Abstract
Mate finding in most moth species involves long-distance signaling via female-emitted sex pheromones. There is a great diversity of pheromone structures used throughout the Lepidoptera, even among closely related species. The conundrum is how signal divergence has occurred. With strong normalizing selection pressure on blend composition and response preferences, it is improbable that shifts to pheromones of diverse structures occur through adaptive changes in small steps. Here, we present data supporting the hypothesis that a major shift in the pheromone of an Ostrinia species occurred by activation of a nonfunctional desaturase gene transcript present in the pheromone gland. We also demonstrate the existence of rare males that respond to the new pheromone blend. Their presence would allow for asymmetric tracking of male response to the new blend and, thus, evolution of an Ostrinia species with structurally different sex pheromone components.
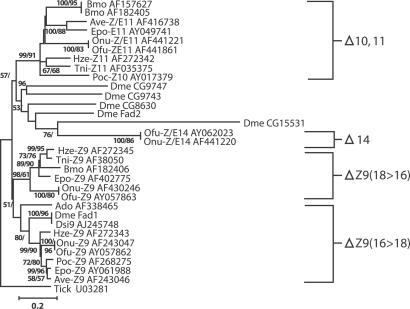
Mate finding in most moth species involves the use of long-distance sex pheromones, which are emitted by females (Arn, H. The Pherolist, www.nysaes.cornell.edu/fst/faculty/acree/pheronet/index.html). These chemical communication systems are highly canalized, with strong selection pressure against novel blends and response preferences. Thus, it is unlikely that shifts to pheromones of diverse structures occur through adaptive changes in small steps (1); rather, structural changes in the signaling system may require a major shift. In moth species studied to date, pheromone production (female) and response (male) are not genetically linked (2), but a model for the evolution of new pheromones based on asymmetric tracking (3, 4) predicts that a large mutational effect in female pheromone production can subsequently be tracked by male response. This model can explain how the Asian corn borer (ACB), Ostrinia furnacalis, evolved to use (Z)- and (E)-12-tetradecenyl acetate (Z/E12-14:OAc) pheromone components (5), whereas all other analyzed Ostrinia species use Z/E11-14:OAcs (6) (Fig. 1). What has been difficult to understand is how a major shift in a pheromone blend like this could occur.
Figure 1.
Pheromone biosynthetic pathways for ACB and ECB from hexadecanoic acid (16:Acid) and proceeding through different routes to the 14-carbon acetate pheromone components.
Moth sex pheromones are produced in specialized female abdominal glands, generally via unsaturated fatty-acid precursors produced by desaturases that exhibit a range of stereo- and regiospecificities (7). In an attempt to understand how these diverse desaturases evolved, we have characterized desaturase genes from various moth species. These include genes for desaturases producing Z9- (8–12), Z10- (10), Z11- (11–13), and E11-isomers (9, 12) of C14 and C16 unsaturated acids. In the course of these studies, we discovered that, in female European corn borer (ECB), O. nubilalis, the pheromone gland contains three different transcripts (cDNA) of desaturase genes, only one of which appears to result in a functional protein product. The functional transcript in ECB is for a Δ11-desaturase used for production of its pheromone components. One of the nonfunctional transcripts is for a Δ14-desaturase, which is used for pheromone production in ACB. Three identical desaturase transcripts also were found in female ACB pheromone glands, but, with this species, only products from the Δ14-desaturase are found in the gland. Behavioral studies with ECB males revealed that there are rare males that respond to both ECB and ACB pheromone blends, which implies that some ECB males could respond to females emitting pheromone components produced by activating the Δ14 desaturase pathway. These data provide evidence for a novel evolutionary mechanism for speciation to occur through a saltational shift in the pheromone communication system.
Materials and Methods
Insect Tissue Collection and Poly(A)+ RNA Isolation.
Female ECB moths were obtained from a laboratory culture reared on wheat germ diet (14). Female ACB moths were obtained from pupae shipped from Shanghai, China (J.-W. Du, Shanghai Institute of Entomology). Fat bodies and pheromone glands were carefully dissected from 2- to 3-day-old female moths and stored at −80°C. Poly(A)+ RNA (mRNA) was isolated and purified from fat bodies and pheromone glands by using an mRNA Isolation kit (Ambion, Austin, TX) according to the procedures recommended by the manufacturer.
cDNA Library Construction.
Poly(A)+ RNA from ECB was denatured and transcribed to single-stranded cDNA by reverse transcriptase with a SMART RACE cDNA Amplification kit (CLONTECH). Poly(A)+ RNA from ACB was dephosphorylated and decapped with a GeneRacer kit (Invitrogen). The decapped mRNA was ligated with GeneRacer RNA Oligo and reverse-transcribed with avian myeloblastosis virus (AMV) reverse transcriptase and GeneRacer Oligo(dT) Primer. Four different cDNA libraries were constructed: ECB pheromone gland cDNA library (ECB-PG-cDNA); ECB fat body cDNA library (ECB-FB-cDNA); ACB pheromone gland cDNA library (ACB-PG-cDNA); and ACB fat body cDNA library (ACB-FB-cDNA).
Cloning of Desaturase cDNA from ECB.
Two degenerate primers, PR1 and PR2 (Table 1; refs. 8–10 and 12) were used to amplify the central region of a desaturase gene from ECB-FB-cDNA library. PR1 and PR2 along with PR3 and PR4 (Table 1) were used to amplify the central regions of desaturase genes from ECB-PG-cDNA library. All of the PCR products amplified from the libraries were ligated to PCR2.1 TOPO vector (Invitrogen) for sequencing. One fragment (ECB-FB1-CR) was obtained from ECB-FB-cDNA library, and three fragments were amplified from ECB-PG-cDNA library: ECB-PG1-CR (amplified by PR1 and PR2), ECB-PG2-CR and ECB-PG3-CR (amplified by PR3 and PR4). These fragments were aligned with known central regions of other desaturase genes to evaluate the possibility of being the desaturase genes.
Table 1.
Primers used for desaturase gene characterization
| Degenerate primers |
| PR1: 5′-ATYACHGCCGGKKMYCAYMG-3′ |
| PR2: 5′-GGRAABDYGTGRTGGWAGTT-3′ |
| PR3: 5′-GGYATYACVGCHGGNGCWCA-3′ |
| PR4: 5′-TGRTARTTRTGGAABSCYTCNCC-3′ |
| PR5: 5′-GGYATYACVGCHGGNGCWCA-3′ |
| PR6: 5′-TGRTARTTRTGGAABSCYTCNCC-3′ |
| PR7: 5′-GACCAYMGNHWSCAYCA-3′ |
| PR8: 5′-GGRWAVRYRTGRTGRTARTT-3′ |
| Primers for ECB-FB1 |
| A1: 5′-GTCACGTGACCACCGCATGCACCACAAATA-3′ |
| A2: 5′-CACAAGCCCTATGACAGCAGCA-3′ |
| A3: 5′-GGGATCAGCATACAAGTCGCTCAGGTCCAG-3′ |
| A4: 5′-GCAGAGTCCTGGAATGATACAGTG-3′ |
| A5: 5′-GGGGGATCCATGGCTCCTAATATTAAGGAC-3′ |
| A6: 5′-CGGGAATTCTTACTCGTCTTTAGGATTA-3′ |
| Primers for ECB-PG1 |
| B1: 5′-GCACCACAAGTACTCGGAGACAGAC-3′ |
| B2: 5′-AACGGAACCTGGCTGGTCAAC-3′ |
| B3: 5′-CACAGATGGCATGATGAAGCAGAT-3′ |
| B4: 5′-GAGGGTCAGTATAATCCTAAGAGGCA-3′ |
| B5: 5′-ATATCTAGAATGCCGCCACAAGGTGCAGAGAGAG-3′ |
| B6: 5′-AATGAGCTCTCACTCTTCTTTTTCGGGGTT-3′ |
| Primers for ECB-PG2 |
| C1: 5′-GAAGTCATCAAACAGGGAAAAT-3′ |
| C2: 5′-TGCATCAACAGCGTCGTCCATAAGTG-3′ |
| C3: 5′-TTCGCCAAGCACAGCAAGGTTC-3′ |
| C4: 5′-TGGCCCACGAGAGCGCAGTATTTTGAA-3′ |
| C5: 5′-GGGGAGCTCAGAATGGTTCCATACGCTACCACAG-3′ |
| C6: 5′-GCCTCTAGATTATTTTAGTTTATCGGCACTG-3′ |
| Primers for ECB-PG3 |
| D1: 5′-CAACTCCGCCGCCCACAAGTG-3′ |
| D2: 5′-ATACTTCCGATCGCTGTTCTTCA-3′ |
| D3: 5′-CCTGGAACCTTAGCAACGGATTCTT-3′ |
| D4: 5′-GTACACGTAACGCAGGAAGAAACACA-3′ |
| D5: 5′-GGGGGAATTCATGGCAGACATAGACGCCACCAAC-3′ |
| D6: 5′-CGGGTCTAGATTAGGGCTTCATTCTGACTTTCA-3′ |
Based on the sequence information of ECB-FB1-CR, gene-specific primers (Table 1) were designed for RACE PCR to amplify the 3′- and 5′-ends. With ECB-FB-cDNA as template, first round 3′-RACE PCR was performed with A1 plus Universal Primer Mix (UPM), and first round 5′-RACE PCR was performed with UPM plus A3. Then, A2 plus Nest Universal Primer (NUP) were used for the second round PCR to amplify the 3′-end, and A4 plus NUP were used for the second round PCR to amplify the 5′-end. Similarly, with ECB-PG-cDNA as template, B1 and B2, C1 and C2, D1 and D2 were used for the first and second round 3′-RACE PCR. Then, B3 and B4, C3 and C4, D3 and D4 were used for the first and second round 5′-RACE PCR. All of the RACE PCR products were cloned to PCR2.1 TOPO vector for sequencing. With the sequencing result of the 5′-end, central region and 3′-end, one full-length cDNA sequence was generated from ECB-FB-cDNA library (ECB-FB1) and three full-length sequences were obtained from ECB-PG-cDNA library (ECB-PG1, ECB-PG2, and ECB-PG3).
Cloning of Desaturase cDNA from ACB.
Similarly, PR1 and PR2 were used to amplify the central region of a desaturase gene from ACB-FB-cDNA library. Three sets of degenerate primer-pairs (PR1 plus PR2, PR3 plus PR4, and PR5 plus PR6) (Table 1) were used to amplify the central regions of desaturase genes from ACB-PG-cDNA library. All of the PCR products were cloned to PCR 2.1 vectors as well for sequencing. Finally, one fragment (ACB-FB1-CR, amplified by PR1 and PR2) was obtained from ACB-FB-cDNA library and two fragments were amplified from ACB-PG-cDNA library: ACB-PG1-CR (amplified by PR3 and PR4) and ACB-PG2-CR (amplified by PR5 and PR6). These three ACB fragments were aligned with those four central regions obtained from ECB libraries, and, surprisingly, the deduced amino acid sequences were identical with each other for the following pairs: ACB-FB1-CR vs. ECB-FB1-CR, ACB-PG1-CR vs. ECB-PG1-CR, and ACB-PG2-CR vs. ECB-PG2-CR. Based on the sequence of ECB-PG3 and other known desaturase genes, another pair of degenerate primers PR7 and PR8 was designed to amplify an additional fragment from ACB-PG-cDNA library. ACB-PG3-CR was obtained, and its deduced amino acid sequence is identical to that of ECB-PG3-CR. With RACE PCR methods described above, one full-length sequence (ACB-FB1) was isolated from ACB-FB-cDNA library, and three full-length sequences were isolated from ACB-PG-cDNA library (ACB-PG1, ACB-PG2 and ACB-PG3).
Functional Assay of Δ9 Desaturases in YEpOLEX System.
YEpOLEX expression system was used for the expression of Δ9 desaturases as described (9–13). Gene-specific primers (Table 1) were designed to amplify the ORFs of ECB-FB1 (A5 and A6) and ECB-PG1 (B5 and B6). The ORFs of desaturase genes were cloned to YEpOLEX plasmid, which then were transformed to mutant ole1 cells (L8-14C strain). Because the mutant cells were devoid of the function of desaturation, any unsaturated fatty acids (UFA) detected from the cells were the products of the desaturase that was expressed by the cloned desaturase genes. Δ9-desaturase clones from ECB fat body and pheromone gland were successfully expressed in this system without any precursor or UFA added to promote yeast growth. The resulting fatty acid methyl esters were analyzed by GC/MS, and double bond positions were confirmed by MS analysis of the dimethyl disulfide (DMDS) adducts (15).
Functional Assay of Δ11 Desaturase in pYES2 System.
Two gene-specific primers, C5 and C6 (Table 1), were designed to amplify the ORF of ECB-PG2 from ECB-PG-cDNA library. The ORF of ECB-PG2 was cloned to pYES2 plasmid (Invitrogen) and transformed to INVSc1 cells for desaturase expression (12). The yeast cells transformed with pYES2-ECB-PG2-ORF were induced at 18°C for 3 days with 0.5 mM myristic acid methyl ester (MAME) added as precursor. The induced cells were collected and extracted with methanol/chloroform for 1 h. The solvent was decanted from the debris and evaporated under nitrogen. The oily residue was extracted twice with 0.5 ml 10% boron trichloride/methanol, and the combined extracts were heated at 100°C for 30 min. The resulting fatty acid methyl esters were extracted with 1 ml hexane, and the solution was concentrated under nitrogen for analyses by GC/MS. The double bond positions in the products were confirmed by MS analysis of the DMDS adducts.
Functional Assay of Δ14 Desaturase in a Baculovirus/Insect Cell Expression System.
Similar to those described above, two gene-specific primers, D5 and D6 (Table 1), were designed to amplify the ORF of ECB-PG3 from ECB-PG-cDNA library. The ORF of ECB-PG3 was cloned to a pFastBac1 vector (Invitrogen), which was then transposed to the baculovirus genome (bacmid DNA) by using a baculovirus expression system as previously described (9). The resulting fatty acid methyl esters were analyzed by GC/MS, and double bond positions were confirmed by MS analysis of the DMDS adducts.
Male Behavioral Responses to ECB and ACB Pheromone Blends.
ECB males were obtained from a laboratory culture of the univoltine-Z race, which uses a 97:3 mix of Z/E11-14:OAcs for its pheromone blend. Pupae were sexed, and males were separated from females and segregated daily by age. The chemicals were obtained from the Pherobank (www.pherobank.nl), and mixtures were prepared in hexane and applied to 5 × 9-mm rubber stoppers (Thomas) at a dose of 30 μg/septum. Individual males were tested in the sustained flight tunnel (16) as previously described for the ECB (17). During each test period, each male was tested to the following blends, in the order indicated: (i) the ACB blend (2:1 Z12/E12-14:OAcs); (ii) a 97:3 blend of Δ12-14:OAcs; (iii) a 2:1 blend of Δ11-14:OAcs; and (iv) the ECB blend (97:3 Z11/E11-14:OAcs).
Comparative DNA and Protein Sequence Analysis.
The phylogenetic relationships among the ECB and ACB desaturase genes were analyzed by comparing mRNA and genomic DNA sequences from representative species of Lepidoptera (ECB and ACB; corn earworm, Helicoverpa zea; cabbage looper, Trichoplusia ni; redbanded leafroller, Argyrotaenia velutinana; light brown apple moth, Epiphyas postvittana; greenheaded leafroller, Planotortrix octo; and silkworm, Bombyx morii), Diptera (fruit flies, Drosophila melanogaster and D. simulans), and Orthoptera (cricket, Acheta domesticus). Sequences were labeled by using the first letter of the genus and first two letters of the species name followed by the gene name and accession number. The accession numbers for the D. melanogaster sequences are not given because these sequences were extracted from the complete genome sequence of this species, which is accessible through the ENTREZ sequence retrieval system at www.ncbi.nlm.nih.gov/Entrez/. We used the program clustal-x (18) to align the deduced desaturase amino acid sequences, which were used for the alignment of nucleotide sequences. Both final alignments were checked for errors by visual inspection. The nucleotide alignment consisted of a set of sequences with 1,434 nucleotide sites (478 aa sites), excluding the start and stop codons. The numbers of nonsynonymous substitutions per nonsynonymous site (dN) were estimated by using the modified Nei-Gojobori method (19) with Jukes and Cantor (20) distances.
Because the divergence time involving many species comparisons were very old (several hundred million years or older), the deduced amino acid sequences were used to reconstruct phylogenetic trees because they are less prone to the confounding effects of multiple substitutions that can occur in nucleotide sequences over long periods of evolutionary time. The trees were reconstructed by using the neighbor-joining (NJ; ref. 21) method with Jones et al. (JTT; ref. 22) distances. JTT distances were estimated in mega2 by computing gamma distances with a shape parameter of 2.4 (23). Phylogenetic trees were also reconstructed with the maximum likelihood (ML) method by using the Müller and Vingron (MV; ref. 24) substitution model with gamma distributed rates (shape parameter = 1.08 estimated by using the ML method) across eight rate categories. For both NJ and ML analyses, we rooted the resultant trees by using the tick (Amblyomma americanum; accession number U03281), because it is the taxon that is most closely related to the insect species analyzed in this study. The reliabilities of internal branches were assessed by using 1,500 bootstrap replicates for the NJ analysis and 10,000 quartet-puzzling steps for the ML analysis; sites with gaps were ignored in both types of analyses. NJ searches were conducted by using the computer program mega2 (25), and ML analyses were conducted by using the computer program tree-puzzle 5.0 (26).
Results
Isolation and Sequencing of cDNAs from ECB and ACB.
A full-length sequence, ECB-FB1, was isolated from ECB-FB-cDNA library. This cDNA spans 1,516 nt and contains an ORF encoding a 355-aa protein, which has high identity to other Z9-desaturases (65% to Trichoplusia ni-Z9 and 87% to Helicoverpa zea-Z9). Three full-length cDNAs (ECB-PG1, ECB-PG2, and ECB-PG3) were obtained from the ECB-PG cDNA library. ECB-PG1 spans 1748 nt with an ORF encoding 351 aa. This deduced aa has high identity to ECB-FB1 (63%) and other insect Z9-desaturases (73% to T. ni-Z9 and 64% to H. zea-Z9). ECB-PG2 spans 1404 nt with an ORF encoding a 329-aa protein with a high identity to other insect Δ11-desaturases (53% to T. ni-Z11 and 61% to H. zea-Z11). ECB-PG3 spans 2116 nt with an ORF encoding a 367-aa protein with low identity to ECB-PG2 (36%) and other Δ11 insect desaturases (34% to T. ni-Z11 and 37% to H. zea-Z11).
Similarly, one full-length cDNA (ACB-FB1) was isolated from ACB-FB-cDNA library, and three full-length cDNAs (ACB-PG1, ACB-PG2, and ACB-PG3) were isolated from ACB-PG-cDNA library. When comparing the deduced amino acid sequences of ACB and ECB clones, it was surprising to find that the ACB clones are identical to the corresponding ECB clones, except for one substitution of an H for a P in amino acid no. 9 in ACB-PG3 (Ofu-Z/E14, Fig. 2).
Figure 2.
Comparison of deduced amino acid sequences of Onu-Z916, Onu-Z918, Onu-Z/E11, Onu-Z/E14, and Ofu-Z/E14 to the Z11-desaturase from the cabbage looper moth (T. ni-Z11) and the redbanded leafroller moth (RBLR-Z/E11). The consensus residues are shaded with solid gray. Ofu-Z/E14 has an AA different from Onu-Z/E14 at amino acid 9 denoted with an asterisk.
Functional Assay of Z9-Desaturases.
The mutant ole1 yeast cells were used to assay the Z9-desaturases isolated from ECB and ACB fat bodies and pheromone glands. The fat-body consensus clones (ECB-FB1 and ACB-FB1) were found to produce a mixture of Z9-16:Acid and Z9-18:Acid (2.2/1), and the desaturases were named Onu916 and Ofu916, respectively. The pheromone-gland Z9-desaturase consensus clones (ECB-PG1 and ACB-PG1) were found to produce a mixture of Z9-16:Acid and Z9-18:Acid (1:1.5), and the desaturases were named Onu918 and Ofu918, respectively.
Functional Assay of Δ11-Desaturases.
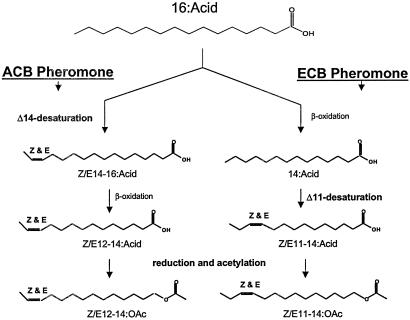
The pYES2 expression system was used to assay the consensus clone ECB-PG2. The data show that the expressed desaturase produces mainly Z11-16:Acid, along with a mixture of Z11 and E11-14:Acids (5:4 ratio) (Fig. 3), and were named Onu-Z/E11. The identical desaturase from ACB-PG2 was named Ofu-Z/E11.
Figure 3.
Z/Ell desaturase expression in pYES2 system-GC/MS analyses of methyl esters (DMDS adducts) with the ion 245 m/z displayed. (A) Control INVSc1 cells with 0.5 mM myristic acid methyl ester (MAME) added. Z11-16 and Z11-18:Acids produced by chain elongation of Z9-14 and Z9-16:Acids. (B) INVSc1 cells transformed with pYES2-ECBG-PG2-ORF complemented with 0.5 mM MAME. New products are Z11- and E11-14:Acid along with additional Z11-16:Acid (amount above dotted line).
Functional Assay of Δ14-Desatursaes.
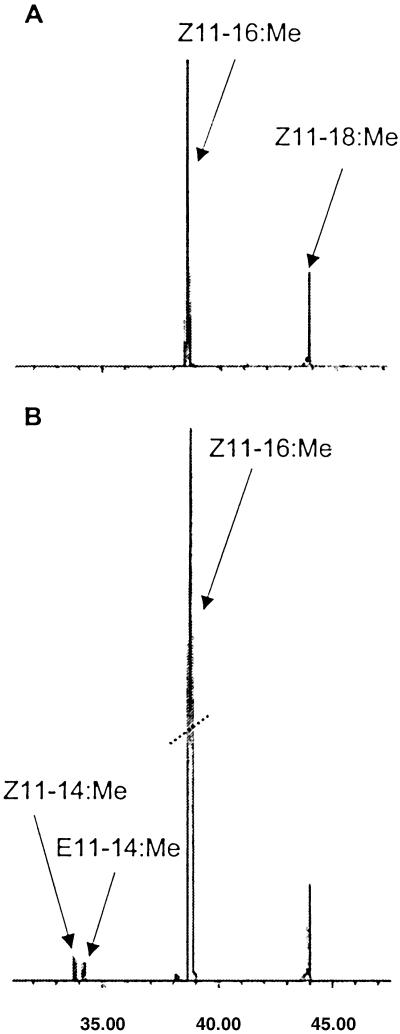
A baculovirus expression system was used to assay the consensus clones ECB-PG3 and ACB-PG3. The results show that the expressed desaturases produce a mixture of Z/E14-16:Acids (10:1 ratio; Fig. 4) and were named Onu-Z/E14 and Ofu-Z/E14. Mass spectral analysis of the DMDS adducts proved the Δ14-unsaturation with major ions at 87 and 255 m/z.
Figure 4.
Z/E14-desaturase expression in a Baculovirus System. GC/MS analyses of methyl esters (DMDS adducts) with ion 217 and 287 displayed. (A) Control Sf21 cells that produce Z9-16, Z9-17, and Z9-18:Acids. (B) Sf21 insect cells infected with Bacmid-pFastBac1-ECBG-PG3-ORF and shown to produce additional Z14-16 and E14-16:Acids.
Male ECB Behavioral Responses to ECB and ACB Pheromone Blends.
Of 136 ECB males tested in the flight tunnel, 119 flew upwind to the normal 97:3 blend of Z/E11-14:OAcs. Of the 119 responders, 5 also flew upwind to the ACB pheromone (2:1 mix of Z/E12-14:OAcs), and 2 of these moths also flew upwind to a 97:3 mix of these Δ12 acetates. None of the 136 ECB moths tested responded to a 2:1 mix of the Δ11 acetates.
Phylogenetic Analysis of Desaturase Genes.
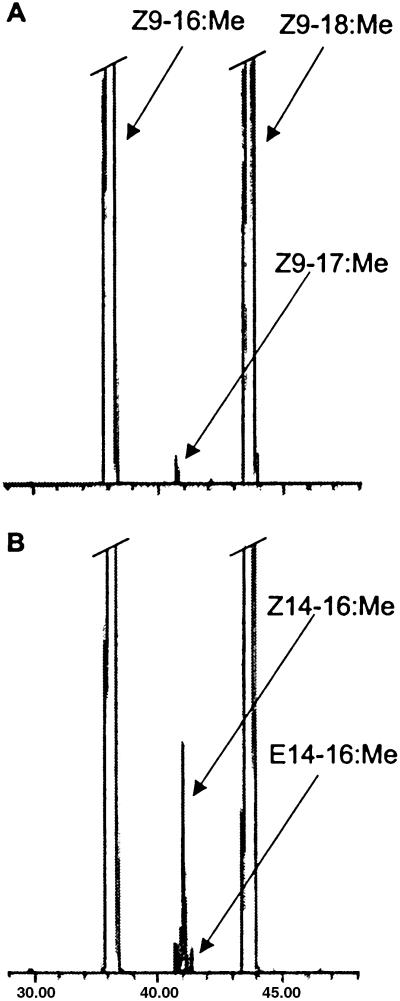
The phylogeny of insect desaturases (Fig. 5) obtained from our analysis of selected amino acid sequences shows that there are four main clusters, which we labeled as Z9(16>18), Z9(18>16), Δ14, and Δ10,11. The deep internal branches in the phylogeny, which represent the ancestors of gene cluster duplicates, are very short and suggest that the duplication events giving rise to the insect desaturase multigene families occurred within a narrow time frame. Consistent with a phylogenetic analysis of desaturases by Knipple et al. (27), the phylogeny clearly shows that the insect desaturase multigene families originated before the split among Lepidoptera, Diptera, and Orthoptera, which is estimated to have occurred ≈320 million yr ago (28). By using the standard distance-regression method (23), our attempts to calculate an emergence date on the basis of either Kimura nucleotide (29) or Poisson amino acid (30) distances resulted in an estimate of more than 700 million yr ago, which is substantially older than the date placed on the emergence of insects (450 million yr ago; ref. 31). However, the existence of substantial rate heterogeneity displayed between clusters confounds any attempt to reliably date the origin of the insect desaturase multigene family, even if only the most conserved regions of the protein sequence are used.
Figure 5.
Phylogeny of insect desaturases. Numbers along branches represent bootstrap (in front of slash) and quartet-puzzling (behind slash) values greater than 50%. Because the ML and NJ trees were highly similar, only the NJ tree is shown here.
An examination of the phylogeny in Fig. 5 also shows that, whereas the internal branches within the different clusters are very short, the terminal branches are much longer, suggesting that the changes associated with speciation occurred very quickly. It also is evident that desaturases in the top three clusters are evolving more rapidly than the more constrained Z9(16>18) cluster at the bottom of the tree. An estimation of the rate of nonsynonymous substitution in each ECB/ACB desaturase gene was obtained by comparing the sequence from those species to the sequence from the corn earworm moth, H. zea, desaturase gene under the assumption that the latter diverged from the former approximately 75 million yr ago (31). The results show that the Z916 gene evolves the slowest (0.667 ± 0.087 × substitutions per site per 109 years), followed by the Z918 gene (1.313 ± 0.014 × substitutions per site per 109 years) and then the Z/E11 gene (2.253 ± 0.213 × substitutions per site per 109 years). The rate for the Z/E14 gene could not be calculated, but, based on the branch lengths and the results of the likelihood ratio test of the molecular clock, it must be substantially higher than what is seen for the Z/E11 gene.
The phylogenetic tree (Fig. 5) shows that there are Drosophila desaturase genes that evolve very fast, similar to the Δ14 genes, but it is not known whether they are functional. It is possible that this cluster is comprised of very fast-evolving pseudogenes, and that the Ostrinia Δ14 desaturase represents a resurrected pseudogene with a rare functional occurrence in the Lepidoptera. Although we can only currently speculate as to how long the Δ14 desaturase gene has been a pseudogene in Ostrinia, it is clear that the gene itself arose from a duplication event that took place before the Lepidoptera diverged from the Diptera (Fig. 5). An examination of Δ14 orthologs in other lepidopteran lineages may shed light on determining exactly how long this gene has been nonfunctional.
Discussion
Discovery of the Δ11 and Δ14 desaturase-gene transcripts in both ECB and ACB pheromone glands provides evidence for an evolutionary mechanism involving a sudden shift in the pheromone systems, based on a model (27) for subfunctionalization of the diverse sex-pheromone desaturases after a gene duplication event early in the evolution of insects. Although both desaturase transcripts were isolated from both species, UFA products from only the Δ11-desaturase are found in ECB pheromone-gland analyses (32), and UFA products from only the Δ14-desaturase are found in ACB pheromone-gland analyses (33). It is not known at this time whether both transcripts are translated in each species, but our results suggest that a switch in pheromone blends was accomplished when a mutational event activated the production of UFAs from the Δ14-desaturase transcript and eliminated the presence or function of the Δ11-desaturase. Rare males possessing the ability to respond to this new blend would spread in the population, and selection could eventually lead to signal coordination (3, 4). This scenario fits well with simulation models (34) showing that sudden major switches in pheromone blend and male response appear more likely than accumulation of small changes.
What evidence is there that the above scenario occurred and why did the new population not use Δ14-16:OAcs instead of Δ12-14:OAcs for its new pheromone blend? Substrate specificity studies (35) with ACB showed that enzymes in the pheromone gland cannot convert 16-carbon acids to 16-carbon acetates, indicating that the mutated females (ACB) with a functional Δ14-desaturase would not be able to reduce the Δ14-16:Acid intermediates to alcohols with the existing reductases. However, the ACB pheromone-gland enzymes can reduce and acetylate Δ12-14:Acids, which are the chain-shortened products of Δ14-16:Acids (Fig. 1). The ACB reductases have a slight preference of E12-14:Acid over Z12-14:Acid but, surprisingly, have a much higher preference for Z11-14:Acid over all other substrates. This preference profile is what would be expected for a population that uses Z11-14:OAc as its main pheromone component, and provides strong support for the previous statement that ACB is derived from an ancestral Ostrinia population that used Z11-14:OAc as it major pheromone component.
Phylogenetic relationships of Ostrinia species, including ECB and ACB, based on morphological data (36) and mtDNA analyses (6) suggest that ACB was derived from a population of Ostrinia that used the common Z/E11-14:OAc pheromone components found with all Ostrinia species (6). The divergence time between the two Ostrinia species is very recent [≈1 million yr ago by using the molecular clock rate of 2.3%/million yr for mtDNA sequences (37) and an estimated 2.3% sequence divergence for mtDNA sequences of ECB and ACB (R. Harrison, personal communication)] and the associated protein sequences are identical, so the isolating mechanism leading to the formation of these species can be inferred to have evolved very quickly. The difficulty has been to determine how one species in a complex of closely related Ostrinia species could evolve the use of pheromone components that are produced by a different desaturase and a different biosynthetic route (Fig. 1). An examination of the desaturase multigene family phylogeny (Fig. 5) suggests that a gene derived from a duplication event that occurred before the split among Diptera, Orthoptera, and Lepidoptera became a pseudogene at some point during the evolution of the Ostrinia. Later, this gene was resurrected in certain populations and became functional as a Δ14 desaturase gene in the stochastic event that generated the new pheromone system and, thus, the ACB species.
Asymmetric tracking of male response is also an important component in the proposed evolution of ACB. In this regard, a male response shift from Z11-14:OAc to Δ12-14:OAcs is more likely than a change from Z11-14:OAc to Δ14-16:OAc relative to possible interaction with the antennal pheromone binding proteins (PBPs) and olfactory receptors. When pheromone molecules contact a male's antenna, they interact with PBPs, which are involved in transporting the hydrophobic pheromone molecules in the aqueous environment of the antennal sensillar lymph to their specific dendritic receptor. Because PBPs have been shown to possess some specificity for different structural classes (38), the longer Δ14-16:OAcs would be less likely to bind than the Δ12-14:OAcs, which are the same length as the Δ11-14:OAcs of the ancestral Ostrinia species. It is interesting that the antennae of ECB and ACB contain a single PBP for the Z and E pheromone components, and its structure is identical in both species (39).
Even with the plausible argument for PBP and receptor affinities between ECB and ACB components, we were surprised to find rare ECB males that responded to the Z/E12-14:OAc blend. Preliminary flight-tunnel assays with ECB males (Z race that respond specifically to a mix of 97:3 of Z/E11-14:OAcs) showed that most males remained quiescent in the presence of an ACB blend (3:2 Z/E12-14:OAc), but 3 of 235 males activated and flew upwind to the odor source (1.5 m distance). A second behavioral study showed that rare ECB males responding to the 2:1 mixture of Z/E12-14:OAcs (ACB blend) and to the 97:3 mixture of Z/E11-14:OAcs (ECB blend) do not respond to a corresponding 2:1 mixture of Δ11-14:OAcs. The existence of these rare males supports the possibility for a major shift in pheromone blend being tracked by male response.
Finally, the present findings could have significance in agriculture relative to the question as to whether the continuous application of pheromone over large areas for insect control can result in the evolution of resistance to this tactic. Traditional thinking is that the potential for resistance to pheromones, particularly where long-term use of pheromone effectively controls pest populations, would depend on genetically based variation in production of and response to pheromones (40). It was not anticipated that a species can harbor a nonfunctional gene that could produce a major shift in pheromone blend if activated and generate a population of individuals that would be unaffected by the pheromone applications.
Acknowledgments
We thank J.-W. Du for supplying ACB pupae, R. Harrison for discussions on this project, K. Cotropia for conducting flight-tunnel bioassays, and D. Knipple for providing a preprint of their publication on the evolution of desaturases in moths and flies. This work was supported by National Science Foundation Grant IBN-9870669.
Abbreviations
- ACB
Asian corn borer
- Z/E12-14:OAc
(Z)- and (E)-12-tetradecenyl acetate
- ECB
European corn borer
- UFA
unsaturated fatty acid
- DMDS
dimethyl disulfide
- NJ
neighbor-joining
- ML
maximum likelihood
- PBP
pheromone binding protein
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF441220 (for Onu-Z9/E14), AF441221 (for Onu-Z/Ell), AF243047 (for Onu-Z916), AF430246 (for Onu-Z918), AY062023 (for Ofu-Z/E14), AF441861 (for Ofu-Z/E11), AY057862 (for Ofu-Z916), and AY057863 (for Ofu-Z918)].
See commentary on page 13368.
References
- 1.Paterson H E H. In: Species and Speciation. Vrba E S, editor. Pretoria, South Africa: Transvaal Museum; 1985. pp. 21–29. [Google Scholar]
- 2.Phelan P L. In: Insect Pheromone Research. Cardé R T, Minks A K, editors. New York: Chapman & Hall; 1997. pp. 563–579. [Google Scholar]
- 3.Phelan P L. In: Insect Chemical Ecology. Roitberg B D, Isman M B, editors. New York: Chapman & Hall; 1992. pp. 265–314. [Google Scholar]
- 4.Löfstedt C. Philos Trans R Soc London B. 1993;240:167–177. [Google Scholar]
- 5.Klun J A, Bierl-Leonhardt B A, Schwarz M, Litsinger L A, Barrion A T, Chiang H C, Zhungxie J. Life Sci. 1980;27:1603–1606. doi: 10.1016/0024-3205(80)90570-6. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa Y, Takanashi T, Kim C, Hoshizaki S, Tatsuki S, Huang Y. Entomol Exp Appl. 1999;91:237–244. [Google Scholar]
- 7.Roelofs W L, Bjostad L B. Bioorg Chem. 1984;12:279–298. [Google Scholar]
- 8.Liu W, Ma P W K, Marsella-Herrick P, Rosenfield C-L, Knipple D C, Roelofs W L. Insect Biochem Mol Biol. 1999;29:435–443. doi: 10.1016/s0965-1748(99)00020-x. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, Jiao H, Murray N, O'Connor M, Roelofs W L. Proc Natl Acad Sci USA. 2002;99:620–624. doi: 10.1073/pnas.221601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao G, Liu W, O'Connor M, Roelofs W L. Insect Biochem Mol Biol. 2002;32:961–966. doi: 10.1016/s0965-1748(01)00176-x. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfield C-L, You K-M, Marsella-Herrick P, Roelofs W L, Knipple D C. Insect Biochem Mol Biol. 2001;31:949–964. doi: 10.1016/s0965-1748(01)00043-1. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Jiao H, O'Connor M, Roelofs W L. Insect Biochem Mol Biol. 2002;32:1489–1495. doi: 10.1016/s0965-1748(02)00069-3. [DOI] [PubMed] [Google Scholar]
- 13.Knipple D C, Miller S J, Rosenfield C-L, Liu W, Tang J, Ma P W K, Roelofs W L. Proc Natl Acad Sci USA. 1998;95:15287–15292. doi: 10.1073/pnas.95.26.15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roelofs W L, Glover T J, Tang X-H, Robbins P S, Löfstedt C, Hansson B, Bengtsson B O, Sreng I, Eckenrode C J. Proc Natl Acad Sci USA. 1987;84:7585–7589. doi: 10.1073/pnas.84.21.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buser H R, Arn H, Guerin P, Rauscher S. Anal Chem. 1983;55:818–822. [Google Scholar]
- 16.Miller J R, Roelofs W L. J Chem Ecol. 1978;4:142–149. [Google Scholar]
- 17.Linn C E, Jr, Young M S, Gendle M, Glover T J, Roelofs W L. Physiol Entomol. 1997;22:212–223. [Google Scholar]
- 18.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Rosenberg H F, Nei M. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jukes T H, Cantor C R. In: Mammalian Protein Metabolism. Munro H N, editor. New York: Academic; 1969. pp. 21–123. [Google Scholar]
- 21.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 23.Gu X, Zhang J. Mol Biol Evol. 1997;14:1106–1113. doi: 10.1093/oxfordjournals.molbev.a025720. [DOI] [PubMed] [Google Scholar]
- 24.Müller T, Vingron M. J Comput Biol. 2000;7:761–776. doi: 10.1089/10665270050514918. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Tamura K, Jakobsen I B, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 26.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 27. Knipple, D. C., Rosenfield, C.-L., You, K.-M. & Jeong, S. E. (2002) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 28.Burmester T. Mol Biol Evol. 2001;18:184–195. doi: 10.1093/oxfordjournals.molbev.a003792. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 30.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford: Oxford Univ. Press; 2000. [Google Scholar]
- 31.Kristensen N P, Skalski A W. In: Lepidoptera: Moths and Butterflies, Handbuch der Zoologie/Handbook of Zoology IV/35. Kristensen N P, editor. Vol. 1. Berlin: Walter De Guyter; 1999. pp. 7–25. [Google Scholar]
- 32.Ma P W K, Roelofs W L. Zool Sci. 2002;19:501–511. doi: 10.2108/zsj.19.501. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C-H, Löfstedt C, Wang X. Arch Insect Biochem Physiol. 1990;15:57–65. [Google Scholar]
- 34.Butlin R K, Trickett A J. In: Insect Pheromone Research. Cardé R T, Minks A K, editors. New York: Chapman & Hall; 1997. pp. 548–562. [Google Scholar]
- 35.Zhao C-H, Fang L, Bengtsson M, Löfstedt C. J Chem Ecol. 1995;21:1795–1810. doi: 10.1007/BF02035148. [DOI] [PubMed] [Google Scholar]
- 36.Mutuura A, Munroe E. Mem Entomol Soc Can. 1970;71:1–112. [Google Scholar]
- 37.Brower A V Z. Proc Natl Acad Sci USA. 1994;95:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maida R, Krieger J, Gebauer T, Lange U, Ziegelberger G. Eur J Biochem. 2000;267:2899–2908. doi: 10.1046/j.1432-1327.2000.01303.x. [DOI] [PubMed] [Google Scholar]
- 39.Willett C S, Harrison R G. Insect Biochem Mol Biol. 1999;29:277–284. doi: 10.1016/s0965-1748(99)00003-x. [DOI] [PubMed] [Google Scholar]
- 40.Evenden M L, Haynes K F. Entomol Exp Appl. 2001;100:131–134. [Google Scholar]