Abstract
Optimal activation of T cells requires effective occupancy of both the antigen-specific T cell receptor and a second coreceptor such as CD28. We used cDNA microarrays to characterize the genomic expression program in human peripheral T cells responding to stimulation of these receptors. We found that CD28 agonists alone elicited few, but reproducible, changes in gene expression, whereas CD3 agonists elicited a multifaceted temporally choreographed gene expression program. The principal effect of simultaneous engagement of CD28 was to increase the amplitude of the CD3 transcriptional response. The induced genes whose expression was most enhanced by costimulation were significantly enriched for known targets of nuclear factor of activated T cells (NFAT) transcription factors. This enhancement was nearly abolished by blocking the nuclear translocation of NFATc by using the calcineurin inhibitor FK506. CD28 signaling promoted phosphorylation, and thus inactivation, of the NFAT nuclear export kinase glycogen synthase kinase-3 (GSK3), coincident with enhanced dephosphorylation of NFATc proteins. These results provide a detailed picture of the transcriptional program of T cell activation and suggest that enhancement of transcriptional activation by NFAT, through inhibition of its nuclear export, plays a key role in mediating the CD28 costimulatory signal.
Maximal activation of T cells by antigen-presenting cells requires two stimulatory signals, one through the antigen-specific T cell receptor (TCR) complex and a second through a coreceptor such as CD28 (1). Resting T cells stimulated through the TCR complex alone do not become fully activated and can become anergic or even apoptotic (2). Simultaneous signaling by the CD28 costimulatory receptor allows for sustained activation, characterized by the production of IL-2 and cell-cycle entry (see ref. 3 for review). Two main models have been suggested for the mechanism of costimulation, one in which CD28 sends a unique and independent signal, and a second in which CD28 acts primarily to increase the density of signaling molecules in the TCR complex and thus amplifies the proximal TCR signaling cascade. As evidence for the first model, CD28 crosslinking has been shown to activate a number of signaling molecules, including phosphoinositide 3-kinase (PI3K) (4). Support for the second model comes from data demonstrating increased aggregation of lipid rafts at the T cell/antigen-presenting cell interface during costimulation (5–7) and association of the CD28 cytoplasmic tail with molecules such as LCK, which are essential to TCR signaling (8, 9).
Here we examine genome-scale gene expression responses in primary human T cells to monostimulation and costimulation through CD3 and CD28. CD28 costimulation resulted in a largely quantitative increase of the gene expression response to CD3 alone but disproportionately affected targets of the nuclear factor of activated T cells (NFAT) family of transcription factors. Furthermore, CD28 signaling significantly inhibited glycogen synthase kinase-3 (GSK3), an NFAT nuclear export kinase. These findings suggest a critical role for NFAT in the integration of the two signals, likely achieved through enhanced nuclear import by increased calcium influx and decreased nuclear export by inactivation of GSK3.
Materials and Methods
Isolation and Stimulation of Primary T Cells.
Primary T cells were isolated (>98% purity by FACS) from whole blood of healthy donors using Ficoll–Paque Plus (Pharmacia Biotech) followed by magnetic depletion of non-T cells (MACS Pan-T Cell isolation kit, Miltenyi Biotec, Auburn, CA). The activation beads were a kind gift of James Riley (University of Pennsylvania) and consisted of 3-μm tosyl-activated polystyrene beads (M450, Dynal, Great Neck, NY) coated with a 1:1 mixture of αCD3 (OKT3) and α major histocompatibility complex I (αMHCI) (W6/32) antibodies (αCD3 beads), a 1:1 mixture of αCD28 (9.3) and αMHCI antibodies (αCD28 beads), and a 1:1 mixture of αCD3 and αCD28 antibodies (costimulatory beads). Studies of responses to higher levels of CD28 agonists used beads coated with either 100% αCD28 antibody or 100% recombinant B7.2 protein (CD86). Proliferation assays (5 × 104 per well) were performed in triplicate for 72 h. Wells were pulsed with 1 μCi of [3H]thymidine for the last 6 h.
Microarray Procedures.
All microarray methods followed closely those described in a previous study (10). Total RNA was amplified by using a linear amplification method (11). More detailed information including data selection and manipulation methods, as well as searchable figures, and all raw microarray data can be found at http://genome-www.stanford.edu/costimulation.
Protein Studies.
IL-2 protein levels were quantified in supernatants by using a luminescence-based ELISA (R & D Systems). For Western blots, purified T cells were lysed in RIPA (150 mM NaCl/20 mM Tris, pH 7.5/0.1% SDS/1% Triton X-100/0.5% sodium deoxycholate/1 mM EDTA) with protease and phosphatase inhibitors. Extract from 106 cells was loaded per lane for SDS/PAGE on 7.5% gels. Antibodies used for Western blots included αNFATc2 (polyclonal, S. Stewart, Stanford University), αHsp90 antibody (BD Transduction Laboratories, Lexington, KY), αGSK3β, and an antibody specific for the serine-9-phosphorylated form of GSK3β (Cell Signaling Technology, Beverly, MA).
Results and Discussion
Overview of Stereotyped Activation Responses.
We characterized the gene expression program in T cells responding to a variety of models of antigen receptor stimulation. Human peripheral T cells were isolated from healthy volunteer donors and subjected to stimulation by “surrogate antigen-presenting cells” consisting of microbeads coated with antibodies to either CD3 or CD28, or with a combination of both antibodies (αCD3/αCD28 “costimulatory” beads). We also examined the responses of T cells to stimulation with two classical pharmacologic mimics of antigen-receptor signaling—the lectin phytohemagglutinin, which nonspecifically clusters cell surface proteins, and a combination of the calcium ionophore ionomycin and the phorbol ester phorbol 12-myristate 13-acetate (PMA), which together mimic many of the signaling actions of the antigen receptor and costimulation (12). Cells were exposed to these stimuli for the indicated times, after which RNA was extracted and analyzed by using DNA microarrays containing 37,632 elements, representing ≈18,000 different genes (13).
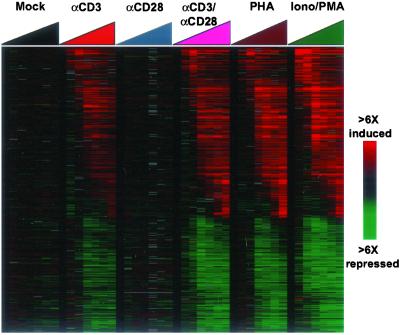
The gene expression programs evoked by αCD3 beads, phytohemagglutinin, costimulatory beads, and ionomycin plus PMA were strikingly similar (Fig. 1). A diverse group of more than 3,000 genes (≈17% of all genes represented in the microarray) showed prominent gene expression changes (≥3-fold on two microarrays), with ≈57% of these being induced and ≈43% being repressed. Although there were significant differences in the magnitudes of the responses elicited by the different treatments, the specific genes responsive to the stimuli and the patterns in which they responded were largely the same. The similarity between changes induced by CD3/CD28 costimulation and those induced by ionomycin and PMA highlights the critical importance of the calcium signal and the protein kinase C pathway in the T cell activation program. Also, whereas studies of gene expression changes during T cell activation have often focused on the genes induced during activation, these results demonstrate that almost an equal number of genes were repressed by these stimuli.
Figure 1.
Gene expression responses of T cells to diverse stimuli mimicking antigen receptor stimulation. Peripheral blood T cells were subjected to six distinct treatments in parallel time series and harvested at seven time intervals (0, 1, 2, 6, 12, 24, and 48 h). Treatments included: no stimulus (mock treated); stimulation with beads coated with αCD3, αCD28, or both; or treatment with 5 μg/ml of phytohemagglutinin or ionomycin [1 μM]/PMA [25 ng/ml]. Array elements that were induced or repressed more than 3-fold compared with baseline on at least two microarrays were included (4,359 cDNA elements representing 2,926 genes). The data are displayed as a self-organizing map that temporally orders the matrix of gene expression data where rows represent genes (unique cDNA elements), and columns represent experimental samples. Colored pixels capture the magnitude of the response for any gene. Shades of red and green represent induction and repression, respectively, relative to the prestimulation specimen (t = 0). Black pixels reflect no change from baseline and gray pixels represent missing data. Supplemental data and enhanced versions of the figures, including searchable clusters and raw microarray data, can be found at our web site (http://genome-www.stanford.edu/costimulation).
Transcriptional changes during T lymphocyte activation have been characterized in the past (14), and it is beyond the scope of this paper to provide a discussion of all of the observed changes and their biological implications. However, several broad features of this program deserve mention. A major theme of the activation response is the importance of the molecular communication between cells. Dozens of genes encoding immune mediators such as cytokines and chemokines, cytokine receptors, cell adhesion molecules, as well as cytotoxic effector molecules, such as granzyme B, granulysin, and fas ligand, were induced in a richly choreographed pattern during this program. Induction of these latter proapoptotic factors would be expected to promote cell death, yet we observed only modest cell death during the 48-h time course of this experiment (data not shown), in agreement with previous reports (2, 15).
The observed regulation of fas, fas ligand, and genes that act downstream of fas suggests how costimulated T cells might protect themselves from their own lethal capacities. Intriguingly, whereas costimulation of T cells dramatically induced expression of fas ligand, fas itself was not induced. Similarly, the antiapoptotic factor BCL2 was induced, whereas its antagonist, BID, was repressed. Caspase 8, which activates BID, was also repressed, whereas the “decoy” protein FLIP, which competes with Caspase 8 for binding to the fas adapter FADD, was induced. Thus, this coordinated transcriptional program may serve to protect T cells against fas ligand-dependent apoptosis while allowing them to “safely” use fas ligand to kill other cellular targets (16).
Activated T cells enter the cell cycle and proliferate and can eventually give rise to memory cells. A significant fraction of the genes that were induced at intermediate and late time points in this study have direct roles in promoting proliferation and progression through the cell cycle. These included cyclins (CCNE1 and -E2), cyclin-dependent kinases (CDK2, -4, and -6), genes directly involved in DNA replication (MCM2, -3, -4, and -6), as well as genes involved in nucleotide biosynthesis (dihydrofolate reductase and ribonucleotide reductases).
Many features of the transcriptional program appear to be related to the increased metabolic demands, macromolecular biosynthesis, and secretion accompanying the physiological mobilization of T cells. For example, we observed a general induction of signal recognition particle subunits, translation initiation factors, chaperones, RNA processing enzymes, and nearly all tRNA synthetase genes, consistent with the increased protein synthesis observed in activated lymphocytes (17). Genes encoding proteins involved in glycolysis and the tricarboxylic acid cycle were also markedly induced, paralleling the dramatic increase in the cell's energy requirements.
Many of the genes whose products are involved in transducing signals from the TCR were repressed on T cell activation. These genes include phospholipase C, LAT, LCK, TRIM, and CD3ζ, as well as genes encoding subunits of the TCR itself. This discovery is consistent with published reports demonstrating rapid internalization and degradation of the TCR on stimulation (18). Moreover, this down-modulation of TCR components might reflect a transition from resting T cells, whose major cell surface communication machinery is dedicated to antigen scanning, to stimulated T cells, whose cell surface proteins are primarily devoted to effector functions and cytokine-mediated communication with other cells.
Mono- and Costimulatory Effects of CD28 Signaling.
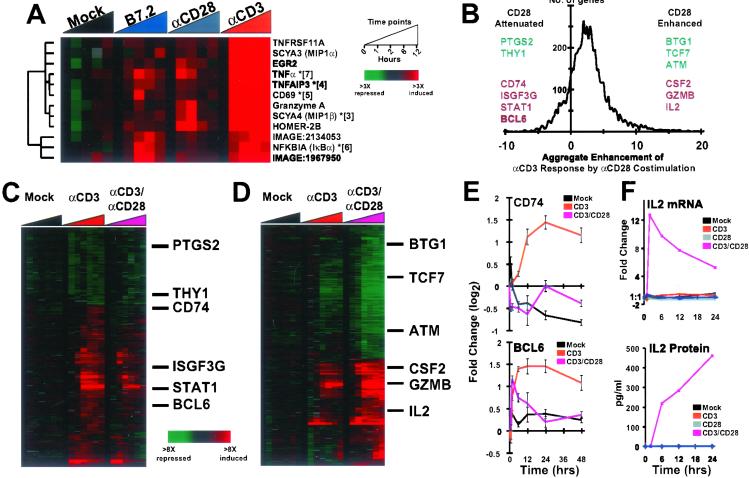
The transcriptional response of T cells treated with beads coated with 50% αCD28 was remarkably subtle (Fig. 1), even though the same surface density of αCD28 antibody induced a significant costimulation response in the context of the costimulatory beads. We also examined responses to stimulation with αCD28 beads coated with 100% antibody and to beads coated with the protein B7.2 (CD86), one of the natural ligands of the CD28 receptor. A search for genes induced by both of these stronger CD28 agonists revealed a few whose expression was modestly, but reproducibly, increased (Fig. 2A and Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org), including TNFα, CD69, EGR2, JUNB, SCYA3, and SCYA4.
Figure 2.
Gene expression responses to CD28 ligands. (A) Genes with detectable responses to monostimulation by CD28 ligands. Geometric means are shown for genes represented by multiple cDNA elements (indicated by asterisks and number of averaged elements). (B) Aggregate enhancement of gene expression responses to αCD3 stimulation by costimulation with αCD28. The abscissa represents the difference in the response between monostimulation with αCD3 and costimulation with αCD3/αCD28 coated beads using a customized distance metric. Representative genes from both ends of the distribution are shown, exemplifying genes whose responses to αCD3 were enhanced or diminished by αCD28. Genes from the bottom 5 and top 10 percentiles of the distribution are depicted in C (348 elements), and D (538 elements), respectively. (E) Example genes from c, plotted as line graphs. Depicted data points represent the geometric mean of independent cDNA elements, with error bars indicating the corresponding standard error. (F) Comparison of IL-2 mRNA and protein levels.
Because all of these genes were induced much more strongly by engagement of CD3 alone than by αCD28 or B7.2 beads, the physiological significance of this CD28 response remains unclear. We therefore examined CD28 responses in a costimulatory context. To measure the effect of costimulation, we defined a simple “CD28 enhancement” parameter to represent the difference between the response to costimulation and the response to αCD3 alone for all responsive genes (see Supporting Materials and Methods in supporting information on the PNAS web site for details). A histogram showing the distribution of genes according to the CD28 enhancement of their response to CD3 stimulation (Fig. 2B) illustrates that for a majority of the genes that respond to CD3 stimuli, activation or repression was enhanced by simultaneous engagement of CD28 (enhancement >0). The behavior of the genes exhibiting the greatest enhancement by CD28 costimulation is illustrated in Fig. 2D. IL-2, which showed virtually no response to CD3 ligation alone, but which was highly induced by CD3/CD28 costimulation, was the most prominent of these genes (Fig. 2F).
For one potentially significant group of genes (Fig. 2C), the transcriptional response to CD3 engagement was blunted by simultaneous engagement of CD28 (in Fig. 2B, these genes have CD28 enhancement <0). Prominently enriched among this group of attenuated genes were many known to be regulated by IFNs (19), or which we observed to be induced in response to IFNγ treatment [P < 10−5, enrichment relative to all well-measured genes assessed by hypergeometric distribution (10); see Fig. 7, which is published as supporting information on the PNAS web site). Among these genes was the “antiproliferative” gene BCL6 (Fig. 2E). BCL6 transcripts increased substantially on stimulation of CD3 but only very slightly on CD3/CD28 costimulation. The CD3-specific activation of BCL6, coupled with the TH2 hyperimmune phenotype that characterizes BCL6 knockout mice (20), suggests the possibility that antigen presentation in the absence of costimulation induces expression of this gene and thereby ablates the proliferation response. Moreover, although costimulation of our mixed population enhanced expression of genes encoding both TH1 and TH2 cytokines (IFNγ, IL-4, and IL-13), the coincident reciprocal down-modulation of these IFNγ responsive genes might reflect the well-known effects of CD28 on promoting TH2 polarized differentiation of naïve CD4+ helper T cells (21).
Expression of CD74, which encodes the invariant chain chaperone of the class II major histocompatibility complex present on antigen-presenting cells, was also attenuated by CD28 costimulation. A processed form of CD74 has been identified as a secreted factor that inhibits IL-2 production by activated T cells (22). The ability of CD28 signaling to reciprocally regulate antiproliferative and stimulatory genes (e.g., repression of BCL6 and induction of IL-2) suggests that multiple mechanisms function in concert to limit T cell proliferation in the absence of a costimulatory signal delivered by a professional antigen-presenting cell or encountered in the context of a potential threat or stress (23).
A Role for NFAT in Integrating the CD28 Signal.
An analysis of the most highly costimulated genes revealed a remarkable enrichment of known targets of NFAT transcription factors (including IL-2, GM-CSF, IL-2Rα, and IFNγ), and this enhancement was statistically significant as early as 6 hours (P < 10−25). Inspection of the genes induced by CD28 monostimulation indicated that four of the 10 characterized genes in this set (TNF, CD69, SCYA3, and EGR2) (24–26) were known NFAT targets (P < 10−4). These observations pointed toward a potentially critical role for NFAT in the integration of the two signals.
The NFAT transcription complex plays a major role in mediating transcriptional activation in response to TCR engagement. In resting T cells, the cytoplasmic calcium-sensitive subunits of NFAT complexes are typically phosphorylated (see ref. 24 for review). On antigen receptor stimulation, calcium enters the cell, activating the phosphatase calcineurin, which then dephosphorylates NFAT and sends the transcription factor into the nucleus (27).
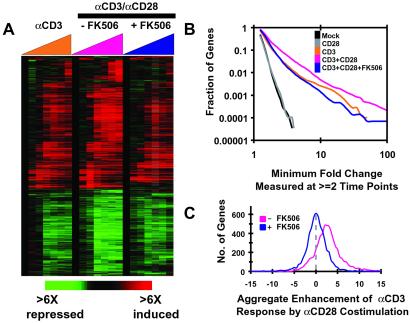
We therefore examined the effect of NFAT inhibition on the genomic expression program in CD3/CD28 costimulated cells (Fig. 3). FK506 blocks dephosphorylation by calcineurin of NFAT transcription complexes and thus prevents their nuclear import (27). Pretreatment of T cells with FK506 before CD3/CD28 costimulation significantly diminished the amplitude of the ensuing changes in gene expression (Fig. 3). For the majority of genes examined, the FK506 treatment reduced the magnitude of the changes in their expression to roughly the level seen after stimulation with αCD3 alone. Interestingly, CD28-dependent blunting of the transcriptional responses to CD3 engagement (e.g., for BCL6) was also generally sensitive to FK506 (Fig. 3D), suggesting that NFAT may repress transcription in certain contexts (28, 29) while enhancing it in others (30).
Figure 3.
FK506 inhibition of gene expression enhancement by CD28. (A) Global overview of T cells stimulated with either αCD3 beads, αCD3/αCD28 costimulatory beads alone, or the latter in combination with FK506. T cells were isolated and stimulated as described in Fig. 1. Cells treated with FK506 were pretreated with 24 nM FK506 for 60 min before addition of costimulatory beads. (B) Distribution of the magnitudes of expression changes. The plot shows the fraction of genes passing a given minimum fold change at two or more time points in the different stimulations. (C) FK506 inhibits aggregate enhancement of transcription by CD28. The customized distance metric from Fig. 2B was used to determine CD28 enhancement of CD3-dependent transcription in the absence or presence of FK506.
The effects of FK506 were evident both in an overview map of the gene expression programs after activation (Fig. 3A) and in the distribution of the magnitudes of expression changes among the genes analyzed in this experiment (Fig. 3B). As shown previously (Fig. 1), few genes responded to mock treatment or treatment with CD28 beads. The magnitude of the responses of genes to CD3 stimulation was generally much smaller than to costimulation. Remarkably, the profile depicted for cells treated with FK506 before costimulation was very similar to that of cells treated with αCD3 alone. This aggregate analysis suggests that the average enhancement of transcriptional activation caused by CD28 costimulation is sensitive to FK506.
We next examined whether the generalized attenuation of transcriptional responses by FK506 was evident at the level of individual genes (Fig. 3C). For this analysis, we measured the difference in activation for each gene between CD3-stimulated and CD3/CD28-stimulated cells in the presence or absence of FK506 pretreatment (Fig. 3C, pink and blue lines, respectively). The distribution of CD28-mediated enhancement is as described for Fig. 2B and demonstrates an average enhancement of gene expression by costimulation. In contrast, in the presence of FK506, the mean of the distribution centers on zero, suggesting that for the majority of genes analyzed, FK506 completely counteracts the transcriptional enhancement caused by CD28 signaling.
The sensitivity of CD28 signaling to FK506 was surprising in light of previously published reports that the CD28 signal was resistant to calcineurin inhibitors, as measured by proliferation and IL-2 production (reviewed in ref. 3). In those studies, the effect was observed mainly in a comparison of cells stimulated jointly with PMA and αCD28 antibody to cells stimulated with a combination of PMA and αCD3 antibody. Although we were able to confirm FK506 resistance in the setting of PMA and soluble αCD28 antibody, cells stimulated with the more physiological costimulatory αCD3/αCD28 beads did not display this resistance (Figs. 2F and 4D and Fig. 8, which is published as supporting information on the PNAS web site).
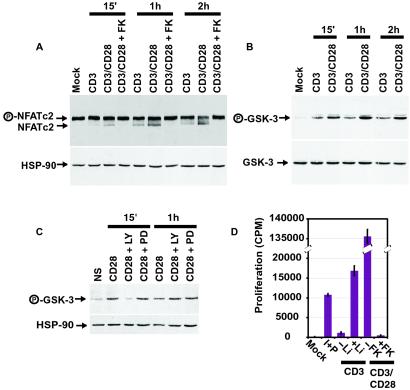
Figure 4.
Enhancement of NFAT activation by CD28 engagement and GSK3 inactivation. (A) Simultaneous engagement of CD28 enhances CD3-dependent dephosphorylation of NFATc2. The phosphorylation state of the NFATc2 bands is as indicated. αHsp90 antibody was used as a loading control. (B) Simultaneous engagement of CD28 enhances CD3-dependent phosphorylation of GSK3. Cells were treated as above. Westerns were probed with antibody to GSK3β as well as an antibody specific for the serine 9 phosphorylated form of GSK3β. Some lots of phospho-GSK3 antibody detect phosphorylated GSK3α as well, and similar results were seen for both isoforms (C and Fig. 12). (C) CD28 engagement alone promotes GSK3 phosphorylation. Cells were isolated as above, and treated for the indicated times with αCD28 beads after pretreatment with the indicated drugs for 15 min. CD28, no inhibitor; LY, 10 μM LY294002; PD, 50 μM PD98059. Westerns were probed with antibody specific for phosphorylated GSK3α. HSP90 was used as a loading control. (D) GSK3 inhibition by lithium enhances proliferation of T cells treated with αCD3 beads. Proliferation assays were preformed as described above. Mock, no stimulus; I+P, 1 μM ionomycin and 25 ng/ml PMA; CD3, αCD3 coated beads with or without 10 mM lithium; CD3/CD28, αCD3/αCD28 costimulation beads with or without 24 nM FK506.
CD28 Signals Activate NFAT and Inhibit Its Nuclear Efflux.
Because these gene expression profiles pointed to NFAT as the critical point of integration of the two signals, we next asked whether we could observe NFAT activation at the protein level. We therefore examined the effects of the CD28 costimulatory signal on the phosphorylation state of the cytoplasmic components of the NFAT complexes. Phosphorylated NFAT has a lower electrophoretic mobility than dephosphorylated NFAT, and thus NFAT dephosphorylation can be assessed from the density ratio of the two bands on a Western blot (Fig. 4A). Both CD3 stimulation and CD3/CD28 costimulation promoted dephosphorylation of NFATc2 (and NFATc1; see Fig. 9, which is published as supporting information on the PNAS web site), as expected. However, a larger fraction of NFATc2 was dephosphorylated in the costimulated cells than in cells stimulated through the TCR alone. This enhanced dephosphorylation was paralleled by an increase in the fraction of nuclear NFATc2 and by an increase in the DNA-binding activity of NFAT as assayed by electrophoretic mobility shift assay (Figs. 10 and 11, which are published as supporting information on the PNAS web site). These results support a model in which CD28's costimulatory effects on transcription are achieved through an enhancement of NFAT activity.
The phosphorylation state of NFAT is controlled by two opposing activities: the phosphatase calcineurin and nuclear protein kinase(s) (31). CD28 may therefore enhance NFAT dephosphorylation by increasing calcium flux and calcineurin activity, as has been demonstrated in murine thymocytes and the human Jurkat T cell leukemia line (32, 33). However, the reported CD28-dependent calcium signals are small and may not fully account for the differences in NFAT dephosphorylation. Rephosphorylation of NFAT provides an additional level of control through enhancement of its nuclear export. An inhibitory role for CD28 on control of nuclear efflux has not been directly tested before. However, CD28 signaling has been shown to activate PI3K (34), which can in turn activate the AKT protein kinase and could thus lead to inactivation of GSK3, a known NFAT kinase (35, 36).
Several lines of evidence support a role for GSK3 as an NFAT kinase. In vitro, GSK3 phosphorylates the serine residues required for cytoplasmic localization of NFATc1 (37). In vivo, overexpression of GSK3 enhances NFAT nuclear export, whereas a dominant negative GSK3 mutant blocks export of NFAT in both lymphocytes and neurons (37, 38). Additionally, constitutively active GSK3 negatively regulates IL-2 production and proliferation by murine T cells (39). Finally, depletion of GSK3 from extracts depletes the NFATc1 and c4 kinases in both neurons and lymphocytes (37, 38). Thus, CD28 signaling could enhance NFAT-dependent transcription by inhibiting GSK3, thereby inhibiting NFAT nuclear export.
To test this model, we stimulated T cells with αCD3 or with αCD3/αCD28 beads and then assayed phosphorylation of serines 9 or 21 of GSK3α/β by using phospho-specific antibodies. Simultaneous signaling through CD3 and CD28 resulted in a higher level of phosphorylated (and thus inactivated) GSK3 than was elicited by stimulation through CD3 alone (Fig. 4B). CD28 costimulation may therefore enhance NFAT activation by inhibiting its rephosphorylation by GSK3.
Even in the absence of αCD3 antibodies, engagement of CD28 stimulated the phosphorylation of GSK3. GSK3 phosphorylation was inhibited at early time points by the PI3K inhibitors LY294002 and wortmannin (Fig. 4C and Fig. 12, which is published as supporting information on the PNAS web site), consistent with a requirement for the PI3K pathway in transducing the CD28 signal. Conversely, this phosphorylation was insensitive to the MEK inhibitor PD98059 (Fig. 4C) or to FK506 (Fig. 12). Thus, engagement of CD28 alone, in the absence of any signal through the TCR, can deliver an intracellular signal. The molecular mechanism by which this signal acts—inhibition of an inactivator of a transcription factor—is such that the transcriptional effects of CD28 signaling alone could be minimal in the absence of a separate signal that activates NFAT dephosphorylation.
Several kinases that phosphorylate NFAT in vitro have been suggested to be physiological NFAT kinases, including GSK3 (37), casein kinase I (40), and p38 MAP kinase (41). As a test of the importance of GSK3 as an NFAT kinase in human peripheral T cells, we used lithium to inhibit GSK3 activity (42). If inhibition of GSK3 is a crucial component of CD28 signaling, then lithium would be expected to partially replace CD28 engagement in CD3-stimulated cells. CD3/CD28 costimulated cells proliferated robustly, whereas T cells stimulated by CD3 engagement alone did not (Fig. 4D). Inhibition of GSK3 by lithium allowed some proliferation by T cells treated with only CD3 beads. A similar effect was seen on IL-2 production: cells treated with αCD3 beads alone produced almost no IL-2, whereas cells treated with αCD3 beads and lithium produced a small amount (Fig. 8).
Cells treated with αCD3 beads and lithium proliferated considerably less than CD3/CD28 costimulated cells. There are at least two plausible reasons for the inability of lithium to compensate completely for the missing CD28 signal. First, lithium is a nonspecific inhibitor with potentially toxic effects on other functions; the concentration of lithium used to inhibit GSK3 also significantly inhibited proliferation of costimulated cells (Fig. 13, which is published as supporting information on the PNAS web site). Second, inhibition of GSK3 might be one of several mechanisms by which the CD28 signal is transduced. As previously noted, CD28 engagement enhances TCR capping and has been reported to enhance calcium flux.
Conclusion and Summary
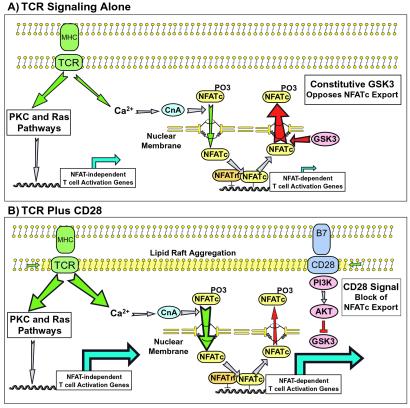
The results presented here are consistent with a model of costimulation in which CD28 signaling lowers TCR thresholds (Fig. 5). Although this effect may be partially mediated by enhanced aggregation of TCRs (6, 7), the genetic, biochemical, and pharmacological data we presented here suggest a novel role for NFAT as the focal point for the integration of the two pathways. Increased NFAT activation on costimulation is likely mediated by enhanced nuclear import of NFAT through increased calcium flux (32), as well as by decreased nuclear export of NFAT through GSK3 inactivation, as shown in this study. Both of these effects are likely mediated by CD28-induced PI3K activation.
Figure 5.
A model of T cell costimulation. Illustrated are the dominant signaling pathways that likely characterize and distinguish TCR stimulation in the absence (A) or presence (B) of CD28-mediated signals. The thickness of each arrow reflects the relative strength of the transduced signal.
Our expression results show that CD3 signaling activates a number of FK506-insensitive transcription factors and by itself causes slight dephosphorylation of NFAT transcription factors. Although this level of NFAT activation is sufficient to activate some NFAT-dependent genes, such as MIP1α and TNFα, rapid rephosphorylation of the transcription factor decreases the nuclear levels of NFAT and prevents full NFAT-dependent transcription. CD28 activation enhances phosphorylation and inhibition of GSK3, thus allowing the nuclear accumulation of dephosphorylated NFAT and transcription of genes that are more dependent on NFAT activity, such as IL-2. The NFκB factor c-Rel has been suggested to be the most relevant target for the costimulatory activity of CD28 (43). However, c-Rel's role as the “integrator” of the CD28 signal remains contentious for several reasons. First, the profound deficit in IL-2 production that characterizes c-Rel knockout mice can be overcome by using the combination of ionomycin and PMA (44). Second, c-Rel is most likely an NFAT target gene (24), and we accordingly observed its induction by CD3 monostimulation and enhanced induction by CD28 costimulation, the latter of which was FK506-sensitive. Finally, we observed a dramatic augmentation in the nuclear accumulation of NFAT within minutes of CD28 costimulation, whereas the increased nuclear influx of c-Rel is known to take hours (45). We thus propose that, whereas both factors are required for efficient costimulation, NFAT is likely critical to the integration of the costimulatory signal, whereas c-Rel plays a permissive role.
The ability of CD28 engagement alone to induce several genes independently of CD3 engagement is intriguing. CD28 ligation in the context of a TCR signal has previously been reported to enhance stability of specific mRNA species (46), all of which contain adenylate/uridylate-rich elements (AREs) in their 3′ untranslated region, which at least partly mediate the increase in mRNA half life (see ref. 47 for review). We found a significant enrichment of multiple ARE motifs (AUUUA) in the 3′ untranslated regions of CD28-induced genes relative to all well-measured genes for which UTR sequences were available (P < 0.01). However, as described above, we found an even stronger enrichment of known NFAT targets in genes induced by CD28 monostimulation (P < 10−4). Furthermore, the genes whose expression was most enhanced by costimulation were not enriched for AREs but were significantly enriched for known NFAT targets. Thus, it is unlikely that CD28 mediates its effect primarily through RNA stabilization in the context of costimulation.
In support of NFAT's role in the integration of costimulatory signals, we have observed that transgenic mice expressing a point mutant in NFATc1 that reduces the rate of nuclear export and leads to constitutive nuclear localization do not require CD28 costimulation for IL-2 production or proliferation (M. Pan and G.R.C., unpublished results). This finding indicates that nuclear retention of NFATc1 is sufficient to replace the costimulatory requirement. Of interest, the constitutive nuclear localization of NFATc1 in NFATc2/NFATc3-deficient mice (28) similarly results in CD28-independent T cell proliferation (48).
T cell activation elicits a complex temporally choreographed gene expression program. The results presented here provide a picture of the molecular program by which the cells activate their cytotoxic potential, protect themselves against apoptosis, and reprogram their responses to physiological signals. This program is expected to enhance cellular energy metabolism, orchestrate recruitment of and communication with other immune cells, and promote entry into and progression through the cell cycle. The results should also provide a useful foundation for further investigations of T cell activation. The differences in transcriptional programs between T cells stimulated through CD3 alone and T cells costimulated through both CD3 and CD28 point toward potential therapeutic targets for augmenting or inhibiting immune responses in clinical settings ranging from autoimmune or inflammatory diseases to the immunotherapy of tumors.
Supplementary Material
Acknowledgments
We thank Scott Stewart for assistance with the gel shift assay and members of the Brown, Crabtree, and Botstein laboratories for helpful discussions. This work was supported by National Institutes of Health Grants CA39612 (G.R.C.) and CA85129-04 (P.O.B. and D.B.), Defense Advanced Research Planning Agency Grant N65236-99-1-5428 (P.O.B.), and National Institute of General Medical Sciences Training Grant GM07365 (A.A.A., M.D., and O.J.R.). P.O.B. is an associate investigator and G.R.C. an investigator of the Howard Hughes Medical Institute.
Abbreviations
- TCR
T cell receptor
- PI3K
phosphoinositide 3-kinase
- GSK3
glycogen synthase kinase-3
- PMA
phorbol 12-myristate 13-acetate
References
- 1.Salomon B, Bluestone J A. Annu Rev Immunol. 2001;19:225–252. doi: 10.1146/annurev.immunol.19.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Van Parijs L, Abbas A K. Science. 1998;280:243–248. doi: 10.1126/science.280.5361.243. [DOI] [PubMed] [Google Scholar]
- 3.June C H, Ledbetter J A, Linsley P S, Thompson C B. Immunol Today. 1990;11:211–216. doi: 10.1016/0167-5699(90)90085-n. [DOI] [PubMed] [Google Scholar]
- 4.Pages F, Ragueneau M, Rottapel R, Truneh A, Nunes J, Imbert J, Olive D. Nature (London) 1994;369:327–329. doi: 10.1038/369327a0. [DOI] [PubMed] [Google Scholar]
- 5.Wulfing C, Davis M M. Science. 1998;282:2266–2269. doi: 10.1126/science.282.5397.2266. [DOI] [PubMed] [Google Scholar]
- 6.Viola A, Schroeder S, Sakakibara Y, Lanzavecchia A. Science. 1999;283:680–682. doi: 10.1126/science.283.5402.680. [DOI] [PubMed] [Google Scholar]
- 7.Wulfing C, Sumen C, Sjaastad M D, Wu L C, Dustin M L, Davis M M. Nat Immunol. 2002;3:42–47. doi: 10.1038/ni741. [DOI] [PubMed] [Google Scholar]
- 8.Hutchcroft J E, Bierer B E. Proc Natl Acad Sci USA. 1994;91:3260–3264. doi: 10.1073/pnas.91.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdorf A D, Lee K H, Burack W R, Allen P M, Shaw A S. Nat Immunol. 2002;3:259–264. doi: 10.1038/ni761. [DOI] [PubMed] [Google Scholar]
- 10.Boldrick J C, Alizadeh A A, Diehn M, Dudoit S, Liu C L, Belcher C E, Botstein D, Staudt L M, Brown P O, Relman D A. Proc Natl Acad Sci USA. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang E, Miller L D, Ohnmacht G A, Liu E T, Marincola F M. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 12.Weiss A, Imboden J, Hardy K, Manger B, Terhorst C, Stobo J. Annu Rev Immunol. 1986;4:593–619. doi: 10.1146/annurev.iy.04.040186.003113. [DOI] [PubMed] [Google Scholar]
- 13.Alizadeh A A, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 14.Crabtree G R. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 15.Van Parijs L, Ibraghimov A, Abbas A K. Immunity. 1996;4:321–328. doi: 10.1016/s1074-7613(00)80440-9. [DOI] [PubMed] [Google Scholar]
- 16.Chang H Y, Yang X. Microbiol Mol Biol Rev. 2000;64:821–846. doi: 10.1128/mmbr.64.4.821-846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagus R, Kay J E. Eur J Biochem. 1979;100:503–510. doi: 10.1111/j.1432-1033.1979.tb04195.x. [DOI] [PubMed] [Google Scholar]
- 18.Minami Y, Samelson L E, Klausner R D. J Biol Chem. 1987;262:13342–13347. [PubMed] [Google Scholar]
- 19.Boehm U, Klamp T, Groot M, Howard J C. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 20.Dent A L, Shaffer A L, Yu X, Allman D, Staudt L M. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 21.Glimcher L H, Murphy K M. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- 22.Eynon E E, Schlax C, Pieters J. J Biol Chem. 1999;274:26266–26271. doi: 10.1074/jbc.274.37.26266. [DOI] [PubMed] [Google Scholar]
- 23.Morel A, Fernandez N, de La Coste A, Haddada H, Viguier M, Polla B S, Antoine B, Kahn A. Cancer Immunol Immunother. 1998;46:277–282. doi: 10.1007/s002620050488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 25.Macian F, Lopez-Rodriguez C, Rao A. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 26.Rengarajan J, Mittelstadt P R, Mages H W, Gerth A J, Kroczek R A, Ashwell J D, Glimcher L H. Immunity. 2000;12:293–300. doi: 10.1016/s1074-7613(00)80182-x. [DOI] [PubMed] [Google Scholar]
- 27.Flanagan W M, Corthesy B, Bram R J, Crabtree G R. Nature (London) 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 28.Ranger A M, Oukka M, Rengarajan J, Glimcher L H. Immunity. 1998;9:627–635. doi: 10.1016/s1074-7613(00)80660-3. [DOI] [PubMed] [Google Scholar]
- 29.Xanthoudakis S, Viola J P, Shaw K T, Luo C, Wallace J D, Bozza P T, Luk D C, Curran T, Rao A. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 30.Peng S L, Gerth A J, Ranger A M, Glimcher L H. Immunity. 2001;14:13–20. doi: 10.1016/s1074-7613(01)00085-1. [DOI] [PubMed] [Google Scholar]
- 31.Neilson J, Stankunas K, Crabtree G R. Curr Opin Immunol. 2001;13:346–350. doi: 10.1016/s0952-7915(00)00225-9. [DOI] [PubMed] [Google Scholar]
- 32.Michel F, Attal-Bonnefoy G, Mangino G, Mise-Omata S, Acuto O. Immunity. 2001;15:935–945. doi: 10.1016/s1074-7613(01)00244-8. [DOI] [PubMed] [Google Scholar]
- 33.Freedman B D, Liu Q H, Somersan S, Kotlikoff M I, Punt J A. J Exp Med. 1999;190:943–952. doi: 10.1084/jem.190.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward S G, Wilson A, Turner L, Westwick J, Sansom D M. Eur J Immunol. 1995;25:526–532. doi: 10.1002/eji.1830250233. [DOI] [PubMed] [Google Scholar]
- 35.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 36.Welsh G I, Proud C G. Biochem J. 1993;294:625–629. doi: 10.1042/bj2940625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 38.Graef I A, Mermelstein P G, Stankunas K, Neilson J R, Deisseroth K, Tsien R W, Crabtree G R. Nature (London) 1999;401:703–708. doi: 10.1038/44378. [DOI] [PubMed] [Google Scholar]
- 39.Ohteki T, Parsons M, Zakarian A, Jones R G, Nguyen L T, Woodgett J R, Ohashi P S. J Exp Med. 2000;192:99–104. doi: 10.1084/jem.192.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, Shibasaki F, Price R, Guillemot J C, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- 41.Gomez del Arco P, Martinez-Martinez S, Maldonado J L, Ortega-Perez I, Redondo J M. J Biol Chem. 2000;275:13872–13878. doi: 10.1074/jbc.275.18.13872. [DOI] [PubMed] [Google Scholar]
- 42.Klein P S, Melton D A. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kane L P, Lin J, Weiss A. Trends Immunol. 2002;23:413–420. doi: 10.1016/s1471-4906(02)02264-0. [DOI] [PubMed] [Google Scholar]
- 44.Kontgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 45.Bryan R G, Li Y, Lai J H, Van M, Rice N R, Rich R R, Tan T H. Mol Cell Biol. 1994;14:7933–7942. doi: 10.1128/mcb.14.12.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindstein T, June C H, Ledbetter J A, Stella G, Thompson C B. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 47.Chen C Y, Shyu A B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 48.Rengarajan J, Tang B, Glimcher L H. Nat Immunol. 2002;3:48–54. doi: 10.1038/ni744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.