Abstract
Quantitative protein bioanalysis in complex biological fluids presents considerable challenges in biological studies and disease diagnosis. The major obstacles are the background signals from both the probe and the biological fluids where the proteins reside. We have molecularly engineered light-switching excimer aptamer probes for rapid and sensitive detection of a biomarker protein, platelet-derived growth factor (PDGF). Labeled with one pyrene at each end, the aptamer switches its fluorescence emission from ≈400 nm (pyrene monomer) to 485 nm (pyrene excimer) upon PDGF binding. This fluorescence wavelength change from monomer to excimer emission is a result of aptamer conformation rearrangement induced by target binding. The excimer probe is able to effectively detect picomolar PDGF in homogeneous solutions. Because the excimer has a much longer fluorescence lifetime (≈40 ns) than that of the background (≈5 ns), time-resolved measurements were used to eliminate the biological background. We thus were able to detect PDGF in a cell sample quantitatively without any sample pretreatment. This molecular engineering strategy can be used to develop other aptamer probes for protein monitoring. Combined with lifetime-based measurements and molecular engineering, light-switching excimer aptamer probes hold great potential in protein analysis for biomedical studies.
Keywords: aptamer, biomarker, platelet-derived growth factor, pyrene, time-resolved fluorescence
Proteins are ubiquitous and essential for life. Detection of proteins in their native environments has always been a critical and challenging task. In the proteomics era, numerous disease-marker proteins are expected to be discovered from various complex biological systems (1–3). Methods for the analysis of proteins have become indispensable tools in new disease-marker discovery and their function studies. Ultimately, assays that allow rapid, simple, sensitive, selective, and cost-effective detection of the proteins discovered are of significant importance for the understanding, diagnosis, treatment, and prevention of many diseases. Key factors, including a highly selective molecular recognition element and a novel signal transduction mechanism, have to be engineered together for successful assay development. Among many molecular recognition elements, synthetic nucleic acid ligands (aptamers) (4–6) have gained increasing attention in this area. Aptamers are single-stranded oligonucleotides selected to bind essentially any molecular targets with high selectivity and affinity through an in vitro selection process called SELEX (selective evolution of ligands by exponential enrichment) (4–6). Besides their excellent binding affinity and selectivity, other characteristics endow aptamers with great potential for use in protein analysis (7, 8). For instance, aptamers can be routinely prepared by chemical synthesis, which allows rapid preparation in large quantity and with excellent reproducibility. Nucleic acid synthetic chemistry also facilitates conjugation of these aptamer sequences to fluorescent dyes, radiolabels, or other biomolecules. Furthermore, aptamer sequences are more stable than proteins under a wide range of conditions and could be repeatedly used without losing their binding capabilities.
To report the binding of an aptamer to its target, a signal transduction mechanism has to be built into the aptamer sequences. Fluorescent techniques offer excellent choices for signal transduction because of their nondestructive and highly sensitive nature. Several fluorescence techniques such as fluorescence anisotropy (7, 8) and fluorescence resonance energy transfer (FRET) (9), as well as fluorescence quenching (9–11), have been used in aptamer assay development. All these signal-transduction techniques have their individual strengths. Nonetheless, they suffer from some limitations that could hamper their effectiveness in complex biological samples. For instance, although fluorescence anisotropy only requires singly labeling of one dye molecule on each aptamer sequence, it entails complicated instrumentation and data interpretation. FRET- or fluorescence-quenching-based probes quantify target concentrations with changes in fluorescence intensity, but these two methods are sensitive to the solution environment. More importantly, they are difficult to apply directly to analyzing proteins in their native environments because of the interference of intense background signal.
When monitoring a protein in its native environment, there are usually two significant background-signal sources. The first one is the probe itself. For example, when a quenching-based FRET molecular probe is used for protein studies, the probe always has some incomplete quenching, resulting in a significant probe background. Moreover, in a native biological environment, there are many potential sources for false positive signals of the molecular probe for protein analysis. The second source of background signal comes from the native fluorescence of the biological environment where the target protein resides. There are many molecular species in a biological environment, some of which will give a strong fluorescence background signal upon excitation. These problems deteriorate assay sensitivity, compromise probe selectivity, and thus hinder the analysis of proteins. Although there have been great efforts in solving these problems in bioanalysis (12), effective solutions to both problems are limited.
We have molecularly engineered a light-switching excimer aptamer probe for protein monitoring in biological fluids using both steady-state and time-resolved fluorescence measurements. This strategy is a combined approach of wavelength switching and time-resolved measurement to solve the significant problems for protein monitoring in its native environment. Our approach is to label molecular aptamer with pyrene, similar to what has been reported in using pyrene for molecular beacons (13). The aptamer sequence that binds with high affinity to the target protein platelet-derived growth factor (PDGF)-BB is labeled with pyrene molecules at both ends. The specific binding of aptamer to its target protein changes the aptamer probe conformation, bringing the two pyrene molecules into close proximity to form an excimer (excited state dimer), which results in a change of fluorescence wavelength from ≈400 nm for the pyrene monomer to 485 nm for the pyrene excimer. This emission wavelength switching solves the probe background-signal problem that occurs with FRET molecular probes. However, this light-switching method alone cannot solve the problem of strong background signal from the multiple species in the biological environment. One special feature of the pyrene excimer is that it has a very long fluorescence lifetime (14) compared with other potential fluorescent species. The lifetime of the pyrene excimer can be 100 ns or longer, whereas that for most of the biological background species is <5 ns. With time-resolved fluorescence measurements, target binding induced excimer signal can be separated from biological background interference. Combining light-switching and time-resolved measurements, we are able to detect picomolar PDGF-BB in a few seconds. Direct detection and quantification of target molecules in complex biological samples such as a cultured cell dish can be carried out without any need of sample clean-up process.
Methods
Chemicals and Reagents. The sequences of oligonucleotides and aptamer probes prepared are listed in Table 1. DNA synthesis reagents were purchased from Glen Research (Sterling, VA). Four aptamer sequences with different lengths (shown in Table 1) were synthesized: ES3, ES4, ES5, and ES6 (excimer probes with 3, 4, 5, and 6 bp in the stem, respectively). All of the aptamer sequences were labeled with pyrene at both ends. PS3 (pyrene monomer with 3 bp in the stem) was an aptamer sequence but only singly labeled with pyrene at the 5′ terminus. ESCRBL was a 39-mer scramble oligonucleotide sequence with pyrene labeled at both ends.
Table 1. Probes and oligonucleotides used in PDGF binding study.
| Name | Sequence |
|---|---|
| ES3 | Pyr-AGGCTACGGCACGTAGAGCATCACCATGATCCT-Pyr |
| ES4 | Pyr-CAGGCTACGGCACGTAGAGCATCACCATGATCCTG-Pyr |
| ES5 | Pyr-ACAGGCTACGGCACGTAGAGCATCACCATGATCCTGT-Pyr |
| ES6 | Pyr-CACAGGCTACGGCACGTAGAGCATCACCATGATCCTGTG-Pyr |
| PS3 | Pyr-AGGCTACGGCACGTAGAGCATCACCATGATCCT |
| ESCRBL | Pyr-GGA ACG TAA TCA ACT GGG AGA ATG TAA CTG ACT GC-Pyr |
Boldface type indicates the bases that form a stem after the probe binds to the protein PDGF.
Recombinant human PDGF-BB, PDGF-AB, and PDGF-AA were purchased from R&D Systems and dissolved in 4 mM HCl with 0.1% BSA and then diluted in a Tris buffer (pH 7.5) before use. Other recombinant human growth factors, including recombinant human EGF and insulin-like growth factor 1, were from Roche (Indianapolis, IN). Human BSA, human hemoglobin, horse myoglobin, chicken lysozyme, human α-thrombin (THR), and other chemicals were from Sigma. A solution of 0.1 M triethylamine acetate (pH 6.5) was used as HPLC buffer A, and HPLC-grade acetonitrile (Fisher) was used as HPLC buffer B. Tris·HCl buffer (20 mM Tris·HCl/20 mM NaCl, pH 7.5) was used for all buffer-solution-based aptamer-binding experiments. Except for the cell media, ultrapure water was used to prepare all of the solutions. The cell media, DMEM (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (Invitrogen), was used for the detection of PDGF in the real samples.
Instruments. An ABI3400 DNA/RNA synthesizer (Applied Biosystems) was used for DNA synthesis. Probe purification was performed with a ProStar HPLC (Varian) where a C18 column (Econosil, 5U, 250 × 4.6 mm) from Alltech Associates was used. UV-Vis measurements were performed with a Cary Bio-300 UV spectrometer (Varian) for probe quantitation. Steady-state fluorescence measurements were performed on a Fluorolog-Tau-3 spectrofluorometer (Jobin Yvon, Edison, NJ). For emission spectra, 349 nm was used for excitation. Time-resolved measurements were made with a single photon-counting instrument (OB900, Edinburgh Analytical Instruments, Livingston, U.K.), where a nitrogen flash lamp was used as the excitation source (λ = 337 nm).
Synthesis and Purification. A solid-phase synthesis method was used to couple pyrene to aptamer sequences at both 3′ and 5′ ends. The synthesis started with a 3′-amino-modifier C7 controlled pore glass (CPG) column at 1-μmol scale. After the synthesis of the aptamer sequence, a 5′-amine was added to the sequence by using 5′-amino-modifier-C6 linker phosphoramidite. The column then was flushed slowly with 15 ml of dimethylformamide (DMF), 15 ml of 20% piperidine in DMF, 15 ml of 3% trichloroacetic acid in dichloromethane, and then another 15 ml of DMF. The CPG contained within the column was released into 1 ml of DMF solution containing 57.7 mg (200 μmol) of pyrene butyric acid, 41.3 mg (200 μmol) of dicyclocarbodiimide, and 24.4 μg (200 μmol) of dimethylaminopyridine. After stirring for 3 h, the solution was centrifuged, and the supernatant was discarded. The pellet was washed three times with DMF, methanol, and water, respectively, before incubated in a 50% solution of methylamine in ammonia at 65°C for ≈10 min. The resulting clear and colorless supernatant was collected. Under UV radiation, an intense green fluorescence was observed from the collected solution. The aptamer solution was desalted with a Sephadex G-25 column (NAP-5, Amersham Pharmacia) and dried in a SpeedVac. The dried product was purified by HPLC using a C18 column with a linear elution gradient with buffer B changing from 25% to 75% in 25 min at a flow rate of 1 ml/min. The second peak in chromatography that absorbed at 260 and 350 nm, and emitted at 400 nm with 350-nm excitation, was collected as the product. The collected product then was vacuum-dried, desalted with a G-25 column, and stored at –20°C for future use.
Results
Design and Synthesis of Light-Switching Excimer Aptamer Probe.Some spatially sensitive fluorescent dyes, such as pyrene (13–16) and BODIPY Fl (17, 18), can form excimers upon close encounter of an excited-state molecule with another ground-state molecule. The excimer emits at a longer wavelength than a monomer does. The formation of excimer between two pyrene molecules that are connected by a flexible covalent chain is useful to probe spatial arrangement of some molecules. Similar to FRET, the stringent distance-dependent property of excimer formation can be used as a unique signal transduction in the development of molecular probes. This technique is especially useful for developing aptamer probes because a variety of aptamers, like those for PDGF-BB (7, 10, 19), cocaine (20), THR (21, 22), and HIV1 Tat protein (23), undergo similar conformational changes upon target binding.
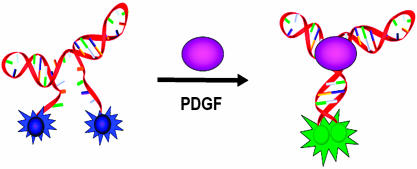
As a proof of principle, the excimer signaling approach was used to develop a probe for PDGF-BB. Identified by Green et. al (19), the PDGF-BB aptamer is a DNA sequence with an open secondary structure in the absence of protein (Fig. 1). When the aptamer binds to PDGF-BB, it changes to a close conformation where the 3′ and 5′ ends hybridize and form a stem. Based on this change, an excimer switching aptamer probe has been developed by labeling both ends with dyes that can form excimers. When the dual-pyrene-labeled aptamer probe is free in solution without the target protein, both pyrene molecules are spatially separated, and only the monomer emission peaks (at 375 and 398 nm) are observed. The binding of the aptamer probe to the target protein brings the pyrene molecules at 3′ and 5′ ends together, allowing the formation of an excimer. Thus, the emission peak at ≈485 nm appears. The change in emission color serves as a rapid way for qualitative analysis, and the excimer fluorescence intensity can be used for highly sensitive real-time quantitation of PDGF in homogeneous solutions.
Fig. 1.
Use of the pyrene excimer to probe PDGF. PDGF aptamer (red) is end-labeled with pyrene molecules (blue) that are separated from each other because of the open structure of the aptamer. The pyrene molecule has monomer emission peaks at ≈378 and 398 nm. After binding to PDGF (purple), the aptamer adapts a close conformation, bringing two pyrene molecules close to each other. Consequently, pyrene excimer (green) forms and green light (≈485 nm) is emitted after photoexcitation.
For labeling oligonucleotide with pyrene, several methods were reported (13, 24). Unfortunately, for dually labeling pyrene on both nucleic acid termini, the reported procedures proceeded in low yields partially because of incompatible solubility of the pyrene derivatives and nucleic acids. A solid-phase coupling method was used to allow multilabeling of the organic dyes to the DNA sequences. Coupling yields as high as 80% were achieved for all probes synthesized. MALDI-TOF MS results confirmed successful preparation of probes.
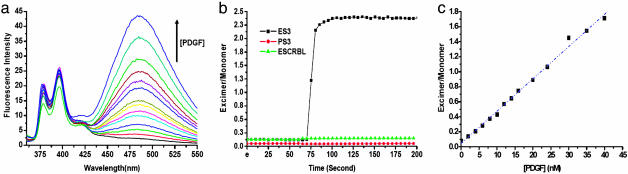
Light-Switching Aptamer Probe for Real-Time Rapid PDGF-BB Detection. A light-switching aptamer probe ES3 (sequence shown in Table 1) was prepared by labeling a 33-nucleotide sequence, specifically selected because of its high affinity to PDGF-BB (19), with pyrene molecules at both 3′ and 5′ ends. This aptamer sequence was obtained through the SELEX process and was reported to have ≈700-fold higher affinity for PDGF when compared with other random DNA sequences (19). Fig. 2a shows the fluorescence emission spectra from solutions of 100 nM ES3 with different concentrations of PDGF-BB proteins. With no target present, monomer emission peaks were observed, and there was no observable excimer emission. Without the target protein, this aptamer sequence assumed an open conformation, spatially separating both pyrene molecules at the 3′ and 5′ ends. Upon addition of PDGF-BB into the ES3 solution, an aptamer–protein complex was formed, causing the 3′ end sequence to hybridize with the 5′ end sequence to form a stable stem. This stem brought both pyrene molecules together, resulting in an intense excimer emission at 485 nm (Fig. 2a). With increasing amounts of target protein in the solution, the excimer intensity increased proportionally.
Fig. 2.
Detection of PDGF in homogeneous solution by steady-state fluorescence. (a) Response of the excimer probe ES3 to different concentrations of PDGF-BB (0–40 nM). (b) Real-time response of ES3 and two pyrene-labeled control sequence to 50 nM PDGF-BB. PS3 is an aptamer sequence with one pyrene labeled at 5′. The ESCRBLE is a random DNA sequence with pyrene labeled at both ends. (c) The fluorescence ratio of excimer over monomer as a function of target protein concentration. [ES3] = [PS3] = [ESCRBLE] = 100 nM.
Two control DNA sequences, PS3 and ESCRBL, were prepared to confirm that the observed excimer emission was a result of the aptamer–protein binding. The first sequence, PS3, was a PDGF aptamer sequence labeled with only one pyrene at the 5′ end. Addition of PDGF into the PS3 solution did not change the emission spectrum of the solution, indicating that two pyrene molecules in close proximity are necessary for the excimer emission and that PDGF-BB itself has no measurable effect on the optical properties of pyrene molecules. The second sequence, ESCRBL, was a random DNA sequence with its 3′ and 5′ ends labeled with pyrene. Because this sequence has no affinity to PDGF, the addition of PDGF should not induce any excimer formation. This experiment was to prove that the excimer emission resulted from aptamer–protein binding. As expected, this dually pyrene-labeled scramble sequence did not give any excimer emission after the addition of PDGF. These two control results confirm that the excimer emission shown in Fig. 2a was because of the aptamer conformation change upon specific binding to PDGF-BB.
Data shown in Fig. 2a reveal one important advantage of the light-switching excimer signaling approach: detection without separation. Because only target-bound probe gives excimer emission, the unbound probe does not have to be separated from the solution for target detection. This detection-without-separation method eliminates tedious washing and separating procedures and allows real-time detection (25). Another advantage of this probe is that it enables ratiometric measurement. As shown in Fig. 2a, protein-bound probe gives three emission peaks, two monomer peaks at 375 and 398 nm, respectively, and an excimer peak at 485 nm. By taking intensity ratio of the excimer peak to either one of the monomer peaks, one could effectively eliminate signal fluctuation and minimize the impact of environmental quenching on the accuracy of the measurement. Real-time response of excimer/monomer ratio is shown in Fig. 2b. It revealed that the binding of the aptamer to PDGF-BB took place within seconds. This result suggests that the PDGF aptamer probe could be used for the rapid monitoring of PDGF in vivo.
The light-switching signal transduction approach affords high sensitivity. As shown in Fig. 2c, the excimer/monomer emission ratio of the probe responded to different concentrations of PDGF-BB proportionally. A linear response was observed with PDGF-BB concentrations ranging from 0 to 40 nM. Based on 3 times standard deviation of 6 measurements of blank samples, the limit of detection for PDGF-BB was in a picomolar range. Such a high sensitivity, together with the emission wavelength switching, fast measurements and detection without separation properties, enabled visual detection of 4 pmol of PDGF (Fig. 3). A clear green color was observed by the naked eye when 4 pmol of PDGF was added to a 100 μl excimer probe solution.
Fig. 3.
Visual detection of 4 pmol of PDGF-BB after illumination with an UV lamp. Solution of the 100 nM excimer probe without (Left) and with (Right)40 nM of PDGF-BB. The total volume of the solution was 100 μl.
Optimization of Aptamer Length. The original sequence of PDGF-BB aptamer identified is a 39-mer sequence (19). When it binds to PDGF-BB, this aptamer forms a three-way helix junction with a three-nucleotide loop at the branch point, where the 3′ and 5′ ends of the aptamer form a 6-mer stem. Theoretical calculations (26) indicated that a significant fraction of the 39-mer aptamer sequence was in a close conformation even without the presence of the target protein (see Figs. 6–8, which are published as supporting information on the PNAS web site). An excimer probe with this sequence would generate excimer emission in the absence of target from this fraction of the aptamer. Because the stem sequence is not important for the high-affinity binding to PDGF-BB (19), to reduce the background emission, the stem was shortened gradually to identify an aptamer sequence that was in fully opened conformation in the absence of target while reserving a good binding affinity. Four aptamer sequences, ES6, ES5, ES4, and ES3, were prepared, all of which were dually labeled with pyrene at both ends of each sequence. ES6 had a full length of the reported sequence, while ES5, ES4, and ES3 had 1, 2, and 3 bases removed from both ends of the reported 39-mer sequence, respectively.
Such a stem modification did not affect the binding of the aptamer to protein. For all four probes, comparable excimer emission intensity was recorded for each probe at the same concentration of PDGF-BB. Conversely, shortening the stem greatly improved signal-to-noise ratio of the aptamer probe by substantially reducing background signal from free probe (see Fig. 9, which is published as supporting information on the PNAS web site). For the ES6 probe without target, a significant excimer emission was observed. This result indicates that a large fraction of this sequence was in closed conformation, as indicated from the secondary structure prediction. Addition of PDGF-BB resulted in a further increase of the excimer emission that was because of the “closing” of the other fraction of the open-structure aptamer due to protein binding. However, the maximum signal change was <3-fold for this probe because of the high excimer emission intensity from the probe itself. By removing one base from both ends of ES6, the excimer emission intensity from probe ES5 was significantly reduced. Negligible excimer emissions were observed for ES4 and ES3. As a result, the aptamer probes with 3-mer stem (ES3) and 4-mer stem (ES4) were found to have the highest signal enhancement (≈40-fold) upon addition of the target protein.
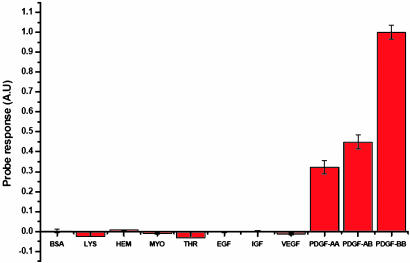
Selectivity of the Probe. To detect PDGF in test samples, the probe has to selectively respond only to PDGF, free from interferences from other biological components. The probe's selectivity was challenged when excess extracellular proteins such as albumin, lysozyme, hemoglobin, myoglobin, and THR were used for a testing. Even at 10 times the PDGF concentration, these proteins did not cause significant signal change (Fig. 4). Further, we tested the selectivity of the excimer probe for proteins and peptides potentially coexisting with PDGF. The results clearly showed that this probe was highly selective for PDGF-BB. It did not respond to the EGF, VEGF, or insulin-like growth factor 1. PDGF-AA and -AB, both of which have shown low binding affinity to the aptamer sequence (19), induced lower signal response. Such an enhanced selectivity comes from both the intrinsic high selectivity of the aptamer sequence and the stringent spatial requirement for the emission from the excimer formation. Partial or weak binding by other species in FRET-based probes will result in substantial signal change, whereas the light-switching probe will have little response for those binding events because the two pyrene molecules have to be in very close proximity to each other. This feature is an additional advantage for the light-switching aptamer excimer probe.
Fig. 4.
Responses of the excimer probe (50 nM) to BSA, lysozyme (LYS), hemoglobin (HEM), myoglobin (MYO), and THR (all 500 nM) and different growth factors [EGF, insulin-like growth factor (IGF), VEGF, PDGF-AA, PDGF-AB, and PDGF-BB, all 50 nM).
Direct Quantitative Detection of PDGF in Cell Media. Results from studies in relative simple and pure buffer systems demonstrate that the excimer aptamer probe has excellent selectivity and high sensitivity. To be more useful in bioassays, this probe should be able to tolerate any interference from biological samples. A dyed cell medium mixed with FBS was used to investigate the feasibility of using this probe in biological samples.
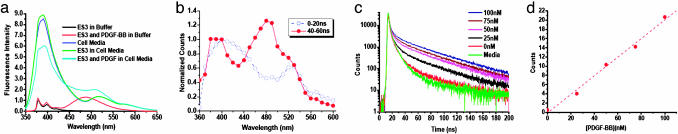
Fig. 5a shows the spectra of the probe in a Tris·HCl buffer solution and in cell media. In the buffer, the probe functioned well, and a strong excimer emission was observed when the target protein was added. Unfortunately, intense background fluorescence, contributing from some indigenous species in the cell media such as proteins, riboflavin, nicotinamide, pyridoxine, tryptophan, and tyrosine as well as phenol red, also was observed. This intense background fluorescence buried the signal response from the probe and made the probe signal indistinguishable from the indigenous background fluorescence. This result indicates that steady-state fluorescence measurement is implausible for direct detection of PDGF in such a complex biological sample. It was also unclear whether this synthetic aptamer sequence retained its binding affinity and selectivity for PDGF-BB in the cell media.
Fig. 5.
Monitoring PDGF in dyed cell media. (a) Steady-state fluorescence spectra of cell media, 200 nM ES3 in cell media, 200 nM ES3 and 50 nM PDGF-BB in cell media, 200 nM ES3 in Tris·HCl buffer, and 200 nM ES3 with 50 nM PDGF-BB in Tris·HCl buffer. (b) Time-resolved fluorescence spectra of 200 nM ES3 and 50 nM PDGF-BB in cell media at different time windows after the excitation pulse, 0–20 ns (blue) and 40–60 (red). (c) Fluorescence decays of 200 nM ES3 in cell media with various concentrations of PDGF-BB. (d) The response of fluorescence intensity to the change of protein concentration.
Most of the background fluorescence has a lifetime of <5 ns. By contrast, the monomer and excimer emission of pyrene have much longer lifetimes (up to 100 ns). Such a big difference allows one to temporally separate the probe fluorescence signal from the intense background signal using time-resolved fluorescence spectroscopy. With a short excitation pulse of ≈1 ns, all chromophores in the solution that absorb at this excitation wavelength, including pyrene and the fluorogenic molecules of the cell media, are excited. By employing a time-resolved detection technique, the decay of the fluorescence signal at different wavelengths can be recorded within a relevant time window (e.g., in the window from 40 to 60 ns). Because background signal is expected to have decayed within the first few nanoseconds after the pulsed excitation, the remaining fluorescence after 20 ns should correspond to the long-lived pyrene fluorescence. As a time-resolved detection technique, time-correlated single-photon counting was used, because it is one of the most sensitive methods (16).
Time-correlated single-photon counting measurement of pure probe and probe with protein solutions suggested that lifetimes of both pyrene monomer and excimer in the cell media were ≈40 ns. This lifetime is one magnitude longer than the lifetimes of most organic fluorophores and fluorescent components in cell media and cells, which further confirmed the possibility of temporal resolution of the excimer signal from intense background fluorescence. Time-resolved emission spectra of 200 nM ES3 in cell media with 50 nM PDGF-BB (Fig. 5b) revealed the change of the emission spectra on a nanosecond scale and demonstrated a clear temporal separation of the signal from background noise. Taken over the first 20-ns decay, the emission spectrum resembled the steady-state emission spectrum of the same sample, where excimer peak was masked by severe background fluorescence from endogenous fluorescence species and scattered light. Because of their short fluorescence lifetimes, the fluorescence and scattered light from the cell media decayed rapidly to 0.1% of its original signal 40 ns after the excitation pulse (see Fig. 10, which is published as supporting information on the PNAS web site). By contrast, the excimer emission decayed slowly, retaining reasonably high emission intensity even after 40 ns of decay, which allows probe signal to be well separated from the background signal. As a result, the fluorescence emission spectrum taken at 40 ns after excitation looked similar to the emission spectra of protein-bound probe in buffer. The temporal separation of intense background from probe signal is evident by comparing the time-resolved fluorescence emission spectrum (40–60 ns) in Fig. 5b with the steady-state fluorescence emission spectrum of the same sample in Fig. 5a. For steady-state measurement, no resolved peak at ≈485 nm could be seen when PDGF was added to ES3 solution in cell media because of the significant amount of background signal. However, in the time-resolved emission spectrum, which was recorded 40 ns after excitation, the long lifetime emission peak at 480 nm was well resolved. This 480-nm peak corresponded to excimer emission from protein-bound probe, which was supported by two observations: (i) its intensity varied with changes of protein concentrations; and (ii) no such peak was observed in the time-resolved spectra of either cell media or a cell-media solution containing PDGF and a single-pyrene-labeled aptamer sequence PS3. Thus, this characteristic emission peak could be immediately used to examine the presence of PDGF in biological samples.
With time-resolved measurement, not only can this probe qualitatively detect target protein in cell media, but it also allows quantitative analysis in situ. When the concentration of PDGF-BB increased, the fluorescence intensity of the excimer peak in time-resolved emission spectrum increased accordingly. Fig. 5c shows the decays of aptamer probe ES3 in the cell media with various concentrations of protein PDGF-BB at 480-nm emission. Fluorescence intensity from the response of each solution could be calculated by integrating photons emitted over an optimized time window, where the background signal decays to a minimum while the excimer signal is still high, to obtain an optimal signal-to-noise ratio for accurate analysis. The highest signal-to-noise ratio at 480 nm was observed from 40 to ≈100 ns after the excitation. Thus, photons emitted between 40 and 100 ns were counted and integrated for each concentration to construct a calibration curve. The resulting fluorescence intensities were proportional to the PDGF-BB concentrations (Fig. 5d). This linear response of fluorescence intensity to PDGF in the cell samples demonstrated the feasibility of direct quantification of target proteins in cell media without any separation or purification. We further used the aptamer probes to determine PDGF secreted from PC3 cells. These cells are prostate cancer cells reported to secrete PDGF in the cell media. PDGF levels were measured from samples that were cultured to confluence and spiked with recombinant human TNF-α to induce PDGF production. TNF-α has the ability to activate different signal transduction pathways that lead to an increase in the growth factor concentration in cells. It was hypothesized that the addition of TNF-α to PC3 cells in culture would increase the levels of PDGF in the conditioned media. Three sets of samples were collected, and their PDGF levels were determined. The cellular sample PDGF levels also were compared with standard ELISA tests for PDGF in cancer samples. Our results demonstrated that the probes and the time-resolved measurements could be used effectively for cancer cell samples.
Discussion
With their unique properties, aptamers are finding increasing applications in therapeutic practices, disease diagnosis, and protein functional studies. These binding elements, once integrated with a novel signal transduction mechanism, can be used as sensitive and selective probes for protein detection. We have demonstrated that the light-switching excimer approach is an excellent signal transduction for aptamer probe development. The generation of the excimer emission requires the conformation change of the aptamer brought about by complexation with a target to bring two pyrene molecules together. This signaling approach has its wide applicability for several reasons. First, there are many aptamers besides PDGF aptamer that undergo conformation changes with target binding event, for example, human THR (21, 22), cocaine (20), and HIV1 TAT protein (23) aptamers. This conformational change can be immediately exploited for the excimer light-switching probe design. For instance, we demonstrated the same properties with a THR aptamer labeled with two pyrene molecules. The probe gave excimer emission that was proportional to different concentrations of THR (see Fig. 11, which is published as supporting information on the PNAS web site). Second, even for an aptamer that does not have an obvious target-induced structure change, it can be designed to change its secondary structure as desired upon target binding with rational structure engineering (11, 23, 27, 28). This strategy has been well demonstrated by Bayer and Smolke (28) who used the aptamer/target-binding event to switch the aptamer structure to convert it to a gene-expression regulator. A simple yet general approach of engineering any aptamer into structure-switching aptamer for real-time signaling applications has also been reported (11, 27). Finally, with a combination of the fast turnaround of automatic SELEX techniques (29) and novel selection approaches (30, 31), virtually any protein target can have at least one conformation-changing aptamer sequence. Thus, numerous excimer-signaling probes may be created quickly for proteomics with a similar approach reported here.
Besides its universal applicability, the excimer light-switching approach offers higher selectivity and excellent sensitivity. A bis-pyrene probe has been recently used for signaling ATP based on ratio changes of the monomer/excimer fluorescence, which is sensitive to its environment (32). However, this probe shows a lower sensitivity (<3.5-fold of signal change at 3 mM target concentration), a narrower dynamic range (0.5–3 mM ATP), and a lack of selectivity (nonspecific binding will cause false positive/negative signal). By contrast, the signal approach reported here requires target-induced structure switching to form excimer, which prevents false signals caused by nucleases degradation or nonspecific binding. Another advantage of using our approach is that it allows ratiometric measurement, which could minimize the environmental effect to afford more precise detection. More importantly, the excimer light-switching approach solves the background problem from both probe itself and the biological environment. With the time-resolved single photon counting technique, this probe is able to detect PDGF-BB in a cell media qualitatively and quantitatively without any need of sample pretreatment. This excimer signaling approach has potential for the development of highly sensitive aptamer probes for clinical, forensic, and environmental applications. The detection-without-separation, high sensitivity, and excellent selectivity of this approach will enable useful applications to construct aptamer probes for protein function studies in an intracellular or intercellular environment.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01 GM66137 and in part by the National Institutes of Health Center of Excellence in Genomic Science under Grant P50 HG002806. C.J.Y. was an American Chemical Society Division of Analytical Chemistry Fellow sponsored by Merck. N.J.T. was supported by National Science Foundation Grant CHE 04-15516.
Conflict of interest statement: No conflicts declared.
Abbreviations: DMF, dimethylformamide; PDGF, platelet-derived growth factor; E, excimer probe; P, pyrene monomer; Sn, n bp in the stem; THR, human α-thrombin; ESCRBL, 39-mer scramble oligonucleotide sequence.
Note. Just after the submission of this work, a pyrene excimer probe for potassium ion was reported (33), which further proves the wide applicability the excimer signaling approach. Such an inorganic ion probe, with the time-resolved fluorescence measurement method described here, should find useful applications for monitoring potassium ion in complex biological environments.
References
- 1.Hanash, S. (2003) Nature 422, 226–232. [DOI] [PubMed] [Google Scholar]
- 2.Tyers, M. & Mann, M. (2003) Nature 422, 193–197. [DOI] [PubMed] [Google Scholar]
- 3.Wulfkuhle, J. D., Liotta, L. A. & Petricoin, E. F. (2003) Nat. Rev. Cancer 3, 267–275. [DOI] [PubMed] [Google Scholar]
- 4.Ellington, A. D. & Szostak, J. W. (1990) Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 5.Robertson, D. L. & Joyce, G. F. (1990) Nature 344, 467–468. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk, C. & Gold, L. (1990) Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 7.Fang, X. H., Cao, Z. H., Beck, T. & Tan, W. H. (2001) Anal. Chem. 73, 5752–5757. [DOI] [PubMed] [Google Scholar]
- 8.Potyrailo, R. A., Conrad, R. C., Ellington, A. D. & Hieftje, G. M. (1998) Anal. Chem. 70, 3419–3425. [DOI] [PubMed] [Google Scholar]
- 9.Li, J. W. J., Fang, X. H. & Tan, W. H. (2002) Biochem. Biophys. Res. Commun. 292, 31–40. [DOI] [PubMed] [Google Scholar]
- 10.Fang, X. H., Sen, A., Vicens, M. & Tan, W. H. (2003) Chembiochem. 4, 829–834. [DOI] [PubMed] [Google Scholar]
- 11.Nutiu, R. & Li, Y. F. (2004) Chem. Eur. J. 10, 1868–1876. [DOI] [PubMed] [Google Scholar]
- 12.Gao, X. H., Cui, Y. Y., Levenson, R. M., Chung, L. W. K. & Nie, S. M. (2004) Nat. Biotechnol. 22, 969–976. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto, K., Shimizu, H. & Inouye, M. (2004) J. Org. Chem. 69, 3271–3275. [DOI] [PubMed] [Google Scholar]
- 14.Birks, J. B. (1970) Photophysics of Aromatic Molecules, Wiley Monographs in Chemical Physics (Wiley, New York).
- 15.Winnik, F. M. (1993) Chem. Rev. 93, 587–614. [Google Scholar]
- 16.Lakowicz, J. R. (1999) Principles of Fluorescent Spectroscopy (Kluwer Academic/Plenum, New York).
- 17.Dahim, M., Mizuno, N. K., Li, X. M., Momsen, W. E., Momsen, M. M. & Brockman, H. L. (2002) Biophys. J. 83, 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagano, R. E., Martin, O. C., Kang, H. C. & Haugland, R. P. (1991) J. Cell Biol. 113, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, L. S., Jellinek, D., Jenison, R., Ostman, A., Heldin, C. H. & Janjic, N. (1996) Biochemistry 35, 14413–14424. [DOI] [PubMed] [Google Scholar]
- 20.Stojanovic, M. N., de Prada, P. & Landry, D. W. (2001) J. Am. Chem. Soc. 123, 4928–4931. [DOI] [PubMed] [Google Scholar]
- 21.Paborsky, L. R., McCurdy, S. N., Griffin, L. C., Toole, J. J. & Leung, L. L. (1993) J. Biol. Chem. 268, 20808–20811. [PubMed] [Google Scholar]
- 22.Hamaguchi, N., Ellington, A. & Stanton, M. (2001) Anal. Biochem. 294, 126–131. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto, R., Baba, T. & Kumar, P. K. R. (2000) Genes Cells 5, 389–396, and erratum (2000) 5, 523. [DOI] [PubMed] [Google Scholar]
- 24.Masuko, M., Ohtani, H., Ebata, K. & Shimadzu, A. (1998) Nucleic Acids Res. 26, 5409–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyagi, S. & Kramer, F. R. (1996) Nat. Biotechnol. 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 26.Zuker, M. (2003) Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nutiu, R. & Li, Y. F. (2003) J. Am. Chem. Soc. 125, 4771–4778. [DOI] [PubMed] [Google Scholar]
- 28.Bayer, T. S. & Smolke, C. D. (2005) Nat. Biotechnol. 23, 337–343. [DOI] [PubMed] [Google Scholar]
- 29.Cox, J. C. & Ellington, A. D. (2001) Bioorg. Med. Chem. 9, 2525–2531. [DOI] [PubMed] [Google Scholar]
- 30.Nutiu, R. & Li, Y. F. (2005) Angew. Chem. Int. Ed. 44, 1061–1065. [DOI] [PubMed] [Google Scholar]
- 31.Jhaveri, S., Rajendran, M. & Ellington, A. D. (2000) Nat. Biotechnol. 18, 1293–1297. [DOI] [PubMed] [Google Scholar]
- 32.Yamana, K., Ohtani, Y., Nakano, H. & Saito, I. (2003) Bioorg. Med. Chem. Lett. 13, 3429–3431. [DOI] [PubMed] [Google Scholar]
- 33.Nagatoishi, S., Nojima, T., Juskowiak, B. & Takenaka, S. (2005) Angew. Chem. Int. Ed. 44, 5067–5070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.