Abstract
The integration of data from multiple global assays is essential to understanding dynamic spatiotemporal interactions within cells. In a companion paper, we reported a data integration methodology, designated Pointillist, that can handle multiple data types from technologies with different noise characteristics. Here we demonstrate its application to the integration of 18 data sets relating to galactose utilization in yeast. These data include global changes in mRNA and protein abundance, genome-wide protein–DNA interaction data, database information, and computational predictions of protein–DNA and protein–protein interactions. We divided the integration task to determine three network components: key system elements (genes and proteins), protein–protein interactions, and protein–DNA interactions. Results indicate that the reconstructed network efficiently focuses on and recapitulates the known biology of galactose utilization. It also provided new insights, some of which were verified experimentally. The methodology described here, addresses a critical need across all domains of molecular and cell biology, to effectively integrate large and disparate data sets.
Keywords: metabolism, yeast, molecular network model, galactose
Systems biology aims to understand the dynamic behavior of molecular networks in the context of the global cell, organ and organism state by exploiting (i) high-throughput interrogation technologies; (ii) increasingly comprehensive databases of biomolecules and their interactions; and (iii) computational predictions of molecular function and interaction (Fig. 1). Use of each of these sources of information has its own drawbacks (1). For example, many current global assays of mRNA and protein abundance/state are systematically biased toward more abundant species and measure only the average content of many thousands of cells. Global assays are also inherently noisy and include significant numbers of false positives and false negatives. Databases tend to combine data from different cell types, different strains of an organism, and different experimental conditions. Moreover, well studied molecules and pathways are systematically overrepresented in databases. As a result, integration of database information with a particular set of experimental data can introduce systematic biases into the model-building process. Likewise, computer predictions tend to be more accurate for members of well characterized molecular families. Therefore, there is a pressing need for data integration methodologies that effectively address both random noise and systematic bias in data.
Fig. 1.
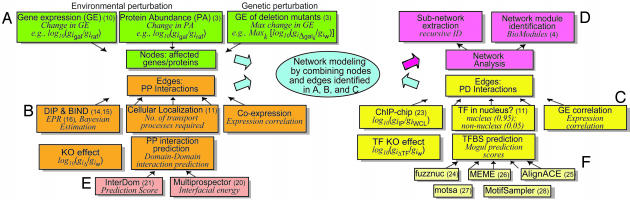
Data integration framework for network modeling. Five data integration problems are shown: Identification of genes affected by environmental and genetic perturbations (A); PP interactions (B); PD interactions (C); domain-domain interactions (E); and TFBS predictions (F). The data sets used for network modeling to study galactose utilization in yeast are presented for illustration purposes. The label for each box indicates the type of data used. The italic note below the label indicates the statistical measure we used to calculate empirical P values (or significances) for each data set. GE, gene expression; PA, protein abundance; KO, (gene) knockout; PP, protein–protein (interaction); PD, protein–DNA (interaction); TF, transcription factor; TFBS, transcription factor binding site; w, wild type; gal, galactose; raf, raffinose; IP, immunopurification enriched; WCL, whole cell lysate enriched. (D) A set of network analysis tools that helps us explore a complex network systematically. These tools allow us to build a subnetwork that includes PP and PD interactions (edges) pertinent to affected genes/proteins (nodes) and given perturbations, and to identify clusters of proteins in the network (see text).
In a companion paper (2), we present a data integration methodology and its software implementation to address these challenges. To present the methodology clearly, we used only simulated data in that paper. Here, we present the application of our methodology (named Pointillist) to 18 types of biological data, which we integrate to arrive at a detailed and comprehensive picture of galactose utilization in yeast. The data we integrate include information from several high-throughput assays, public databases, and computational predictions. They pertain to gene expression, protein abundance, protein–protein (PP) interactions, and protein–DNA (PD) interactions. As such, our data sources provide a comprehensive test of the efficacy of Pointillist and our conclusions may be applicable to studies of other molecular biological systems.
The yeast galactose utilization pathway has been studied extensively and intensively for >40 years, and is one of the best understood eukaryotic molecular systems (3). As such, it provides us with an ideal opportunity to evaluate the extent to which the network model we arrive at captures all aspects of the system of interest. We show that Pointillist not only captures many known features of the system, but it also provides additional insights, some of which we confirmed experimentally.
Materials and Methods
Supporting Information. For further details, see Supporting Text, Figs. 5–11, and Tables 1–6, which are published as supporting information on the PNAS web site.
Network Analysis. Using a subnetwork extraction algorithm (see Supporting Text for details), we constructed a parsimonious subnetwork, which includes only those PP and PD interactions relevant to the affected genes, rather than the whole PP and PD interactome. This algorithm selects only the nodes/edges that provide closed connection paths to the nodes for the affected genes (Fig. 9 A and B). The number of intermediate nodes is selected with an iterative algorithm that searches for the least number of intermediate nodes that result in a maximally connected network. However, the structure of this subnetwork is still too complicated to allow visual exploration (Fig. 10A). To facilitate exploration, we then clustered the nodes in the network into a set of functional modules. To incorporate known functional information into the network, we first inserted an additional edge between any two proteins sharing the same GO Biological Process category (Fig. 10B). We then used the Cytoscape BioModules tool (4) to identify modules in the network (Fig. 11). In this study, BioModules generated 20 functional modules (Table 4). To focus on galactose-mediated interactions between metabolic pathways, our network kept only nine modules (Table 5) that have metabolism-related functions (see Fig. 3 for the resulting network).
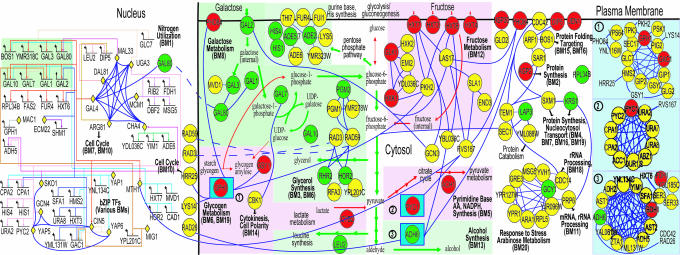
Fig. 3.
The final network resulting from applications of our integration method to the 18 types of evidence for yeast galactose utilization and a set of network analysis tools. This network model recapitulates many known features of galactose metabolism (e.g., GAL regulon induction; see text). Also, it provides a number of insights into regulatory interactions between different metabolic modules. For example, we note a possible mechanistic explanation of how fructose metabolism (BM12) is down-regulated in galactose: Gal4p contributes to a decrease in fructose uptake by repressing the fructose transporter Hxt7p via Mth1p (see text). The legends for nodes and edges in the network are as follows: (i) TFs in nucleus are represented by yellow diamonds; (ii) all other proteins (circles) are located according to their subcellular localizations (plasma membrane, cytosol, and nucleus); (iii) circle colors represent increase (green), decrease (red), and no significant change (yellow) in expression when the carbon source is changed from raffinose to galactose; (iv) the three numbered squares shown at the right represent complexes; and (v) blue edges represent PP interactions and multicolored edges in the nucleus represent PD interactions. Short colored arrows in the cytosol represent the increase (green) and decrease (red) in pathway fluxes. Biomodules are labeled with BM, and the short black arrows represent communication between the 9 modules selected for this subnetwork and the remaining 11 modules (Fig. 11).
Yeast Strains. Yeast strains were derived from BY4741 (MATA his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) or BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). Genes were genomically tagged with the sequence encoding the IgG binding domains of Staphylococcus aureus protein A, or 13 copies of the c-myc epitope (from pFA6–13MYC (5) by homologous recombination using a previously described PCR-based integrative transformation procedure into BY4741 or BY4742 (6). Strains with no apparent growth defects and containing appropriately sized fusion proteins were used. HXT7-TAP and MTH1-TAP strains were obtained from the yeast TAP-fusion library (Open Biosystems, Huntsville, AL). GAL4-MYC has been described (7).
Microarray Experiments. For the time course study of galactose induction, wild-type cells (see Yeast Strains) were grown in YEPR (1% yeast extract/2% peptone/2% raffinose) to an OD600 of 0.6. Cells were collected for “time 0” immediately before the addition of prewarmed YEP containing sufficient galactose for a final concentration of 2%. Cells were then collected at 5, 10, and 30 min and 1, 2, 4, 6, and 9 h. For all microarray expression studies, total RNA was first isolated by using the hot acid phenol method (8), followed by the extraction of poly(A)+ RNA using the Poly(A) Pure kits (Ambion). Cy3- and Cy5-labeled cDNAs were generated by a reverse transcription reaction as described (3). Microarray hybridization was performed exactly as described (9) with only one modification: the images were processed by using the microarray spotfinding and quantitation software analyzer dg (MolecularWare, Cambridge, MA).
Chromatin Immunoprecipitation (IP). Conventional chromatin IP experiments were performed as described (7) except that PCRs were of 15 μl and contained 0.2 μl of either IP-enriched DNA or unenriched DNA; 5 pmol of each primer; 0.1 mM each of dATP, dGTP, dCTP, and dTTP; and 0.7 unit of TaqDNA polymerase (Fermentas, Hanover, MD). Reactions were 26 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s. PCR products were separated on 7% polyacrylamide gels and visualized with ethidium bromide.
Hxt7p-TAP and Gal2p-pA Abundance Assay. Tagged strains were grown to an OD600 of ≈0.6 in YEPR. The cells were harvested and transferred to YEP containing 0.5% glucose, 5% glucose, 3% glycerol, 3% ethanol, or 2% galactose and grown for the indicated times. Cells were harvested, and whole cell protein lysates were prepared. Equal amounts of protein from each of the resulting lysates were separated by SDS/PAGE, transferred to nitrocellulose membranes, and blocked with TBS containing 5% dried skim milk and 0.1% Tween 20. The protein A and TAP moieties or Gsp1p were detected with affinity-purified rabbit IgG (Cappel, Irvine, CA) or anti-Gsp1p, respectively, and visualized with horseradish peroxidase-conjugated secondary antibodies (anti-rabbit-HRP) and enhanced chemiluminescence (ECL).
Results and Discussion
Application to the Galactose Utilization System in Yeast. The goal of Pointillist is to integrate a variety of data sets, such as those illustrated in Fig. 1, into a unified biochemical network. A network is represented as a graph whose nodes are biomolecular species (e.g., genes, mRNA, proteins, lipids, and metabolites), and the edges connecting these nodes are the interactions among the biomolecules. These edges can be directed (e.g., PD interactions) or undirected (e.g., protein complex formation). A cellular process such as cell fate specification or the cell cycle can then be modeled and visualized as a series of interactions that change the graph topology over time and include positive and negative feedbacks. To demonstrate the utility and efficacy of our methodology (Pointillist) (2), we integrated 18 types of evidence, derived from 15 data sources, to reconstruct the well studied galactose utilization network in Saccharomyces cerevisiae (3). The data sources included four sets of galactose-specific high-throughput experiments, seven computational interaction prediction tools, and information culled from four curated general-purpose databases, as described below. We grouped these data into five distinct classes (see Fig. 1 for an overview) and integrated each class of data separately using Pointillist.
Identification of Affected Genes by System Perturbations. We first identified genes and proteins affected by two types of perturbations, one environmental and one genetic: (i) carbon source change from raffinose to galactose and (ii) deletion of known galactose pathway genes. The available data included time-course gene expression profiles from 5 min to 9 h after induction (10), relative protein abundances 9 h after galactose addition (3), and steady-state gene expression change in galactose after the deletion of eight known galactose metabolism related genes [the three regulatory genes GAL3, 4, and 80, and the structural genes GAL1, 2, 6 (Lap3), 10, and 7].
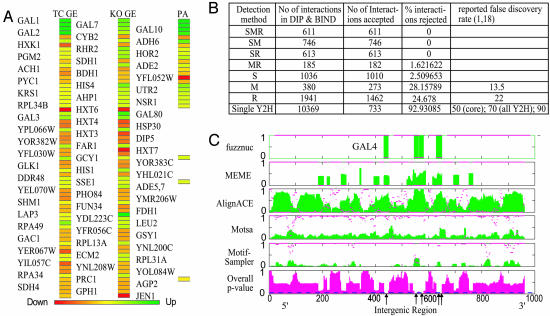
Pointillist identified 69 affected genes from the integration of these three types of evidence. Fig. 2A shows expression changes of these 69 genes in response to galactose exposure and in response to deletions of galactose pathway genes above (red, decrease; green, increase; yellow, no change). Note that the selected set is not limited to the simple case where all three sources of data support each other (e.g., GAL1, 2, 7, and 10), but also includes cases where the data are incomplete (e.g., GAL3 and GAL80, for which no protein data were available in our data set) and contradictory (e.g., LAP3, for which the knock out data suggest a strong role not supported by the time-course data, and for which no protein abundance data are available).
Fig. 2.
Integration results. (A) The final set of selected genes. The colors represent the increase (green), decrease (red), and no change (yellow) after two perturbations (TC stands for time-course and see Fig. 1 legend for the other abbreviations). These selected genes show how metabolic fluxes are redistributed to optimally use galactose from Leloir pathway mainly to alcohol synthesis via glycolysis (Figs. 3 and 6B). (B) Integration results for determination of PP interactions. The detection methods used to identify PP interactions in DIP and the corresponding selection and removal rates by our integration method are shown. Detection method abbreviations are S, small-scale experiments; M, multiple yeast two hybrid assays; R, paralog analysis. Our method selected 99.1% (2,980) of 3,006 interactions detected by small-scale experiments (S), which are generally considered as true PP interactions in DIP (see text). (C) TFBS prediction results for the upstream (putative regulatory) sequence of GAL2, which is reported to have five binding sites for Gal4p. The results for each search algorithm are shown in a separate row. Green bars indicate the averaged score at each position, whereas magenta dots indicate P values from one-tailed t test. The integrated overall P values are shown in the bottom row (magenta bars). The integration method predicted all reported Gal4p binding sites correctly (arrows), and performed better than individual algorithms by effectively summarizing the supportive, complementary, or contradictory nature of the predictions (see text).
Gene Ontology (GO) biological processes (11, 12) for the 69 genes suggest that both environmental and genetic perturbations affect mostly metabolic genes. Fig. 3 shows metabolic reactions whose activities are increased (green) and decreased (red) by genes encoding proteins with metabolic roles. This network suggests that cells exploit galactose availability by directing some of the metabolic flux from the Leloir pathway (3) to alcohol synthesis (ADH6) via glycolysis, and to glycerol (GPP1/RHR2 and GPP2/HOR2), purine base (ADE4 and ADE5,7), and amino acid syntheses (HIS1 and HIS4) via pentose phosphate pathways. In contrast, several pathways seem to be down-regulated: glycogen synthesis (GSY1 and GPH1), gluconeogenesis (GLK1), the citrate cycle (PYC1 and SDH4), pyruvate metabolism (ACH1), and fructose utilization (HXT3,4,6,7 and HXK1). These results agree with those reported by Ostergaard et al. (13), who measured metabolite amounts and performed Metabolic Flux Analysis in galactose-limited conditions (Fig. 6B). The remaining selected genes in our list perform general functions such as transport (PHO84, AGP2, and DIP5), protein synthesis/degradation (RPL34B and RPL31A; PRC1), protein folding (SSE1 and HSP30), RNA processing/synthesis (NSR1), cell cycle regulation (FAR1), and stress response (DDR48 and GCY1).
Using similar types of data, Ideker et al. (3) identified 997 genes by simply selecting genes altered in at least one experiment (the union method). Our integration method has reduced the number of candidate genes by an order of magnitude. All but nine genes (LEU2, ECM2, RPA34, YOL084W, SHM1, YNL208W, PRC1, YFL052W, and AGP2) in our list of 69 genes were included among the 997 genes identified by Ideker et al. (3). However, it is interesting to note that the significance values of these nine genes are close to the cutoff threshold Ideker et al. (3) used to select their 997 genes (see Supporting Text). Thus, our methodology has selected an order of magnitude fewer genes. Moreover, the nine genes selected by Pointillist on the basis of all available evidence could not be identified on the basis of the single significance measure used by Ideker et al. (3). Also, the GO tree constructed by using our 69 genes is more focused on metabolism than Ideker et al.'s 997 genes (see GO term frequencies and P values for carbohydrate and galactose metabolisms in Fig. 6C).
Determination of PP Interactome. PP interactions were identified by integrating the following five types of data (Fig. 1B): (i) the full set of PP interactions in DIP (14) and BIND (15), including data from yeast two hybrid (16) and TAP-tag assays (17), as well as paralog interaction analysis (18); (ii) the combined subcellular localization data from SGD (GO cellular components) (11) and GFP databases (19); (iii) gene expression correlations estimated from ≈1,300 gene expression profiles from ExpressDB (http://salt2.med.harvard.edu/cgi-bin/ExpressDByeast/EXDStart) with additions from our time-course and deletion gene expression profiles; (iv) changes in gene expression levels due to gene deletions, and (v) domain–domain (DD) interactions computationally predicted by multiprospector (20) and interdom (21). multiprospector computes the interfacial energy between two protein domains after determining the structures by using threading. interdom predicts DD interactions from orthologous and other known PP interactions stored in databases. We used the Pfam database (22) to obtain the domains that each yeast protein contains when mapping DD interactions to PP interactions (see Supporting Text for the details of the estimation of P values for each of the five data sets).
From these various sources, Pointillist identified 16,985 PP interactions. Fig. 2B shows the numbers of PP interactions in DIP that were selected or removed as false positives by our integration method. It indicates several important points about the integration method and its reliability. First, our method selected 99.1% (2,980) of 3,006 interactions detected by small-scale experiments (S), which are generally considered as true PP interactions in DIP (18). Thus, our integration method indeed identifies virtually all known PP interactions. Second, our method rejected 92.9% of PP interactions detected by any single yeast two-hybrid assay, 24% of PP interactions predicted by paralog analysis (R), and 28.1% of PP interactions detected by multiple yeast two-hybrid assays (M). Deane et al. (18) reported a false positive rate of 22% for paralog analysis, which is close to our rejection rate of 24%. On the other hand, our 28% rejection rate for the intersection of multiple yeast two-hybrid experiments is twice the previously described false positive rate (18). In contrast, Pointillist selected 99.8% of PP interactions detected by more than two different detection methods (MR, SR, SM, and SMR; Fig. 2B). This finding suggests that intersection-based approaches to data integration, although having an inherently high false negative rate, can produce data sets virtually free of false positives when they integrate different detection technologies, but not when integrating multiple data sets from the same technology.
Determination of PD Interactome. We determined PD interactions by integrating the following five types of data (Fig. 1C): (i) chromatin IP-chip data for 113 transcription factors (TFs) in YPD media (23), supplemented with Gal4p chromatin IP-chip in galactose media (10); (ii) subcellular localization data from SGD (11) and GFP databases (19); (iii) gene expression (ExpressDB) correlation between TFs and their target genes; (iv) expression changes resulting from deletion of 23 galactose-metabolism related genes (3); (v) the overall P values of five computational TF-binding site (TFBS) prediction tools (Fig. 1F). fuzznuc (24) scans a given sequence for known TFBS motifs (Table 6). alignace (25), meme (26), motsa (27), and motifsampler (28) search for statistically overrepresented patterns in putative cis-regulatory DNA. For each gene, we searched the entire upstream sequence from the transcription start site to the end of the coding region of the preceding gene. For an example case where the TFBSs are already known (Gal4p binding within the upstream region of GAL2), Fig. 2C shows the individual predictions of these algorithms (green bar, top five graphs), the computed P values for the individual algorithms (magenta dots, top five graphs), and the overall P values (magenta bars, bottom graph) after integration by Pointillist. All five reported Gal4p-binding sites (29, 30) were correctly identified, as marked by arrows in the bottom row (Fig. 2C). For the computational component of the overall PD data interaction process, we used the lowest of the overall P values of all predicted TFBS for each TF on each gene (see Supporting Text for details of how individual P values for the other four types of data were estimated).
We identified 8,555 PD interactions by integrating the five types of data. A total of 3,982 of these PD interactions are in common with the 3,985 PD interactions selected in Lee et al. (23) using a P value cutoff of 0.001. Thus, our integration method captures virtually every interaction identified by Lee et al. (23). Importantly, our integration method found that the Lee et al. cutoff was stringent and included additional predicted interactions that would be missed on the basis of chromatin IP-chip data alone. This finding is consistent with earlier results by Bar-Joseph et al. (31), who showed that they could identify false negatives in chromatin IP-chip data by additionally considering gene expression correlations. The final trustworthiness weights (2) were 0.534 (chromatin IP-chip), 0.019 (cellular components), 0.014 (gene expression correlation), 0.123 (deletion effects), and 0.31 (TFBS predictions), respectively. Thus, computational TFBS predictions, although they are not condition specific, provide the second most useful contribution to identifying PD interactions.
Network Predictions. To demonstrate the usefulness of Pointillist in generating specific, testable hypotheses, we built a network model for the selected 69 genes by combining the identified network nodes and edges (affected genes/proteins, and PP and PD interactions). The nodes in the network were then clustered into distinct functional groups by using Cytoscape BioModules (see Materials and Methods). Fig. 3 shows the nine metabolic modules identified and their interactions (see Materials and Methods). This network captures the known genetic regulatory interactions involved in galactose utilization. For example, Gal4p is a transcriptional driver of four structural GAL genes (GAL1, 2, 7, and 10) and two regulatory GAL genes (GAL3 and 80). The network also recapitulates the known redistribution of metabolic fluxes in the presence of galactose: (i) metabolic fluxes in galactose (green arrows) flow from the Leloir pathway mainly to alcohol synthesis (via glycolysis) and also to glycerol and purine-base/amino acid syntheses (via pentose phosphate pathways); (ii) metabolic fluxes active in raffinose metabolism (fructose metabolism, the citrate cycle, and glycogen synthesis) are down-regulated (red arrows). Thus, both intra- and intermodule interactions are correctly delineated with Pointillist.
In addition to revealing many previously studied features of galactose utilization, our network model leads to insights into information paths connecting galactose metabolism with other metabolic processes. For example, our model suggests that galactose results in down-regulation of the fructose metabolism BioModule (Fig. 3, BM12). By following information paths in the model, we predict that the galactose-responsive transcriptional activator, Gal4p (a nuclear component of the primarily cytoplasmic galactose metabolism BioModule; BM8), contributes to this effect by activating the expression of MTH1 encoding the transcriptional repressor Mth1p, which interacts with and reduces the expression of target genes including hexose transporter, Hxt7p. To test this possibility, we grew strains containing Hxt7p-TAP or Gal2p-pA fusion proteins in raffinose and monitored protein levels at various times after a shift to glucose (high and low), galactose, or ethanol. As predicted, the levels of Hxt7p-TAP were reduced in galactose (Fig. 4C). The reduction rate under these conditions was slower than that observed when cells were transferred to media containing high concentrations of glucose (5%). Hxt7p-TAP remained relatively stable when these cells were transferred to media containing either low levels of glucose (0.5%), or high levels of ethanol.
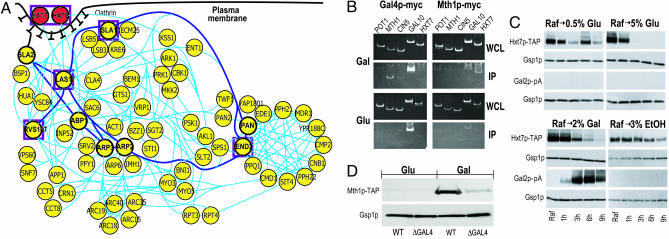
Fig. 4.
Theoretical prediction and experimental verification. (A) A subnetwork of proteins related to the galactose-mediated endocytosis and degradation of Hxt6p and Hxt7p. This network provides a detailed view of processes related to vesicle-mediated protein degradation including: (i) endocytosis (SlaI/2p, Myo3/5p, Lsb3/5p, Las17p, Rvs167p, End3p, Ark1p, Prk1p, etc.); (ii) actin cortical patch assembly (Ent1p, Arc15/19p, Hua1p, BspI, etc.); (iii) vesicle movement along actin filament (Act1p); (iv) protein–vacuole transport (Arp6p). Thus, this network not only captures existing models for endocytosis (33), but also hypothesizes additional proteins and their interactions. (B) Chromatin IP showing galactose-specific binding of Gal4p-myc to MTH1, CIN5, and GAL10 (Left) and of Mth1p-myc to HXT7 (Right). Strains with myc-tagged versions of Gal4p or Mth1p were grown in glucose or galactose, cells were lysed, and chromatin was sheered and immunoprecipitated with antibodies to the myc epitope. DNA fragments in IP and whole cell extract (WCE) fractions were amplified by PCR and resolved on acrylamide gels. (C) Western blots assaying the abundances of Hxt7p-TAP, Gal2p-pA, and Gsp1p (loading control) after shifting cells from growth in raffinose to growth in glucose, galactose, or ethanol. Strains with TAP- or pA-tagged versions of Hxt7p or Gal2p, respectively, were grown to mid-logarithmic in rich medium containing 2% raffinose. The cells were harvested and incubated for 9 h in rich medium containing different carbon sources at the indicated concentrations. Whole cell lysates from these cultures were probed with affinity-purified rabbit IgG or anti-Gsp1p, and visualized with anti-rabbit-HRP secondary antibodies and ECL. (D) Wild-type and ΔGAL4 strains containing MTH1-TAP were grown overnight in YEP containing 2% glucose, and then transferred to YEP containing either 2% glucose or 2% galactose and grown for 14 h to mid-log phase. Equal amounts of protein from each culture were separated by SDS/PAGE and analyzed by immunoblotting with antibodies to the TAP tag (Open Biosystems) or to Gsp1p (to monitor protein loads). Levels of Mth1p-TAP increase in the presence of galactose, whereas the levels of Gsp1p remain unchanged. Robust Mth1p-TAP induction depends on GAL4, suggesting that Gal4p is a positive regulator of MTH1.
To further test this hypothesis, we determined whether MTH1 and HXT7 are galactose-specific targets of myc-tagged versions of Gal4p and Mth1p, respectively, by chromatin IP (Fig. 4B). As predicted, Gal4p-myc bound to MTH1 in the presence of galactose (7), but not in the presence of glucose. Gal4p-myc also bound to its known target GAL10, as expected, but not to HXT7 or POT1, genes not implicated as targets. As predicted, Mth1p-myc bound the target HXT7 in galactose and not in glucose. Interactions were not detected with other genes tested. To further characterize the effect of galactose on the expression of MTH1, we analyzed Mth1p-TAP levels in wild-type and ΔGAL4 strains containing TAP-tagged versions of Mth1p, after growth in the presence or absence of galactose (Fig. 4D). Levels of Mth1p-TAP increased in response to galactose in a GAL4-dependent manner. Together, these data support our predicted Gal4p-, Mth1p-mediated effect of galactose on fructose utilization. Interestingly, Gal4p-myc also has a galactose-specific interaction with CIN5, which, like MTH1, encodes a transcriptional repressor. Our network predicts that CIN5 is regulated by Gal4p and targets HXT3 and HXT6, thus suggesting that CIN5 is also a candidate mediator of the fructose module repression. Thus, a hypothesis derived from the experimental network arising from the Pointillist-integrated data was formulated, tested, and verified.
Our model also provides a hypothesis for how glycogen synthesis (Fig. 3; BM6) is decreased in galactose. Mth1p, induced by Gal4p, binds the promoter region of GAC1 (Fig. 3; ref. 23). Moreover, GAC1 expression is decreased in galactose, potentially because of inhibition by Mth1p. Gac1p is an important regulator of protein phosphatase I (Glc7p), which is involved in the synthesis of glycogen (32). Interestingly, our network predicts that Cin5p (induced by Gal4p) also binds the promoter region of GAC1, which suggests the presence of an alternate path for the decrease of glycogen synthesis involving Gal4p and Cin5p. Our network also captures a partial view of Hxt6/7p endocytosis and degradation in galactose (Hxt6/7p interactions with Las17p-Rvs167p and Sla1p-End3p in Fig. 3). For a more detailed picture, we built a subnetwork (Fig. 4A) around these proteins (enclosed by purple squares) as well as Sla2p, Pan1p, Abp1p, and Arp2/3p (circled in bold), which are reported to be involved in degradation of hexose transporters (33). This network provides an extensive view of vesicle-mediated protein degradation: (i) endocytosis (SlaI/2p, Myo3/5p, Lsb3/5p, Las17p, Rvs167p, End3p, Ark1p, Prk1p, etc.); (ii) actin cortical patch assembly (Ent1p, Arc15/19p, Hua1p, BspI, etc.); (iii) vesicle movement along actin filaments (Act1p); and (iv) protein–vacuole transport (Arp6p). It can be seen that this network not only captures Gourlay et al.'s model (33) (thick blue lines in Fig. 4A), but also hypothesizes additional proteins and their interactions.
Finally, our network model suggests the following mechanisms whereby the various modules interact with each other to coordinate metabolic and cellular activities. (i) The glycogen synthesis module (BM6, 19) interacts with the pseudohyphal growth (BM17) and cell cycle regulation (BM10) modules. This finding implies that decreased glycogen synthesis may result in cell cycle arrest by increasing the expression of FAR1 (one of the 69 affected genes; BM10). (ii) Gcy1p (BM20) is up-regulated in galactose and interacts with mRNA/rRNA processing via Prp6p. Therefore, Gcy1p may be involved in the regulation of RNA processing (e.g., degradation) in addition to response to stress and arabinose metabolism. (iii) A cluster of proteins, including the up-regulated Lap3p (Gal6p), interact with the protein degradation module (middle right of Fig. 3). Thus, Lap3p, which is known to act as a negative regulator of Gal4p (34), may be involved in protein degradation.
Conclusions
We reported integration of 18 different types of data for the galactose pathway, demonstrating the efficacy of our data integration approach. The application of our methodology to these data also generated a number of insights and hypotheses. For most of our reconstructed network, we found supportive evidence in the literature. Our network model also resulted in a number of unique hypotheses. For the example case of fructose metabolism down-regulation in galactose-rich media, we experimentally verified the predictions of our network model. Pointillist is a useful tool for model building in systems biology and for enormously reducing the dimensionality of the large integrated data sets. It can also be applied to entirely different scenarios (e.g., mass spectrometry) without the need for the costly and time-consuming process of collecting training sets. This feature is particularly useful considering the rapid rate of progress in high-throughput technologies, where, by the time curated training data are available, improved technologies supercede them.
Supplementary Material
Acknowledgments
We thank Jeffrey Skolnick (Center of Excellence in Bioinformatics, University at Buffalo) for providing Multiprospector data and Vesteinn Thorsson (Institute for Systems Biology) for providing microarray data (3). A.F.S. holds the Grant I. Butterbaugh Professorship at the University of Washington. This work was supported in part by National Science Foundation Grant BES-0223056 (to L.H.) and National Institutes of Health/National Institutes of General Medical Sciences Grant R01 GM067228 (to J.D.A.).
Conflict of interest statement: No conflicts declared.
Abbreviations: PP, protein–protein; PD, protein–DNA; IP, immunoprecipitation; TF, transcription factor; TFBS, TF-binding site.
References
- 1.Mrowka, R., Patzak, A. & Herzel, H. (2001) Genome Res. 11, 1971–1973. [DOI] [PubMed] [Google Scholar]
- 2.Hwang, D., Rust, A. G., Ramsey, S., Smith, J. J., Leslie, D. M., Weston, A. D., de Atauri, P., Aitchison, J. D., Hood, L., Siegel, A. F. & Bolouri, H. (2005) Proc. Natl. Acad. Sci. USA 102, 17296–17301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ideker, T., Thorsson, V., Ranish, J. A., Christmas, R., Buhler, J., Eng, J. K., Bumgarner, R., Goodlett, D. R., Aebersold, R. & Hood, L. (2001) Science 292, 929–934. [DOI] [PubMed] [Google Scholar]
- 4.Prinz, S., Avila-Campillo, I., Aldridge, C., Srinivasan, A., Dimitrov, K., Siegel, A. F. & Galitski, T. (2004) Genome Res. 14, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longtine, M. S., McKenzie, A., Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 6.Aitchison, J. D., Rout, M. P., Marelli, M., Blobel, G. & Wozniak, R. W. (1995) J. Cell Biol. 131, 1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 8.Ausubel, F., Brent, R., Kingston, R., Moore, D., Seidman, J., Smith, J. & Struhl, K. (2000) Current Protocols in Molecular Biology (Wiley, New York).
- 9.Smith, J. J., Marelli, M., Christmas, R. H., Vizeacoumar, F. J., Dilworth, D. J., Ideker, T., Galitski, T., Dimitrov, K., Rachubinski, R. A. & Aitchison, J. D. (2002) J. Cell Biol. 158, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weston, A. D., Baliga, N. S., Bonneau, R. & Hood, L. (2003) Cold Spring Harbor Symp. Quant. Biol. 68, 345–357. [DOI] [PubMed] [Google Scholar]
- 11.Dwight, S. S., Balakrishnan, R., Christie, K. R., Costanzo, M. C., Dolinski, K., Engel, S. R., Feierbach, B., Fisk, D. G., Hirschman, J., Hong, E. L., et al. (2004) Brief Bioinform. 5, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B., Mungall, C., et al. (2004) Nucleic Acids Res. 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostergaard, S., Olsson, L. & Nielsen, J. (2001) Biotechnol. Bioeng. 73, 412–425. [DOI] [PubMed] [Google Scholar]
- 14.Xenarios, I., Salwinski, L., Duan, X. J., Higney, P., Kim, S. M. & Eisenberg, D. (2002) Nucleic Acids Res. 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfarano, C., Andrade, C. E., Anthony, K., Bahroos, N., Bajec, M., Bantoft, K., Betel, D., Bobechko, B., Boutilier, K., Burgess, E., et al. (2005) Nucleic Acids Res. 33, D418–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. & Sakaki, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 18.Deane, C. M., Salwinski, L., Xenarios, I. & Eisenberg, D. (2002) Mol. Cell. Proteomics 1, 349–356. [DOI] [PubMed] [Google Scholar]
- 19.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. & O'Shea, E. K. (2003) Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- 20.Lu, L., Arakaki, A. K., Lu, H. & Skolnick, J. (2003) Genome Res. 13, 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng, S. K., Zhang, Z., Tan, S. H. & Lin, K. (2003) Nucleic Acids Res. 31, 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 24.Rice, P., Longden, I. & Bleasby, A. (2000) Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, J. D., Estep, P. W., Tavazoie, S. & Church, G. M. (2000) J. Mol. Biol. 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 26.Bailey, T. L. & Elkan, C. (1994) Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- 27.Reiss, D. J. & Schwikowski, B. (2004) Bioinformatics 20, Suppl. 1, I274–I282. [DOI] [PubMed] [Google Scholar]
- 28.Thijs, G., Lescot, M., Marchal, K., Rombauts, S., De Moor, B., Rouze, P. & Moreau, Y. (2001) Bioinformatics 17, 1113–1122. [DOI] [PubMed] [Google Scholar]
- 29.Bram, R. J., Lue, N. F. & Kornberg, R. D. (1986) EMBO J. 5, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nehlin, J. O., Carlberg, M. & Ronne, H. (1989) Gene 85, 313–319. [DOI] [PubMed] [Google Scholar]
- 31.Bar-Joseph, Z., Gerber, G. K., Lee, T. I., Rinaldi, N. J., Yoo, J. Y., Robert, F., Gordon, D. B., Fraenkel, E., Jaakkola, T. S., Young, R. A., et al. (2003) Nat. Biotechnol. 21, 1337–1342. [DOI] [PubMed] [Google Scholar]
- 32.Feng, Z. H., Wilson, S. E., Peng, Z. Y., Schlender, K. K., Reimann, E. M. & Trumbly, R. J. (1991) J. Biol. Chem. 266, 23796–23801. [PubMed] [Google Scholar]
- 33.Gourlay, C. W., Dewar, H., Warren, D. T., Costa, R., Satish, N. & Ayscough, K. R. (2003) J. Cell Sci. 116, 2551–2564. [DOI] [PubMed] [Google Scholar]
- 34.Ostergaard, S., Walloe, K. O., Gomes, S. G., Olsson, L. & Nielsen, J. (2001) FEMS Yeast Res. 1, 47–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.