Abstract
Helix–hairpin–helix (HhH) is a widespread motif involved in sequence-nonspecific DNA binding. The majority of HhH motifs function as DNA-binding modules with typical occurrence of one HhH motif or one or two (HhH)2 domains in proteins. We recently identified 24 HhH motifs in DNA topoisomerase V (Topo V). Although these motifs are dispensable for the topoisomerase activity of Topo V, their removal narrows the salt concentration range for topoisomerase activity tenfold. Here, we demonstrate the utility of Topo V's HhH motifs for modulating DNA-binding properties of the Stoffel fragment of TaqDNA polymerase and Pfu DNA polymerase. Different HhH cassettes fused with either NH2 terminus or COOH terminus of DNA polymerases broaden the salt concentration range of the polymerase activity significantly (up to 0.5 M NaCl or 1.8 M potassium glutamate). We found that anions play a major role in the inhibition of DNA polymerase activity. The resistance of initial extension rates and the processivity of chimeric polymerases to salts depend on the structure of added HhH motifs. Regardless of the type of the construct, the thermal stability of chimeric Taq polymerases increases under the optimal ionic conditions, as compared with that of TaqDNA polymerase or its Stoffel fragment. Our approach to raise the salt tolerance, processivity, and thermostability of Taq and Pfu DNA polymerases may be applied to all pol1- and polB-type polymerases, as well as to other DNA processing enzymes.
Many gene regulatory proteins contain small, discrete structural motifs that use either α-helices or β-strands for specific DNA binding. Among these are the helix-turn-helix, zinc finger, leucine zipper, and helix–loop–helix motifs (1). The motifs may arise in different molecular contexts, which is believed to occur because of either divergent evolution by means of gene duplication and insertion or structural convergence by means of the effects of selective pressures on protein function. Other proteins bind DNA by using sequence-nonspecific interactions. They belong to various protein families, such as nucleases, N-glycosylases, ligases, polymerases, and helicases that are essential for the protein-mediated synthesis and DNA repair. These proteins have recently been shown to have a common structural motif, called helix–hairpin–helix (HhH; refs. 2 and 3). Doherty et al. (4) have analyzed in detail the structures of HhH motifs in several protein families and concluded that they exist as separate units. Shao et al. (5) have shown that HhH motifs are typically involved in the formation of a larger structure of five α-helices called (HhH)2 domain. (HhH)2 is a pseudo-twofold unit composed of two HhH motifs linked by a connector α-helix. This compact structure with a well defined hydrophobic core mirrors the symmetry of the DNA-double helix and facilitates strong DNA binding. Protein–DNA contacts do not involve DNA bases but rather a sugar-phosphate chain, making proteins with HhH motifs able to bind DNA in the nonsequence-specific manner.
A unique number of HhH motifs has been identified in DNA topoisomerase V (TopoV; refs. 6–8). The 684 C-terminal amino acids (from a total of 984 amino acids) are organized into 12 repeats of ≈50 amino acids long each (9). All repeats consist of two similar HhH motifs. We demonstrated earlier that Topo V proteins lacking different parts of an HhH superdomain remain fully active in relation to supercoiled DNA but become sensitive to salts (9, 10). Thus, HhH motifs play a crucial role in Topo V interactions with DNA, which anchor the enzyme on the substrate at high levels of salts. They also can confer high processivity on Topo V in a very broad range of salt concentrations (6–8).
Prompted by this effect of HhH motifs on the activity and processivity of Topo V at high concentrations of salts and by the discovered domain structure of the motifs (10), we designed hybrid proteins consisting of the Stoffel fragment of Taq DNA polymerase or Pfu DNA polymerase and different TopoV's HhH subdomains fused with either the NH2- or the COOH terminus of polymerases. By using the purified hybrid enzymes, we investigated in detail the effect of HhH motifs on the polymerization rates, processivity, and thermostability of hybrid DNA polymerases at high salt concentrations.
Materials and Methods
DNA Polymerases.
All plasmids for chimeric proteins were constructed by common subcloning techniques, and the DNA polymerases were expressed and purified as described in Figs. 5–14 and Tables 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org. Taq DNA polymerase and its KlenTaq variant were purchased from Roche Molecular Biochemicals and from GeneCraft (Munster, Germany), respectively. The Stoffel fragment was obtained from Applied BioSystems, and Pfu DNA polymerase was obtained from Stratagene.
Primer Extension Assay.
An enzymatic polymerization in primer extension reactions can be described by Scheme 1:
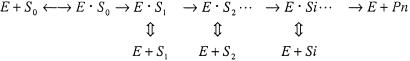 |
in which each product of a consecutive step of the polymerization, except the last one, is, in turn, a substrate (Si) for the next step. Rates of accumulation of these intermediate substrates Si and their complexes with the enzyme [or products of each step of polymerization, Pi; (Si) ≡ (Pi)] can be expressed by equations
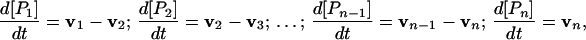 |
1 |
where vi are rates of appearance of a product for each step of polymerization. The total rate of accumulation of the products calculated for the sum of concentrations of all extended species in reaction in Scheme 1,
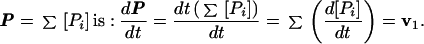 |
2 |
That is, dP/dt is equal to the rate of attachment of the first nucleotide (v1). Therefore, the initial rates for nucleotide incorporation into DNA can be obtained from time courses for accumulation of extended products by extrapolating dP/dt to t = 0.
A primer extension assay was developed with a fluorescent duplex substrate containing a primer-template junction (PTJ). The duplex was prepared by annealing a 5′-end-labeled with fluorescein 20-nt long primer with a 40-nt long template:
DNA polymerase reaction mixtures (15–20 μl) contained 1 mM each dATP, dTTP, dCTP, and dGTP, 4.5 mM MgCl2, 0.2% each of detergents Tween 20 and Nonidet P-40, fixed concentrations of PTJ-duplex, other additions, as indicated, and appropriate amounts of DNA polymerases in 30 mM Tris·HCl buffer, pH 8.0 (25°C). The background reaction mixtures contained all components except the polymerases. Primer extensions were carried out for a preset time at 75°C in a PTC-150 Minicycler (MJ Research, Cambridge, MA). For each reaction, 5 μl of samples were removed, typically, after 3, 6, and 9 min of incubation and chilled to 4°C, followed by the immediate addition of 20 μl of 20 mM EDTA. The samples were desalted by centrifugation through Sephadex G-50 spun columns, diluted, and analyzed on an ABI Prism 377 DNA sequencer (Applied Biosystems). For each sample, raw data were extracted from the sequencer trace files with the program CHROMAS V.1.5 (Technelysium, Australia), and the fluorescent signals were analyzed by our nonlinear regression data analysis programs written in FORTRAN. The programs applied Powell (11) algorithms to approximate the signals by a number of Gaussian peaks and calculate integral fluorescent intensities for each product peak (see Fig. 5). The total amount of fluorescent products for each time of incubation was determined, and the initial rates of extension were calculated from the progressive accumulation of the products, as described (12).
Processivity Assays for DNA Polymerases.
The microscopic processivity parameter, pn, for a position n of PTJ can be determined from signals of 5′-end-labeled extended products that appeared in a template-directed DNA synthesis, which was carried out under single-hit conditions of the assay as follows (13):
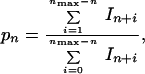 |
3 |
where In+i are gel-band intensities for oligonucleotides extended by subsequent additions of deoxynucleotides. The single-hit conditions occur in the initial period of a primer extension reaction if the PTJ is in great excess for DNA. Because during the initial period of primer extension reaction (Δt)In+i = (dIn+i/dt)⋅Δt, pn can also be expressed through initial rates of appearance of extended products:
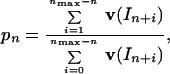 |
4 |
where
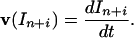 |
For processivity assays, the primer extension reactions were carried out and analyzed as described above, but after determination of the amount of extended products, the initial rates for appearance of each extended primer were calculated (12). Then, processivity for each position of the template was determined by using Eq. 4, and the processivity equivalence parameter, Pe, was calculated for each reaction (see Fig. 7).
Studies of Thermostability of DNA Polymerases.
Proteins in 25 μl of 20 mM Tris·HCl buffer (pH 8.0 at 25°C) containing 1 M potassium glutamate and 1 M betaine were incubated in a PTC-150 Minicycler (MJ Research) at 100°C. Samples (4 μl) were removed at defined times of incubation and assayed for primer extension activity.
Results
Design of Chimeric DNA Polymerases.
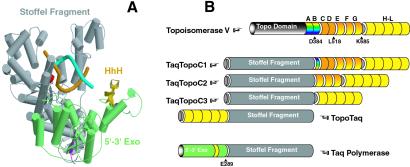
We designed five protein chimeras containing either the Stoffel fragment of Taq polymerase or whole size Pfu polymerase and a different number of HhH motifs derived from Topo V: TopoTaq, containing HhH repeats H-L of Topo V (10 HhH motifs; repeats are designated as in ref. 9) fused with the Stoffel fragment of Taq polymerase; TaqTopoC1, TaqTopoC2, and TaqTopoC3, consisting of the Stoffel fragment fused, respectively, with Topo V's repeats B-L (21 HhH motifs), repeats E-L (16 HhH motifs), and H-L (10 HhH motifs); and PfuC2-containing repeats E-L at the Pfu polymerase's COOH termini. Repeats H-L and F-L with a half of the repeat E are dispensable for the topoisomerase activity of Topo V (10). The composition of Taq polymerase and HhH domains and their positions in the Taq chimeras are shown in Fig. 1B.
Figure 1.
Schematic representation of chimeric polymerases. (A) Domain organization of TaqDNA polymerase in which helices are represented by cylinders and β-strands by arrows. This structure has been modeled by using two available x-ray structures of Taq polymerase (in “open” and “closed” conformations; for details, see Text, which is published as supporting information on the PNAS web site). The polymerase and inactive 3′-5′ exonuclease domains are colored gray, and the 5′-3-exonuclease domain is colored green. Several amino-terminal and carboxyl-terminal amino acids are colored magenta and red, respectively. The only HhH motif in the 5′-3′ exonuclease domain is colored gold. DNA strands are colored cyan and orange. (B) Cartoon illustration of chimeric constructs. HhH repeats of Topo V are colored yellow (H–L), orange-yellow gradient (E–G), orange (C and D), and rainbow (A and B). Arrows indicate cleavage positions that result in C1–C3 domains (in case of Topo V) and the Stoffel fragment (in case of Taq polymerase).
The x-ray structure of Topo V and its HhH repeats is unknown, and we can only assume that H-L repeats in TopoTaq and TaqTopoC3 (Fig. 1) exist as well defined structural domains. However, the ability of individual Topo V repeats to adopt an HhH-fold is supported by 3D modeling using the structural information obtained for other proteins with HhH domains (for details, see supporting information on the PNAS web site). The gross structures of C1–C3 domains are likely preserved in chimeras, because the domains are relatively resistant to proteolysis, both in Topo V and when expressed as individual proteins (Topo34; ref. 10).
HhH Domains Confer DNA Polymerase Activity on Chimeras in High Salts.
To assess the influence of salts on interaction of DNA polymerase with PTJ, the primer extension reactions were carried out at low concentrations of substrate, when the initial rates were proportional both to total protein and to PTJ concentrations. (Because at [PTJ] ≪ Kmapp, the equation for the initial rate becomes: v1 = (kapp/Kmapp)⋅[Et]⋅[PTJ], where Kmapp and kapp are apparent Michaelis and catalytic constants, respectively (for details, see supporting information on the PNAS web site). We varied concentrations of NaCl, KCl, and KGlu to assess inhibition of Stoffel fragment, KlenTaq, Taq polymerase, and the chimeras by salts and to estimate the effects of HhH motifs.
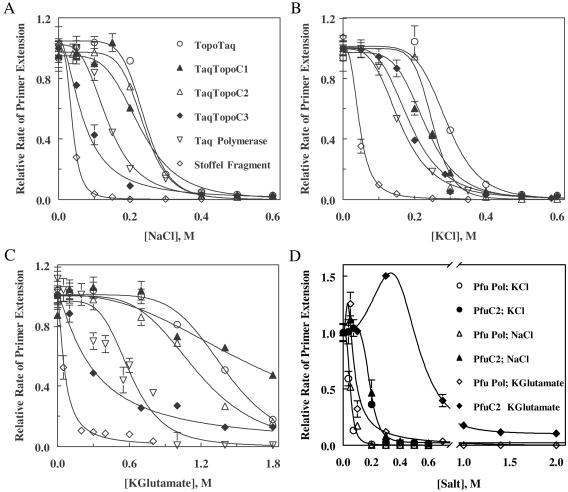
Fig. 2 shows sigmoid curves indicating the cooperative inhibition of the enzymes by salts. Experimental values of initial polymerization rates were analyzed by nonlinear regression using the following function:
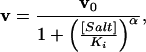 |
5 |
where v and v0 are initial primer extension rates with and without salt, respectively; Ki is an apparent inhibition constant, and α is a parameter of cooperativity. Values for Ki and α are listed in Table 1, which is published as supporting information on the PNAS web site. The dependencies in Fig. 2 can be explained if α refers to the number of ions of any kind, bound either to the protein or to DNA, that prevent formation of a productive polymerase–DNA complex. However, the results presented below suggest that anions play a major role in the inhibition.
Figure 2.
Activity of TaqDNA polymerase, the Stoffel fragment, Pfu polB, and the hybrid polymerases in salts. Initial rates of primer extension reactions for the proteins were determined as described in Materials and Methods, and the dependencies of the rates for enzymes with Taq polymerase catalytic domain on salt concentrations were plotted for NaCl (A), KCl (B), and potassium glutamate (C). Solid lines are theoretical curves of inhibition by salts calculated by using Eq. 5. The dependencies of the rates for enzymes with Pfu polymerase catalytic domain are collected in D.
For Taq polymerase, Ki for NaCl and KCl are essentially the same, but the substitution of Cl− for Glu− increases Ki fourfold. Hence, Taq polymerase is sensitive to anions. The parameter of cooperativity is very similar for all salts tested and suggests that as many as four anions simultaneously bound to the protein are involved. The Stoffel and KlenTaq fragments of TaqDNA polymerase have almost equal sensitivities to Cl−, which is about four times higher than that of Taq polymerase. KGlu inhibits the fragments only ≈1.5–2 times less efficiently than the chlorides, implying that only the HhH motif in the 5′-3′ exonuclease domain of Taq polymerase might be responsible for the resistance of the enzyme to Glu−.
TopoTaq has higher values of Ki in salts, as compared with Taq polymerase, and may require six or seven anions to be bound for the inhibition (Table 1). As a result, TopoTaq is active at much higher salt concentrations than TaqDNA polymerase. For example, 20% inhibition of primer extension reaction occurs at ≈200 mM NaCl in the case of TopoTaq vs. ≈90 mM NaCl for TaqDNA polymerase. This hybrid also displays little distinction between Na+ and K+ and is less sensitive to Glu− vs. Cl−.
Remarkably, 21 and 16 HhH motifs at the COOH terminus of the Stoffel fragment in TaqTopoC1 and TaqTopoC2, respectively, also increase their resistance to salts. Twenty percent inhibition occurs at ≈160 mM NaCl with TaqTopoC1 and at ≈190 mM NaCl with TaqTopoC2 (Fig. 2). Similar to Taq polymerase, these chimeras show no difference in the inhibition by KCl vs. NaCl (with α ≈ 5), and Glu− is much more preferable than Cl−. However, α for TaqTopoC1 and TaqTopoC2 in KGlu is lower than that of Taq polymerase or TopoTaq, which suggests that only two glutamate ions are involved in the inhibition.
Although inhibiting TaqTopoC3 by KCl is similar to that of TaqTopoC1 or TaqTopoC2 (with α ≈ 5, and Ki similar to that of TaqDNA polymerase), replacement of K+ by Na+ results in a much stronger inhibition of the TaqTopoC3 polymerase activity and, at the same time, decreases the apparent number of inhibiting ions to ≈2. Consequently, just 30 mM NaCl inhibits the enzyme by 20%. TaqTopoC3 has about fivefold relative decrease in sensitivity to KGlu with respect to NaCl (but not to KCl), which is similar to other hybrids. However, in the case of KGlu, no cooperativity at all was found; that is, only one Glu− ion per molecule can be involved in the inhibition.
Surprisingly, the sensitivity of Pfu DNA polymerase to salts was almost identical to that of Stoffel or KlenTaq fragments of DNA polymerase from Thermus aquaticus, possibly indicating the close functional similarity of charged amino acid residues in the active sites of these enzymes from different structural families. Attachment of Topo V HhH domains to the C terminus of Pfu polB significantly increased the resistance of polymerase activity to salts (Fig. 2D). Both Pfu DNA polymerase and the chimera PfuC2 demonstrated virtually indistinguishable curves for KCl vs. NaCl, suggesting no role for cations in inhibition. Interaction of both Pfu polymerase and PfuC2 with KGlu was more complicated because of activation at low concentrations of Glu−, as we also observed for other archaeal polB DNA polymerases (unpublished data). However, the TopoV domains greatly increased the resistance of Pfu polB activity to high levels of KGlu.
Activation of TopoTaq in Betaine.
We found that betaine at 1 M concentration significantly increases the initial rate of primer extension reaction in the presence of 0.3 M NaCl (see Fig. 8). Further increase of betaine concentration up to 2 M at 10.5 μg/ml TopoTaq did not affect the primer extension activity. Higher betaine concentrations inhibited the polymerase reaction (data not shown). The activation by betaine only occurred if TopoTaq was inhibited by NaCl and brought the polymerase activity back to the level of the noninhibited enzyme.
The reason that betaine activates TopoTaq in 0.3 M NaCl is of interest. Because binding DNA to proteins involves both electrostatic and nonelectrostatic interactions, it seems possible to eliminate the electrostatic interactions at high salt concentrations. We previously observed that the very slow dissociation of Topo V from DNA at a low salt concentration (50 mM NaCl) prevents cycling of the enzyme between DNA molecules (7). However, at 0.3 M NaCl, the binding becomes less tight, so that Topo V is able to cycle between DNA molecules. Accordingly, the activity of Topo V prebound to dsDNA substrates is strongly inhibited by ssDNA added at 0.3 M NaCl (14). Such behavior of Topo V suggests that the charge interactions are predominant in the binding of DNA to the HhH domains. However, this nonspecific inhibition by ssDNA could be abolished if 2.2 M betaine is added to 0.3 M NaCl, resulting in selective binding of Topo V to dsDNA.
The inhibition of DNA synthesis by anions (Fig. 2; Table 1) presumes the combined effect of salts on electrostatic interaction of DNA substrate with both Taq catalytic domain and HhH structures of Topo V. The lower activity of TopoTaq in 0.3 M NaCl alone, as compared with that in the presence of 0.3 M NaCl and 1–2 M betaine, can be attributed to binding of the single-stranded region of PTJ by the HhH domains, which facilitates dissociation of the substrate DNA from the catalytic domain. In the presence of betaine, the TopoTaq HhH domains may preferentially bind the double-stranded region of the duplex and provide proper orientation of PTJ with respect to the polymerase catalytic site.
Rees et al. (15) have found that both electrostatic properties of DNA and B-conformation of dsDNA were retained even at very high concentrations of betaine (≈5.5 M). They observed that, in the absence of salts, betaine did not affect the preferential interaction of RNase A with an ssDNA much and, even at very high concentrations, did not disrupt electrostatic interactions of DNA with the protein. Their results (15) also indicate that, at very high concentrations of betaine, RNase A starts binding mostly dsDNA.
Overall, our data support earlier conclusions that prevention of nonspecific protein–nucleic acid interactions might be one of the so-far unaccounted for physiological roles of zwitterionic compounds like betaine or other modified tetraalkylammonium compounds (14–16).
Role of HhH Domains in Processive DNA Synthesis.
DNA polymerases can perform a sequence of polymerization steps without intervening dissociation from the growing DNA chains. This property, called processivity, strongly depends on the nature of the polymerase, the sequence of a DNA template, and other reaction conditions, such as salt concentration, temperature, or presence of specific proteins.
von Hippel and coworkers (17) introduced a model to describe the processive synthetic process in DNA replication quantitatively. They defined the microscopic processivity parameter, pi, which is the probability that a polymerase positioned at the PTJ at template position i will not dissociate from the DNA in translocation to position i+1. This parameter can be determined experimentally by measuring the fraction of extended primers that reaches position n but does not terminate there under single-hit conditions of an assay; i.e., when a DNA polymerase dissociating from the PTJ does not bind to extended products (13, 17, 18). To characterize processive DNA synthesis on heterogeneous templates, the geometric mean microscopic processivity parameter was introduced (17). This parameter is useful for comparison of the same number of nucleotide attachments in different reactions. However, any premature termination of extension within the defined length of DNA synthesis, as in the case of primer extensions in high salts, renders this parameter 0. To by-pass this limitation, we define here the processivity equivalence parameter, Pe (see Fig. 7). Pe is a value of processivity of a DNA polymerase reaction for an infinite homopolymer template that would produce the same average substrate extension per polymerase-binding event as a reaction on a heterogeneous template characterized by nonconstant values of pi. The modified parameter allows one to conveniently assess the general effect of salts on processivity in primer extension, as it eliminates the length restriction inherent to the geometric mean microscopic processivity parameter.
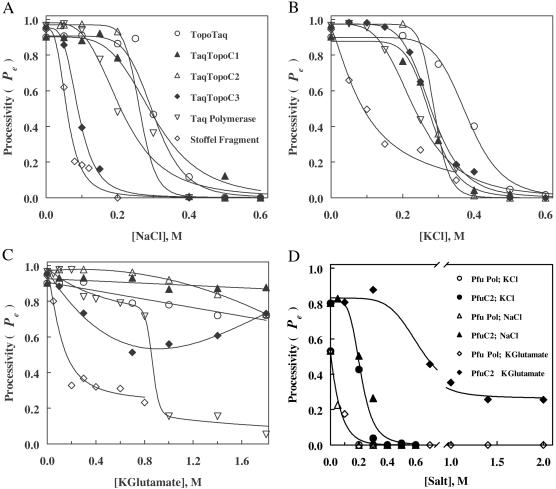
Plots of Pe for Taq polymerase, its fragments, and the Taq polymerase-Topo V hybrids are shown in Fig. 3 A–C. In the absence of added salts, at 75°C, all proteins have high processivity (Pe = 0.90–98). Lower values of Pe are observed for Pfu and PfuC2 DNA polymerases (Fig. 3D). Clearly, Cl− completely inhibits the processive synthesis of DNA. In contrast, inhibition of the processive synthesis by KGlu is weaker and more complicated. Although the rates of primer extension become very low (Fig. 2C), the processivity of the reaction is not suppressed completely (Fig. 3C). Because of the functional dependence of the polymerization rate on processivity (see supporting information on the PNAS web site), a PTJ duplex, once bound to the polymerase, makes some inhibitory anion-binding sites inaccessible to Glu− but not to Cl−. Inhibition of processivity by KGlu also demonstrates that anions rather than cations play a major role in modification of DNA polymerase activity.
Figure 3.
Processivity of TaqDNA polymerase, the Stoffel fragment, Pfu polB, and the hybrid polymerases in salts. Processivities of enzymes in primer extension reactions were determined as described in Materials and Methods, and the dependencies of Pe for enzymes with Taq polymerase catalytic domain on salt concentrations were plotted for NaCl (A), KCl (B), and potassium glutamate (C). The dependencies of Pe for enzymes with Pfu polymerase catalytic domain are collected in D.
Thermostability of DNA Polymerase Chimeras.
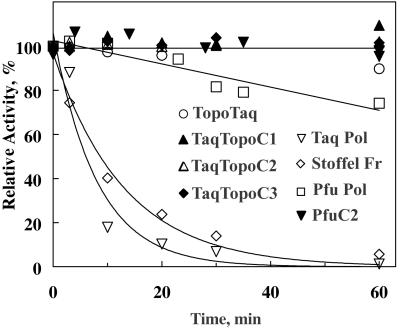
Introduction of additional protein domains into TaqDNA polymerase could have adverse effects on the enzyme stability, especially at high temperatures (19). To determine the effect of the fused Topo V HhH domains on the stability of the catalytic polymerase domains, we incubated Taq polymerase, the Stoffel fragment, Pfu polymerase, and the hybrid polymerases at high temperature (100°C) in the presence of 1 M potassium glutamate and 1 M betaine. As shown in Fig. 4, under these conditions, all constructed chimeras with the amino acid sequences of Topo V HhH repeats have extremely high thermostability, allowing them to stay at least 1 h at 100°C without any detectable loss of activity. In contrast, Taq polymerase, the Stoffel fragment, and even highly resistant Pfu polymerase show significant inactivation; therefore, the effect of stabilization can be specifically attributed to the attached TopoV domains.
Figure 4.
Thermostability of TaqDNA polymerase, the Stoffel fragment, Pfu polB, Taq polymerase-Topo V, and Pfu polB chimeras at 100°C in 1 M potassium glutamate and 1 M betaine.
DNA Sequencing with TopoTaq at High Salt Concentrations.
We attempted to use TopoTaq chimera for direct cycle sequencing at higher salt concentrations. The hybrid enzyme was able to perform both primer extension and chain termination with ddNTPs at 0.25 M NaCl, whereas unmodified TaqDNA polymerase was totally inefficient under these conditions (see Fig. 9). Because in all other respects a regular sequencing protocol (developed for TaqDNA polymerase; ref. 20) was used, we assume that NaCl did not cause any significant changes in dNMP/ddNMP incorporation. Also, it seems that the ability to incorporate 7-deaza-dGTP (used in this protocol) was not impaired, in agreement with our suggestion that salts do not inhibit the catalysis of chain elongation by active sites of DNA polymerases but do interfere with the ability of the proteins to remain bound to DNA substrate during synthesis.
Discussion
Salt Resistance.
Inhibition of Taq and Pfu polymerases and their derivatives by the salts reveals complicated cooperative binding of DNA by polymerase catalytic domains with HhH repeats. Such interaction between the polymerase domains and Topo V domains provides full DNA polymerase activity of the chimeras at high salt concentrations. With the exception of TaqTopoC3, introduction of C-terminal domains of Topo V into the hybrid proteins significantly extends the range of salt concentrations for the polymerase activity. The effect is caused by the increase of both Ki and α, allowing chimeras to maintain their full activity at high salt concentrations. Such behavior indicates that, in the catalytic domain, the incorporation of dNMPs is not affected even at high concentrations of salts; rather, binding of DNA substrate is impaired, because of competition between DNA and the anions for the positively charged amino acid residues of the proteins. Raising the number of HhH motifs from 11 to 23 at the COOH terminus of the Stoffel fragment makes the hybrid enzymes progressively more resistant to salts.
Although any TopoV HhH domain can potentially participate in DNA binding, we presume that the domains with partial positive charge at neutral pH would more easily interact with negatively charged DNA. Positively charged HhH motifs may initiate DNA binding and position DNA for further interaction with other domains. The dependencies of charge on pH calculated for individual HhH domains revealed very small changes in the charge at pH 5.5–8.5 (for charge distribution along TopoV's HhH motifs, see Fig. 10, which is published as supporting information on the PNAS web site). The strong positive charge of the C-terminal domains I-L should provide preferential interaction of DNA with those domains that are responsible for Topo V activities at high salt concentrations (10). In addition, domains H-L form a separate structure that is resistant to proteolysis and binds DNA without the rest of Topo V molecule (10). A 3D model illustrating a possible position of DNA bound to the L domain of TopoV is shown in Figs. 11–13. The model uses molecular contacts similar to those found in complexes of E. coli helicase RuvA and human polymerase β with DNA.
Nonetheless, it is TopoTaq that has the highest resistance to Cl−. Although both TopoTaq and TaqTopoC3 contain the same C3 HhH domain (also known as Topo34; ref. 10), TaqTopoC3 does not show significant increase in salt resistance. It seems that, in TopoTaq, the C3 domain at the NH2 terminus of the Stoffel fragment (Fig. 1) is in a favorable orientation to interact specifically with dsDNA. This conclusion is further supported by modeling experiments described in Text, which is published as supporting information on the PNAS web site. In addition, a significant homology of Topo V's HhH motifs to the entire 5′ → 3′ exonuclease domain of Taq polymerase may create a better environment for the smooth integration of the C3 domain with the Taq polymerase domain (see Fig. 14).
In contrast, at the COOH terminus, in TaqTopoC3, the C3 domain is remote from dsDNA. Yet, it still contacts PTJ, tethering the polymerase domain to DNA and providing the difference in salt inhibition, as compared with the Stoffel and KlenTaq fragments. Most likely, in this case, the C3 domain makes nonspecific electrostatic contacts with the single-stranded part of PTJ. In TaqTopoC1, and C2 constructs, both C1 and C2 domains include the C3 domain, but they also contain additional HhH repeats, which form rather loose structures as shown by limited proteolysis of Topo V (10). These additional repeats may extend the “HhH arm” sufficiently far to reach dsDNA (Fig. 1).
Processivity.
The pattern of inhibition of the processive DNA synthesis by Taq and Pfu polymerases and chimeric constructs reveals separate roles of catalytic polymerase domains and HhH domains in maintenance of processivity. The inhibition demonstrates that, at least in TopoTaq, the HhH domains support proper orientation of the PTJ substrate in polymerase active site in addition to tethering the whole protein to DNA substrate, as is likely the case with other constructs. Without salts added, the Stoffel fragment has processivity that is very close to that of Taq polymerase. Addition of N-terminal amino acids that produce KlenTaq slightly decreases the processivity, probably because of the interference of those amino acids with the optimal PTJ position. The published structure of the catalytic domain of Taq polymerase (21) suggests that some anion-binding sites that stabilize the position of PTJ can easily interact with anions in the solution (especially, residues Arg-677 and -746 and Lys-508). However, such interactions do not completely destabilize the productive polymerase-PTJ complex, because the other four basic amino acid residues in proximity to the substrate (Arg-487, -587, -728, and Lys-540) are less accessible to anions. We presume that the original HhH domain in Taq polymerase stabilizes the proper position of PTJ in the protein-substrate complex, but when the domain gets dissociated at higher salt concentrations, PTJ remains bound, as in TaqDNA polymerase fragments. The dissociation produces an abrupt decrease of Taq polymerase processivity in the presence of KGlu, shown in Fig. 3C. Constructs TopoTaq, TaqTopoC1–C3, and PfuC2 do not seem to have their HhH domains dissociated from the substrate even at very high glutamate concentrations; in contrast, the increase of the salt level apparently helps to organize tighter binding of HhH domains with the substrate, as is the case with TopoTaqC3 (Fig. 3C).
It is important to point out the difference in effect of the fused HhH domains on the processivity of Taq and Pfu chimeras. The processivity of PfuC2 is considerably higher than that of the original Pfu polymerase, both without salt added and at increased salt concentrations (Fig. 3D). In contrast, it seems that the processivity of Taq chimeras (Fig. 3 A–C) never exceeds that of the unmodified Taq polymerase (corresponding to the reported average length of extended products 20–40 nucleotides; ref. 22). Rather, the HhH domains raise the processivity of the chimeras in high salts to the level of the core polymerase. As the rate of nucleotide incorporation by the Taq catalytic domain is not affected by salts if the PTJ is bound to the active site (Fig. 2 A–C), the processivity is determined by the rate of dissociation of the polymerase–DNA complex. It is likely that, at low salt concentrations, the dissociation of TopoV HhH domains from DNA substrates occurs with similar or even lower rates as that of the Taq polymerase catalytic domain. Yet, no increase in processivity within the precision of our measurements could be found in experiments with short-substrate DNA, as the processivity of the catalytic domain alone is very high. However, at high salt concentrations, the detachment of the salt-resistant TopoV domains clearly becomes the rate-limiting step for the dissociation of the entire polymerase-DNA complex. In contrast, the rate of dissociation of Pfu polymerase catalytic domains seems to be much higher than that of TopoV HhH domains and contributes to low processivity of DNA synthesis by this polymerase together with its 3′ → 5′ exonuclease activity. The hybrid PfuC2 has improved binding to DNA, and its lower processivity in DNA extension, as compared with Taq polymerase chimeras, can be attributed to the balance of the polymerase/exonuclease reactions in the core Pfu polB domain.
Conclusion.
Harnessing of other DNA processing enzymes, for example, reverse transcriptases or restriction enzymes, by topologically linked HhH domains to DNA in the presence of salts also should be possible. A potential problem of such tasks lies in possible interference of the attached polypeptide segments with functions of the catalytic domains in hybrid proteins. Thus, most attempts to modify the substrate or cofactor specificity of the enzymes rely on the site-directed mutagenesis of one or a few residues. An interesting concept, however, lies in the exchange of defined structural subdomains, whose proper folding is more likely to be preserved (reviewed in refs. 23 and 24). The TopoTaq chimera is a good example of such an approach.
However, the incorporation of topologically linked HhH repeats even into unmodified enzymes should not necessarily perturb the structurally defined catalytic domains; rather, they may act in concert, as is the case with PfuC2. In this regard, it is important that there are many naturally occurring proteins, in which one or more HhH motifs are positioned either at the NH2 or COOH termini (see Table 2). Such proteins are primary candidates for HhH-domain exchange or additions.
The ability to engineer hybrid proteins with enhanced DNA-binding properties can be useful for many practical applications. TaqDNA polymerase (25) carries out fast and processive synthesis of DNA, it has high thermostability, but its activity is inhibited at elevated salt concentrations. Pfu polymerase has a high fidelity, but it is salt-sensitive and is not processive. Moreover, it seems that the majority of commercially available thermostable DNA polymerases show little or no activity at NaCl or KCl concentrations over 80 mM (20, 26). Their resistance to salts could be increased by fusion with properly selected Topo V HhH domains.
Supplementary Material
Acknowledgments
We thank Anna Pavlova for help with manuscript preparation, and VTI Ltd. for excellent laboratory services. This work was supported in part by Department of Energy and National Institutes of Health Grants DE-FG02-98ER82577, 00ER83009, R44GM55485, and R43HG02186 (to S.A.K and A.I.S) and by a grant from the Howard Hughes Medical Institute (to A.I.S.).
Abbreviations
- HhH
helix–hairpin–helix
- Topo V
topoisomerase V
- PTJ
primer-template junction
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Harrison S C. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 2.Thayer M M, Ahern H, Xing D, Cunningham R P, Tainer J A. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelletier H, Sawaya M R, Wolfle W, Wilson S H, Kraut J. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 4.Doherty A J, Serpell L C, Ponting C P. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao X, Grishin N V. Nucleic Acids Res. 2000;28:2643–2650. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slesarev A I, Stetter K O, Lake J A, Gellert M, Krah R, Kozyavkin S A. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 7.Slesarev A I, Lake J A, Stetter K O, Gellert M, Kozyavkin S A. J Biol Chem. 1994;269:3295–3303. [PubMed] [Google Scholar]
- 8.Slesarev A I, Belova G I, Lake J A, Kozyavkin S A. Methods Enzymol. 2001;334:179–192. doi: 10.1016/s0076-6879(01)34467-1. [DOI] [PubMed] [Google Scholar]
- 9.Belova G I, Prasad R, Kozyavkin S A, Lake J A, Wilson S H, Slesarev A I. Proc Natl Acad Sci USA. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belova G I, Prasad R, Nazimov I V, Wilson S H, Slesarev A I. J Biol Chem. 2002;277:4959–4965. doi: 10.1074/jbc.M110131200. [DOI] [PubMed] [Google Scholar]
- 11.Press W H, Flannery B P, Teukolsky S A, Vetterling W T. Numeric Recipes: The Art of Scientific Computing. Cambridge, U.K.: Cambridge Univ. Press; 1986. [Google Scholar]
- 12.Boeker E A. Biochem J. 1982;203:117–123. doi: 10.1042/bj2030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washington M T, Johnson R E, Prakash S, Prakash L. J Biol Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 14.Kozyavkin S A, Pushkin A V, Eiserling F A, Stetter K O, Lake J A, Slesarev A I. J Biol Chem. 1995;270:13593–13595. doi: 10.1074/jbc.270.23.13593. [DOI] [PubMed] [Google Scholar]
- 15.Rees W A, Yager T D, Korte J, von Hippel P H. Biochemistry. 1993;32:137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- 16.Melchior W B, Jr, Von Hippel P H. Proc Natl Acad Sci USA. 1973;70:298–302. doi: 10.1073/pnas.70.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairfield F R, Newport J W, Dolejsi M K, von Hippel P H. J Biomol Struct Dyn. 1983;1:715–727. doi: 10.1080/07391102.1983.10507477. [DOI] [PubMed] [Google Scholar]
- 18.von Hippel P H, Fairfield F R, Dolejsi M K. Ann NY Acad Sci. 1994;726:118–131. doi: 10.1111/j.1749-6632.1994.tb52803.x. [DOI] [PubMed] [Google Scholar]
- 19.Villbrandt B, Sobek H, Frey B, Schomburg D. Protein Eng. 2000;13:645–654. doi: 10.1093/protein/13.9.645. [DOI] [PubMed] [Google Scholar]
- 20.Innis M A, Myambo K B, Gelfand D H, Brow M A. Proc Natl Acad Sci USA. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Mitaxov V, Waksman G. Proc Natl Acad Sci USA. 1999;96:9491–9496. doi: 10.1073/pnas.96.17.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perler F B, Kumar S, Kong H. Adv Protein Chem. 1996;48:377–435. doi: 10.1016/s0065-3233(08)60367-8. [DOI] [PubMed] [Google Scholar]
- 23.Nixon A E, Ostermeier M, Benkovic S J. Trends Biotechnol. 1998;16:258–264. doi: 10.1016/s0167-7799(98)01204-9. [DOI] [PubMed] [Google Scholar]
- 24.Beguin P. Curr Opin Biotechnol. 1999;10:336–340. doi: 10.1016/S0958-1669(99)80061-5. [DOI] [PubMed] [Google Scholar]
- 25.Kaledin A S, Slyusarenko A G, Gorodetskii S I. Biokhimiya. 1980;45:644–651. [PubMed] [Google Scholar]
- 26.Abu Al-Soud W, Radstrom P. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.