemented with sorbitol or NaCl) were maintained under controlled growth conditions. Root length was measured for all seedlings prior to the heat stress treatment and 1 d following heat stress. At least six different plates were used for each condition with approximately 30 seedlings per plate.
To measure survival rate in seedlings subjected to osmotic stress, heat stress, and their combination, surface-sterilized seeds were germinated on MS plates containing different concentrations of sorbitol (0, 50, 100 mm). Each plate was divided into two halves, and approximately 100 seeds of wild-type or MBF1c-expressing lines were spread on each of the different halves. Plates were maintained horizontally in a growth chamber as described above. Seedlings were untreated, acclimated at 38°C for 1.5 h, and treated at 45°C for 2 h, or directly treated at 45°C for 2 h without acclimation. Following heat stress, plates were incubated at 21°C for 2 to 4 d and scored for survival rate. To examine the effects of ethylene-signaling inhibitors on the response of wild-type and transgenic plants to abiotic stresses, water, or AVG (10 μm) or STS (10 μm) prepared in water, were applied to plates by spraying 30 min prior to the heat stress treatment. At least six different plates were used for each condition with approximately 200 seedlings per plate. A Student's t test was used to determine statistical significance.
This work was supported by the National Science Foundation (grant nos. NSF–0431327 and NSF–0420033) and the Nevada Agricultural Experimental Station (publication no. 03055517).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ron Mittler (ronm@unr.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070110.
References
- Bariola PA, MacIntosh GC, Green PG (1999) Regulation of S-like ribonuclease levels in Arabidopsis: antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol 119: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E (2000) Responses to abiotic stresses. In W Gruissem, B Buchannan, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 1158–1249
- Brendel C, Gelman L, Auwerx J (2002) Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism. Mol Endocrinol 16: 1367–1377 [DOI] [PubMed] [Google Scholar]
- Busk PK, Wulf-Andersen L, Strom CC, Enevoldsen M, Thirstrup K, Haunso S, Sheikh SP (2003) Multiprotein bridging factor 1 cooperates with c-Jun and is necessary for cardiac hypertrophy in vitro. Exp Cell Res 286: 102–114 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3: 117–124 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak M, Horvath GV, Davletova S, Torok K, Sass L, Vass I, Barna B, Kiraly Z, Dudits D (1999) Plants ectopically expressing the iron-binding protein, ferritin, are tolerant to oxidative damage and pathogens. Nat Biotechnol 17: 192–196 [DOI] [PubMed] [Google Scholar]
- De Paepe A, Vuylsteke M, Van Hummelen P, Zabeau M, Van Der Straeten D (2004) Transcriptional profiling by cDNA-AFLP and microarray analysis reveals novel insights into the early response to ethylene in Arabidopsis. Plant J 39: 537–559 [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad B, Irizarry RA (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerling M, Kumar A (2003) The perfect ocean for drought. Science 299: 691–694 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs R, Collin R, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jindra M, Gaziova I, Uhlirova M, Okabe M, Hiromi Y, Hirose S (2004) Coactivator MBF1 preserves the redox-dependent AP-1 activity during oxidative stress in Drosophila. EMBO J 23: 3538–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA (2000) Analysis of variance for gene expression microarray data. J. Comput Biol 7: 819–837 (R) [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Hall DJ, Knight MR, Vierling E (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QX, Jindra M, Ueda H, Hiromi Y, Hirose S (2003) Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development 130: 719–728 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Del Pozo O, Meisel L, Lam E (1997) Pathogen-induced programmed cell death in plants: a possible defense mechanism. Dev Genet 21: 279–289 [DOI] [PubMed] [Google Scholar]
- Mittler R, Herr EH, Orvar BL, van Camp W, Willekens H, Inze D, Ellis BE (1999) Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyper-responsive to pathogen infection. Proc Natl Acad Sci USA 96: 14165–14170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Shulaev V, Lam E (1995) Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell 7: 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat AS (2002) Finding new ways to protect drought-stricken plants. Science 296: 1226–1229 [DOI] [PubMed] [Google Scholar]
- Näär AM, Lemon BD, Tjian R (2001) Transcriptional coactivator complexes. Annu Rev Biochem 70: 475–501 [DOI] [PubMed] [Google Scholar]
- Penna S (2003) Building stress tolerance through over-producing trehalose in transgenic plants. Trends Plant Sci 8: 355–357 [DOI] [PubMed] [Google Scholar]
- Peters DP, Pielke RA Sr, Bestelmeyer BT, Allen CD, Munson-McGee S, Havstad KM (2004) Cross-scale interactions, nonlinearities, and forecasting catastrophic events. Proc Natl Acad Sci USA 101: 15130–15135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R (2002) Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J 31: 319–330 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hongjian L, Mittler R (2003) Growth suppression, abnormal guard cell response, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R (2004. a) The zinc-finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004. b) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosegrant MW, Cline SA (2003) Global food security: challenges and policies. Science 302: 1917–1919 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF (2001) Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res 29: 1524–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Harashima S, Ueda H, Hirose S (1998) Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol Cell Biol 18: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Li FQ, Ueda H, Hirose S (1997) Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc Natl Acad Sci USA 94: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Tsuji T, Hirose S, Yamazaki K (2004) Three Arabidopsis MBF1 homologs with distinct expression profiles play roles as transcriptional co-activators. Plant Cell Physiol 45: 225–231 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Yamazaki K (2004) Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim Biophys Acta 1680: 1–10 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2004) SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 17306–17311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ (2003) Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]