Abstract
Background
Recent studies of ancestral maize populations indicate that linkage disequilibrium tends to dissipate rapidly, sometimes within 100 bp. We set out to examine the linkage disequilibrium and diversity in maize elite inbred lines, which have been subject to population bottlenecks and intense selection by breeders. Such population events are expected to increase the amount of linkage disequilibrium, but reduce diversity. The results of this study will inform the design of genetic association studies.
Results
We examined the frequency and distribution of DNA polymorphisms at 18 maize genes in 36 maize inbreds, chosen to represent most of the genetic diversity in U.S. elite maize breeding pool. The frequency of nucleotide changes is high, on average one polymorphism per 31 bp in non-coding regions and 1 polymorphism per 124 bp in coding regions. Insertions and deletions are frequent in non-coding regions (1 per 85 bp), but rare in coding regions. A small number (2–8) of distinct and highly diverse haplotypes can be distinguished at all loci examined. Within genes, SNP loci comprising the haplotypes are in linkage disequilibrium with each other.
Conclusions
No decline of linkage disequilibrium within a few hundred base pairs was found in the elite maize germplasm. This finding, as well as the small number of haplotypes, relative to neutral expectation, is consistent with the effects of breeding-induced bottlenecks and selection on the elite germplasm pool. The genetic distance between haplotypes is large, indicative of an ancient gene pool and of possible interspecific hybridization events in maize ancestry.
Background
Direct analysis of genetic variation at the DNA sequence level at many loci became possible in recent years due to improvements in sequencing technology. High throughput genotyping methods, including DNA chips, allele-specific PCR and primer extension approaches make single nucleotide polymorphisms (SNPs) especially attractive as genetic markers [1-3].
If a whole-genome scan is to be undertaken, trait mapping by allele association requires high marker density [4-7] which could be provided by SNPs. Recent detailed analysis of allelic diversity at the maize Dwarf8 gene, which indicated association with flowering time [8], is an example of association approach using candidate genes. SNPs may also be used for mapping expressed sequence tags (ESTs) in defined segregating populations and for the integration of genetic and physical (contig) maps, which contain EST-derived landmarks.
While polymorphic simple sequence repeats (SSRs, [9]) are excellent molecular markers, because of their multiallelism and the resulting high informativeness, they may not be frequent enough for association studies. Size homoplasy of SSR alleles, as well as allele reversion could also be a problem in some applications [10,11].
In contrast to humans [12], few systematic whole genome searches for single nucleotide polymorphisms have been undertaken in plant species, with the exception of Arabidopsis http://www.arabidopsis.org/cereon/. However, it has been established that plants differ widely in the level of intra-specific sequence diversity. For a recent review, see [13] Maize is generally considered highly polymorphic, and it has been suggested that active transposon systems contributed to the creation of diversity [14]. For example, in one of the early studies of the sh1 locus, [15] detected 16 nucleotide changes in 540 bp, while in the 3'-untranslated region 10 changes occurred in 270 bp. Several other maize loci including Adh1 [16,17], Adh2 [18], Opaque-2 [19], b [20], glb1 [21] have been studied systematically. The range of nucleotide diversity π reported for maize genes is wide, from 0.47 (per 1000 bp) for the promoter region of tb1 [22] to 37, for synonymous substitutions at glb1 [21], a difference of almost two orders of magnitude.
Reduced allelic diversity is expected in domestication related genes. This was found in c1, an anthocyanin-biosynthesis regulatory locus [23]. Wang [22] and White [24] recently examined domestication – related changes in nucleotide diversity along the length of two maize genes. In the case of teosinte branched, tb1, a significant reduction of diversity occurred in the promoter region, but not in the coding region [22]. The sequences of terminal ear 1 (te1) alleles showed evidence of linkage disequilibrium, and only a small number of haplotypes was identified in cultivated maize, in contrast to maize progenitors.
A recent study involved 21 loci along chromosome 1 of maize, and indicated high level of diversity in landraces, only somewhat reduced in U.S. inbreds [25]. As has been previously found in Drosophila [26], diversity was correlated with recombination rate. Linkage disequilibrium was found to decline within 100–200 bp [25].
Further studies of the nature, frequency and distribution of sequence variation in the agronomically relevant maize germplasm, would allow better understanding of the range diversity and the nature of genetic changes associated with domestication and selection for agronomic performance. To this end, we surveyed sequence diversity at 18 loci. Gene segments were amplified from 36 maize elite inbred lines and sequenced. The frequency and the nature of polymorphisms were examined in detail. Structure of SNP haplotypes and short-range linkage disequilibrium within loci were also analysed.
Results and Discussion
Experimental approach
To identify and characterise patterns of DNA sequence polymorphisms in or near maize genes, we sequenced 22 maize amplicons from up to 36 diverse maize genotypes, representing the major heterotic groups of cultivated maize germplasm mainly of U.S. origin (Table 1). This germplasm set provides an excellent representation of the allelic diversity in agronomically relevant maize, as evidenced by the fact that RFLP alleles present in a modest subset of these lines (#3, 5, 16, 26–28, 30, 31, 36, Table 1) represent allelic diversity of 94.7 % of the 345 maize lines tested (data not shown). To maximise the amount of observed sequence diversity, and thus to increase the number of informative SNPs discovered, we analysed primarily the 3'-untranslated regions of the selected maize genes. PCR primers were designed to amplify a 300–500 bp segment of each gene. In some cases parts of the last intron and exon were also included. The amplicons were derived from 17 different ESTs, eight of which have exact maize GenBank homologs, and from one well-characterised gene sequence (see Additional file 1).
Table 1.
List of maize germplasm
| Number | Name | Classification |
| 1 | 647 | Central Cornbelt male/Non-Stiff Stalk |
| 2 | 684 | INRA Flint |
| 3 | B_1 | Reid / Iowa Stiff Stalk Synthetic |
| 4 | B_6 | Central Cornbelt male/Non-Stiff Stalk |
| 5 | B73 | Reid / Iowa Stiff Stalk Synthetic |
| 6 | B84 | Stiff Stalk Synthetic |
| 7 | C_5 | Reid / Iowa Stiff Stalk Synthetic |
| 8 | C_9 | Early Cornbelt male, Iodent-NSS mix |
| 9 | C123 | Lancaster Non Stiff Stalk |
| 10 | CO109 | Derived from a Canadian open pollinated population |
| 11 | D_6 | Central Cornbelt male/Non-Stiff Stalk |
| 12 | D_7 | Early Cornbelt male, Iodent-NSS mix |
| 13 | D_9 | Early Cornbelt male, Iodent-related |
| 14 | D71-4HT | Unknown pedigree |
| 15 | DE811 | Lancaster × Stiff Stalk Synthetic |
| 16 | E_2 | Central Cornbelt male/Non-Stiff Stalk mixed Parentage |
| 17 | F_1 | Lancaster-derived |
| 18 | F_2 | Reid / Iowa Stiff Stalk Synthetic |
| 19 | F_3 | Reid / Iowa Stiff Stalk Synthetic |
| 20 | F_4 | Lancaster-derived |
| 21 | F_5 | Reid / Iowa Stiff Stalk Synthetic |
| 22 | F_6 | Reid / Iowa Stiff Stalk Synthetic |
| 23 | F_7 | Reid / Iowa Stiff Stalk Synthetic |
| 24 | F_9 | Reid / Iowa Stiff Stalk Synthetic Early |
| 25 | G_1 | Early Lancaster line |
| 26 | H_1 | Southern US / Pioneer Propriety Synthetic Population |
| 27 | H_3 | Non Stiff Stalk |
| 28 | H_4 | Central Cornbelt male/Non-Stiff Stalk |
| 29 | H_5 | Central Cornbelt male/Non-Stiff Stalk |
| 30 | H60 | Lancaster Non Stiff Stalk |
| 31 | H98 | Lancaster, OH43-related |
| 32 | H99 | Derived from IllSyn60C population |
| 33 | I_1 | European Flint |
| 34 | IVANA | Unknown pedigree |
| 35 | J40 | Reid / Iowa Stiff Stalk Synthetic |
| 36 | MO17 | Lancaster-derived |
| 37 | TX601 | Tuxpeno |
| 38 | WF9HT | Reid |
Types and frequency of polymorphisms
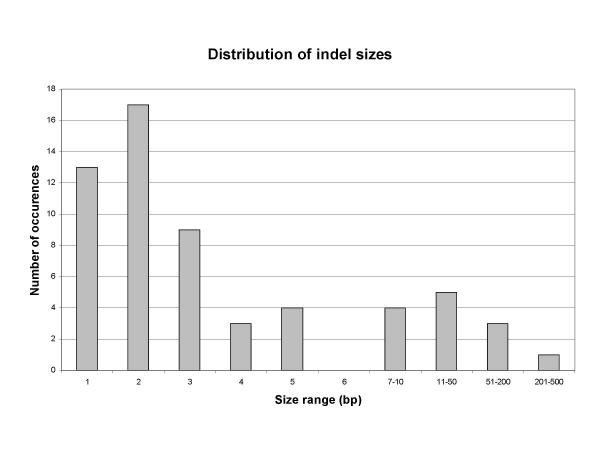
Multiple nucleotide changes and insertions / deletions of various lengths were identified, and the results are summarised in Table 2. The distribution of various types of polymorphisms at individual loci is shown in Additional file 2. Single nucleotide changes occur on average every 60.8 bp, and indels occur every 126 bp. The frequency of nucleotide substitutions is almost three times higher in non-coding regions than in coding sequences. Most of the nucleotide changes in the protein-coding regions are silent – only 5 out of 18 changes detected result in amino acid substitution. The difference in the distribution of indels is even more striking, only one 3 bp indel was found in 2.35 kb of coding sequences, while indels occur on average every 85 bp in non-coding regions (54 indels varying in size from 1 bp to over 400 bp were identified). The number of observed insertion / deletion events per locus varies widely, from 0 to 11 (median 1.5 indels per locus). Figure 1 shows size distribution of indels. Among the 55 indels reported here, dinucleotide indels are most frequent. Previous indel analysis in a larger data set (655 indels in 215 loci) have shown that single base insertions / deletions are most common [27]. The difference may be due to the fact that a few simple sequence repeat (SSR)-like variants, generated by a different mutational mechanism [28] contribute several 2-nt. indels found in the present data. Some nested indels are observed.
Table 2.
Summary of polymorphism analysis
| Parameter | Value | Comments |
| Number of loci screened | 18 | |
| Total length of amplicons, bp. | 6935 | 2349 coding, 4586 non-coding |
| Number of bases of sequence screened, bp. | 213999 | |
| Number of all sequence variants (SNPs and Indels) | 169 | 1 per 41 bp |
| Number of nucleotide substitutions | 114 | |
| Transitions / Transversions ratio | 1.53 | |
| Frequency of polymorphic sites per bp | 0.0164 | 1 per 60.8 bp |
| Frequency of polymorphic sites per bp (coding) | 0.0077 | 1 per 130.5 bp |
| Frequency of polymorphic sites per bp (non-coding) | 0.021 | 1 per 47.7 bp |
| Number of Indels | 55 | |
| Overall indel frequency | 0.0079 | 1 per 126.1 bp |
| Frequency of indels per bp (coding) | 0.0004 | 1 per 2349 bp |
| Frequency of indels per bp (non-coding) | 0.0118 | 1 per 85 bp |
| Mean nucleotide diversity (π) | 0.0063 | |
| Mean nucleotide expected heterozygosity | 0.26 | |
| Mean haplotype expected heterozygosity | 0.56 |
Figure 1.
Distribution of insertion /deletion sizes Number of observed insertion / deletion polymorphisms (indels) of each size class is shown.
SNPs as genetic markers
SNPs were evaluated individually and on the basis of haplotypes (see Additional file 2). The SNP expected heterozygosity is 0.263 (see Additional file 2). Exclusion of indels from the calculation does not produce significant change in the heterozygosity values. In comparison, the SSR expected heterozygosity has been estimated at H = 0.77 [29]. Therefore, individual SNPs are not very informative as molecular markers for genetic diagnostics. If the expected heterozygosity is calculated on the basis of haplotypes, rather than individual SNPs, the value is over twice as high, 0.561. The haplotype expected heterozygosity is comparable to the heterozygosity of RFLP markers (H = 0.58, [29]). Haplotype analysis, while increasing informativeness, it would increase the cost of genotyping, relative to the analysis of single SNPs. This increase would be proportional to the number of SNPs needed to define each haplotype. Usually 2–4 SNPs will be required to tag the haplotypes [30].
The high frequency of polymorphism in maize translates into a large number of SNPs and indels potentially available for use as genetic markers. These markers may be discovered by direct sequencing of gene-adjacent sequences, as described here, or by computer analysis of available EST sequences derived from multiple genotypes [31]. It is in principle feasible to obtain several SNP markers in the vicinity of each maize gene, a subset of which will completely define haplotypes. Such a collection of SNPs may enable whole genome scanning linkage disequilibrium-based approaches [6] to trait dissection and gene mapping in maize, if the amount of linkage disequilibrium in the relevant populations is sufficient.
Gene diversity and divergence dates
The overall level of sequence polymorphism in maize is high, more than double the inter-specific polymorphism rate in mouse (M. castaneous / M.domesticus, [1]), and about an order of magnitude higher than in humans [32]. In maize, expected heterozygosity per nucleotide site (π) values ranging from low 0.47 in the promoter region of a domestication gene, tb1 [22], to 37, for synonymous sites in Globulin-1 (glb1) locus were found [21,24]. For comparison, π in humans is from 0.3 to 1.1 [32]. In our maize study, π averages 6.3 (per bp, ×1000, non-coding regions only), on the low side of the previously reported range for silent sites. This may be explained by the difference in germplasm selections. Most of the earlier studies included a diverse set of maize accessions from North and Central America, while we concentrated on U.S. elites. Gaut [33,34] estimated the synonymous rate of substitution at 4.7–7.0 × 10-9 substitutions per synonymous site per year. The mean between-haplotype distance we observed is 11.5 nucleotide substitutions per 1000 nt of non-coding sequence, excluding indels, corresponding to 0.8–1.2 my. This number is derived primarily from silent sites, at which the substitution rate may be lower than in synonymous sites [24]. Previous estimates for the age of maize gene pool, derived from the most divergent haplotypes of te1, are quite similar, 1.2–1.4 my [24]. As expected, estimates for individual loci deviate considerably from the mean. For example, the two most distant haplotypes of stearoyl-ACP-desaturase differ by 7 nucleotide substitutions over 228 nt of the 3'-untranslated region of the gene, translating to 2.2–3.2 my divergence, which is close to the estimated divergence time between Tripsacum and maize, 2.3–2.6 my [24]. Two divergent Adh1 haplotypes (15 substitutions per 1025 bp) produce numbers close to the te1 estimates, 1–1.5 my, and slightly lower than the previous estimates for Adh of 1.9 my [35]. The individual gene-derived numbers have to be treated with caution, because they are obtained from short sequence segments and thus are burdened with significant error. Despite lower heterozygosity per nucleotide site (π) in elite maize, highly diverse haplotypes have been maintained in elite lines. Selection for heterosis, which is related to genetic diversity between parents [36-38] may have contributed to this effect.
S-adenosylmethionine synthase was the only gene completely monomorphic within the 254 bp (86% 3'-UTR) examined. A reduced diversity was also observed at the Glutamyl-tRNA reductase precursor locus, where one common (p = 0.935) and one rare allele were found, and nucleotide diversity π is 1.9 (per bp, ×1000). However it would be premature to speculate about any functional significance of the apparently reduced diversity at these loci, without first examining larger segments of the genes for polymorphism.
Insertions/deletions occurring on the background of a common haplotype, and therefore presumably of more recent origin, can occasionally be found. The mean difference between haplotypes is strongly affected by the exclusion of indels: 15 differences/ 1000 bp vs. 11.5 nt/1000 bp if indels are disregarded, underscoring the significant contribution of indels to maize genetic diversity.
Haplotype structure and allele distribution
To evaluate the allele distribution in the set of germplasm selected for this study, we applied Tajima D statistics [39,40], which was developed to test neutrality of mutations. Tajima D is based on the comparison of two estimators of Θ = 4Neμ (where Ne is the effective population size and μ is the mutation rate), one based on the number of segregating sites and one based on the number of pairwise differences between sequences in the sample [41].
Departures from neutrality expectation can be dues to a number of factors, including population expansion, bottleneck or heterogeneity of mutation rates [42], therefore neutrality is not an expectation in the set of germplasm analysed here. While the Tajima test in the strict sense does not apply to non-random collections of germplasm such as the maize lines selected for this study, it is still a convenient indicator of the pattern of allele distribution. Negative Tajima D values indicate an excess of low frequency alleles relative to neutral mutation – drift equilibrium. Positive Tajima D indicates a deficit of low frequency alleles relative to expectation. This could be due to a population bottleneck, population subdivision or balancing selection. These factors are likely to be operational in maize elite lines.
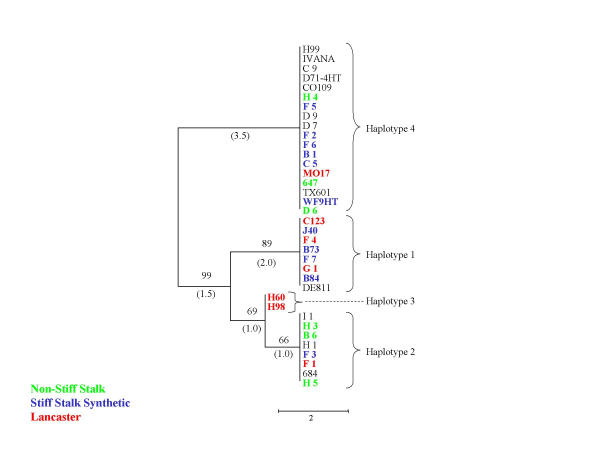
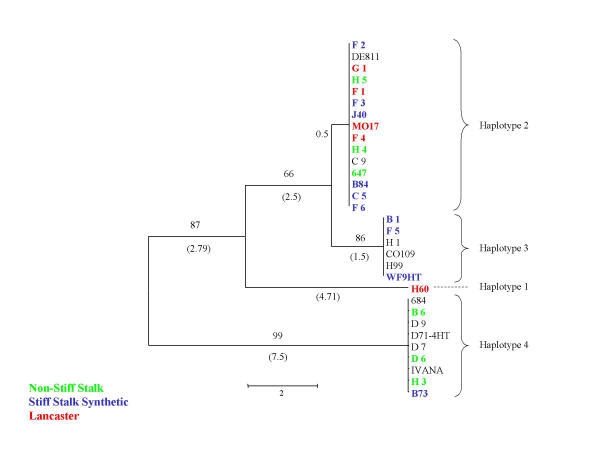
There is no indication of the overall strong bias of Tajima D among the loci examined here (see Additional file 2). Tajima D values range from -1.5 to 2.6, 0.31 on average (0.1 without indels). A strongly positive Tajima D value at the stearoyl-ACP desaturase locus (D = 2.58) indicates that the number of alleles at intermediate frequency is higher than expected, possibly as a result of population subdivision [39,40]. Another locus behaving in a similar fashion is the glycine-rich RNA binding protein. To test whether haplotypes are unequally distributed among Stiff Stalk, Non-Stiff Stalk, and other types of germplasm, we calculated the Tajima D value separately for these subsets of germplasm (data not shown). In the case the two previously mentioned genes which show high positive Tajima D values, the variation was mainly within populations, and Tajima D remained positive for each type of germplasm. No obvious bias in the distribution of haplotypes between heterotic groups was observed (Fig. 2,3,4). It is likely that such patterns would only be revealed upon sampling of a larger set of genetic loci [43,44]. In general, higher genetic similarity is observed within heterotic groups than between heterotic groups, irrespective of the genetic marker system used [44].
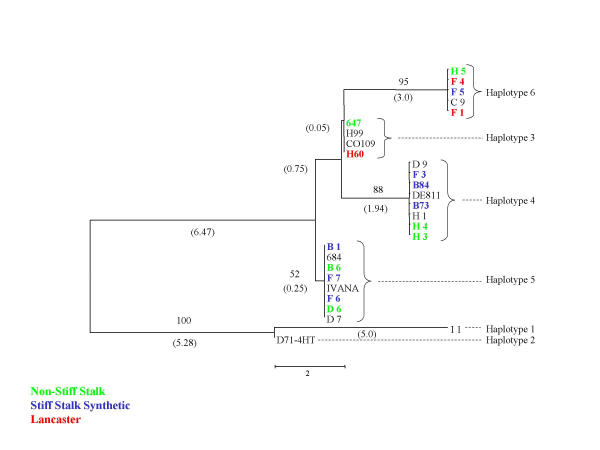
Figure 2.
Neighbor-joining trees representing Adh1 haplotype relationships. Level of support for branch points is indicated in %, and branch length expressed as nucleotide differences are shown in parentheses. Genotypes correspond to those of Table 1, and color indicates major heterotic groups: stiff stalk (blue), non stiff stalk (green) and Lancaster (red).
Figure 3.
Neighbor-joining trees representing stearoyl-ACP desaturase haplotype relationships. Level of support for branch points is indicated in %, and branch length expressed as nucleotide differences are shown in parentheses. Genotypes correspond to those of Table 1, and color indicates major heterotic groups: stiff stalk (blue), non stiff stalk (green) and Lancaster (red).
Figure 4.
Neighbor-joining trees representing acetyl-CoA C-acyltransferase haplotype relationships. Level of support for branch points is indicated in %, and branch length expressed as nucleotide differences are shown in parentheses. Genotypes correspond to those of Table 1, and color indicates major heterotic groups: stiff stalk (blue), non stiff stalk (green) and Lancaster (red).
At each of the loci, the sequence diversity is organised into a relatively small number (two to eight) distinct haplotypes, many of which were represented multiple times among the 36 inbred maize lines analysed. Figure 2,3,4 and Table 3 show examples of the haplotype relationships. The three most common haplotypes account for over 80% of allelic diversity at 16 out of the 18 loci examined. For example, at the stearoyl-ACP desaturase locus (Fig 3) there are three common haplotypes relatively distant from each other, and a rare one which differs by only one nucleotide change from one of the common haplotypes. Eighteen inbreds, from three heterotic groups share haplotype 4, while two Lancaster-type inbreds, H60 and H98 have rare haplotype 3.
Table 3.
Haplotypes at the alcohol dehydrogenase (Adh1), stearoyl-ACP-desaturase and acetyl-CoA C-acyltransferase loci Adh1 haplotypes are based on concatenation of all three segments of Adh1 sequenced
| Adh1 | |||||||||||||||||||||||||||||||||||||||||||
| SNP Position in AF123535 and genotype | |||||||||||||||||||||||||||||||||||||||||||
| Haplotype | Frequency | 16920 | 16933 | 16935 | 16951 | 16978 | 17873 | 17880 | 17904 | 18071 –072 |
21129 | 29729 –730 |
21178 | 21179 | 21180 | 21181 –185 |
21199 –532 |
21359 | 21513 –514 |
21576 | 21649 –50 |
21650 –736 |
21737 | ||||||||||||||||||||
| 1 | 0.031 | G | G | A | A | C | C | A | C | - | G | I | C | G | C | - | - | T | T | C | I | - | G | ||||||||||||||||||||
| 2 | 0.031 | A | T | G | G | T | C | A | C | - | G | I | C | G | C | - | - | T | T | C | I | - | G | ||||||||||||||||||||
| 3 | 0.125 | A | T | G | G | T | C | G | T | - | T | - | T | T | G | I | I | C | - | T | - | - | - | ||||||||||||||||||||
| 4 | 0.250 | A | T | G | G | T | C | G | T | - | T | - | T | T | G | I | I | C | - | T | - | I | C | ||||||||||||||||||||
| 5 | 0.250 | A | T | G | G | T | C | G | T | - | T | - | T | T | G | I | I | T | - | T | - | - | - | ||||||||||||||||||||
| 6 | 0.312 | A | T | G | G | T | G | G | T | I | T | - | T | T | G | I | I | C | I | T | - | - | - | ||||||||||||||||||||
| Stearoyl-ACP-desaturase | |||||||||||||||||||||||||||||||||||||||||||
| SNP Position in GenBank accesion AF498430 and genotype | |||||||||||||||||||||||||||||||||||||||||||
| Haplotype | Frequency | 71 | 82 | 86 | 87 | 153 | 156 | 171 | 190–194 | 200 | |||||||||||||||||||||||||||||||||
| 1 | 0.222 | A | A | T | A | C | G | A | - | T | |||||||||||||||||||||||||||||||||
| 2 | 0.222 | G | A | T | A | C | A | A | I | C | |||||||||||||||||||||||||||||||||
| 3 | 0.055 | G | A | T | A | C | A | A | I | T | |||||||||||||||||||||||||||||||||
| 4 | 0.500 | G | C | A | T | T | A | C | - | T | |||||||||||||||||||||||||||||||||
| Acetyl-CoA C-acyltransferase | |||||||||||||||||||||||||||||||||||||||||||
| SNP Position in GenBank accesion AF498472 and genotype | |||||||||||||||||||||||||||||||||||||||||||
| Haplotype | Frequency | 38 | 73 | 100 | 111 | 143 | 161 | 239 | 245 | 247 | 250 | 251 | 257 | 266 | 270 | 279 | 290 | 295 | 330–391 | 449 | |||||||||||||||||||||||
| 1 | 0.030 | C | C | A | G | C | G | A | C | G | C | G | C | T | A | T | T | A | - | T | |||||||||||||||||||||||
| 2 | 0.470 | C | T | T | G | C | C | A | C | C | T | G | C | A | A | T | C | A | - | C | |||||||||||||||||||||||
| 3 | 0.230 | C | T | T | G | T | C | A | C | G | T | G | C | A | A | T | C | A | - | C | |||||||||||||||||||||||
| 4 | 0.260 | T | T | A | T | C | G | G | T | C | C | A | T | A | C | C | C | T | I | C | |||||||||||||||||||||||
Allelic sites at polymorphic sites are shown, together with their nucleotide positions. The insertion – deletion is represented as I. Missing data are denoted "?". Promoter region is separated from the rest of the gene by "^" (Adh1 only).
The expected number of haplotypes may be calculated using coalescent theory [45,46], even though such calculations involve many assumptions. Mean number of predicted haplotypes for all loci examined was calculated to be 6.01 (st. dev 2.4), while 3.4 (st. dev. 1.1) was observed. These means are significantly different at P < 0.001 level (two-tailed t-test). Two loci, when examined individually, showed a statistically significant difference between the calculated and the lower observed number of haplotypes, at 0.05 confidence level.
Haplotype structure of a few Z. mays genes has been recognised previously [18,24], but the predominance in maize elite lines of a few diverged haplotypes in linkage disequilibrium, has not been obvious until now. In teosinte, no clear haplotype structure has been identified [24].
Selinger and Chandler [20] found three distinct clades in the phylogenetic tree of maize b gene alleles, with strong separation between clades, indicating that the alleles within clades may have arisen recently when compared with the divergence of the three clades. Both Z. mays and Z. mays parviglumis sequences appear in the three clades. One possible interpretation of this finding is that the three clades may have diverged before the divergence of the genus Zea. An alternative hypothesis, that the nucleotide substitution rates at the upstream region of b are much higher, is favored by Selinger and Chandler. Our study indicates that at least one aspect of the evolutionary pattern seen by these authors, the presence of highly divergent haplotypes, is widespread in elite maize inbreds, favoring the hypothesis of early separation of the three clades.
Phylogenetic analysis of the maize terminal ear (te1) sequences did not resolve all Z. mays sequences into a single clade. Members of the Zea subspecies, with the exception of Z. huehuetenangensis are intermixed within clades [24]. This observation has been made for other maize genes [18,21,23] and has been interpreted as indicative of introgression among Zea taxa [24], or of lineage sorting [24]. The lack of resolution of species within the genus Zea into single clades was also found for c1 and Adh2 [14]. In contrast, glb1 and Adh1 appear to have a different evolutionary history, with Zea luxurians alleles forming a distinct clade [14,17,21].
These observations, together with our data which showed a widespread distribution of highly diverged haplotypes, seem to indicate that interspecific gene flow in the genus Zea amy have been significant. It is tempting to speculate that incongruent evolutionary histories of different loci are related to the origins of alleles either within a single Zea species, or within two or more species, followed by an inter-specific introgression event(s) [47]. Recent surprising finding that some alleles at the bz locus differ in their gene complement and in the composition of intergenic repetitive DNA segments appears to lend further credence to this hypothesis [48].
The observed haplotypes predate domestication of corn, and their distribution at different genetic loci may help understand the process of domestication, including the resulting population subdivision and selective pressures. It is tempting to speculate that selection for high yield, and consequently heterosis in open pollinated varieties and, more recently, between heterotic groups, favoured presence of highly divergent haplotypes at many loci, while in the same time bottleneck effects limiting the number of haplotypes. As a result of these competing processes, despite strong selection a relatively high fraction of diversity (77%, [25]) is retained in elite germplasm as few highly divergent haplotypes.
Linkage disequilibrium
The presence of a small number of haplotypes shared by multiple individuals is indicative of linkage disequilibrium (LD). Population bottlenecks and inbreeding increase LD [49]. Thus, elite germplasm may be expected to have extensive linkage disequilibrium.
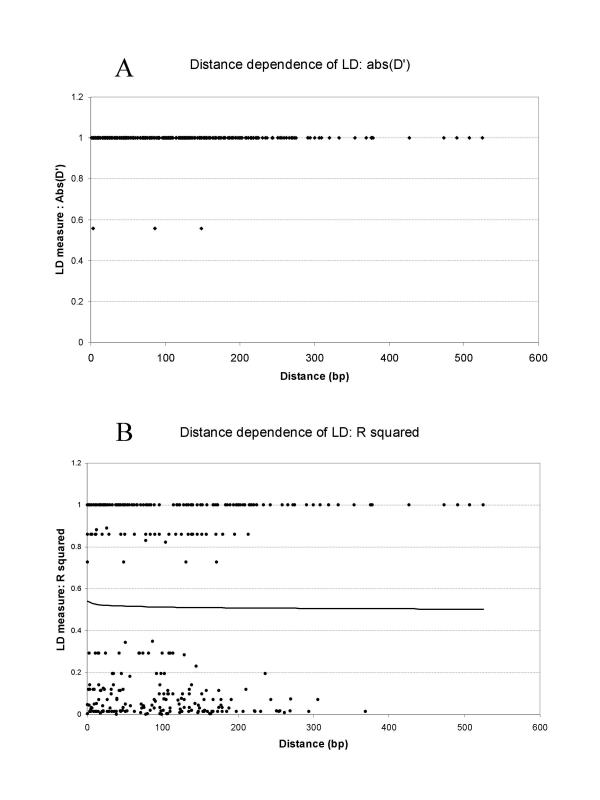
Linkage disequilibrium measures D' and r2 were calculated for SNP loci within each gene (Figure 5). No decline in the value of D' was found within the range of 300–500 bp analysed. Also, the r2 measure of linkage disequilibrium does not appear to be declining significantly. D' is an accepted measure for the analysis of distance dependence of linkage disequilibrium [30,50], but r2 has also been used frequently. As a control, LD was also calculated for all pairs of SNP loci between the 18 genes examined. The genes are not known to be linked genetically, and therefore no significant LD was expected between genes. In agreement with the expectation, only 0.3% of between gene pairs of SNP loci showed significant LD at P < 0.01. In contrast, 36.3 % of within gene pairs of SNP loci showed significant LD at P < 0.01. We conclude that the linkage disequilibrium observed within genes is not an artefact.
Figure 5.
Composite plot of linkage disequilibrium as a function of distance. Two measures of linkage disequilibrium, absolute value of D' (A) and r2 (B) are shown as a function of distance for all loci examined. LD values between all pairs of SNP were plotted. Logarithmic trend line is included in plot (B). Of the 344 pairwise comparisons, 161 were significant at P < 0.01, with Bonferroni correction, and 126 were significant at P < 0.001 level.
It remains to be determined at what distances, on average, LD declines in this population. In contrast to our result, in recent studies, LD was found to decline rapidly in maize [25,51]. However, both authors examined broad-based sets of germplasm – breeding germplasm and diverse landraces, respectively. Significant differences exist between the two studies. [51], unlike [25], found large differences in the rate of LD decay with between loci. Also, overall rate of decay in LD is less in the former study [51], based on a somewhat narrower population of individuals. In conclusion, appropriate choice of germplasm may allow one to adjust resolution of association studies, and, consequently, the number of genetic markers required. Elite germplasm may be preferred for initial lower resolution analysis, followed by higher resolution study in a broader germplasm collection.
Evidence for haplotype recombination
At the Adh1 locus there are only two haplotypes, one common and one rare (D = 0.06) within the promoter region (nt. 2–345, X04050, Table 3), This reduction of diversity may indicate the possibility of selection. The remaining segments of the gene analysed here, including a portion of the 5'-untranslated leader, first exon and first intron (nt. 1030–1386, X04050) and 3'-untranslated sequences (nt. 4196–4552, X04050) show the presence of five haplotypes. Due to the distinctness of the rare haplotype 1, carried by a European Flint inbred I_1 (Figure 2, Table 3), it is possible to identify haplotype 2, represented by inbred D71-4HT, as a likely product of recombination between haplotype 1 and one of the other haplotypes. This recombination occurred in the DNA segment bordered by nucleotides 345 and 1030 (X04050, exon 1 of Adh1 starts at nt. 1195), contributing to the reduction of diversity in the promoter region (Table 3, haplotype 2). Further analysis of the sequences between nt. 345 and 1030 may help localise the site of recombination. This picture bears some resemblance to the observations of Wang on teosinte branched 1 locus, in that one finds a reduction in diversity in the promoter region and also a recombination event close to the beginning of transcription, even though Adh1 is usually considered a neutral gene [22]. We are currently analysing haplotype structure and linkage disequilibrium in a large region surrounding the Adh1 gene (M. Jung, private communication).
Conclusions
In contrast to previous results obtained in ancestral maize populations, the analysis of maize elite inbred lines demonstrated the presence of a small number of highly diverse haplotypes and strong linkage disequilibrium between SNP loci extending at least to 500 bp. This population structure may result from bottlenecks and selection associated with plant breeding, and has implications for the design of genetic association studies in maize.
Methods
Plant material
Inbred maize lines primarily representative of U.S. public and proprietary corn germplasm were obtained from Pioneer Hi-Bred International (Johnston, IA), Table 1. Twelve lines used in a previous study [29] were obtained from G. Taramino and include lines 5, 6, 9, 10, 14, 15, 30–32, 34, 36, 37. Line WF9HT was from M. Williams (DuPont Co). Leaves from two-week old greenhouse-grown plants was harvested for DNA extraction.
DNA extraction
Leaf material (fresh, frozen at -80, or lyophilised) was ground with glass beads (150 microns, Sigma G9018) into a fine powder using mortar and pestle, in the presence of liquid nitrogen. The DNA was then extracted using Plant DNAzol (Life Technologies, Inc.) following the manufacturer's recommendation with one modification: after the initial room temperature incubation the tissue homogenate was centrifuged at 10,000 g for 10 min, and the supernatant was collected and used for the chloroform extraction step.
Gene sequences and primer design
Twenty-two DNA segments derived from 18 different genes were PCR amplified from a set of maize inbred lines. Gene specific primer pairs for the polymerase chain reaction (PCR) were designed using the PRIMER3 program (http://www.genome.wi.mit.edu, S. Rozen, H. J. Skaletsky, 1998) Primer3. Primer3 code is available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html. The sequences of the genes were derived from the 3'-ends of 17 maize ESTs, and from three regions of the maize Adh1 gene (see Additional file 1). Including Adh1, nine of the sequences correspond to known maize genes, nine are new maize EST sequences, with good protein-level homology to known plant genes. All sequences have been deposited in GenBank (see Additional file 1).
The expected product sizes were 300–500 bp on average, usually corresponding to the 3' untranslated region of the gene. In the case of Adh1 three independent amplicons were analysed. A T3 tag (5'-AATTAACCCTCACTAAAGGG-3') was added to the 5' end of the forward primer, and a T7 tag (5'-GTAATACGACTCACTATAGGGC-3') was similarly added to the reverse primer, to facilitate direct PCR product sequencing.
PCR amplification
DNA Amplifications were performed in a 50 μL volume. The reactions contained 100 ng of genomic DNA, 10 pmole (0.2 μM) of each primer, 200 μM of each dNTP, 2 mM MgCl2, 5% DMSO, 1.25 units AmpliTaq Gold (PE/Applied Biosystems, Foster City, CA) and 1 × PE Buffer II (PE/Applied Biosystems, Foster City, CA).
The reactions were incubated in a Perkin Elmer 9700 thermocycler with the following cycling conditions: 95°C for 10 min., 10 cycles of 1 min. at 94°C, 1 min. at 55°C, 1 min. at 72°C, 35 cycles of 30 sec. at 95°C, 1 min. at 68°C, followed by a final extension of 7 min. at 72°C.
PCR products were analysed on agarose gel, purified using a Qiaquick PCR purification kit (Qiagen, Inc. Valencia, CA), and quantitated prior to DNA sequencing.
DNA sequencing
PCR products were sequenced directly using T3 and T7 primers. Sequencing reactions were performed using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq FS DNA polymerase (PE Applied Biosystems, Foster City, CA) and analysed on ABI 377 (PE Applied Biosystems, Foster City, CA) sequencers. Any sequence ambiguities were resolved by repeated sequencing of the PCR products from both ends. The sequences derived from all inbred lines were aligned in Sequencher (Gene Codes Corp., Ann Arbor, MI). The base changes at all polymorphic positions were identified by inspection for each of the inbred lines and catalogued in an Excel (Microsoft Corp.) spreadsheet.
DNA sequence accession numbers
GenBank accession numbers (18 loci, all genotypes examined) are included in Additional file 2. The aligned and concatenated DNA sequences used in the analysis are available as additional data files in the text format (Additional file 3), Nexus format (Additional file 4) and MEGA format (Additional file 5). The list of the sequences included and the coordinates of individual loci within the above listed file is available in Additional file 6.
Data analysis
Conserved haplotypes, that is DNA sequences containing identical allelic variants at all identified polymorphic sites at a locus, but derived from separate individuals, were identified visually or by alphabetical sorting of the list of sequence variants at a locus (see Table 3 for an example). Number of transitions (S), number of transversions (V) and number of insertion / deletion polymorphisms (indels) were counted directly or calculated by using Arlequin 1.1 [41]. Linkage disequilibrium measures D' and R2 were calculated with DNAsp [45] and with Tassel (Buckler IV, E.S., http://brooks.statgen.ncsu.edu/buckler). Insertions / deletions and sites with excess missing data were excluded from the LD calculations. Estimation of expected number of haplotypes, given the estimated value of Theta and recombination using coalescent process simulations were also performed with DNAsp [45].
Frequencies of polymorphic sites per bp (Table 2) were calculated by dividing the total number of polymorphic sites of a given type (SNPs, indels, or both) by the length of the DNA sequence examined. Genetic parameters, including nucleotide expected heterozygosity, number of haplotypes, haplotype expected heterozygosity, mean number of differences between pairs of haplotypes, and Tajima D were calculated using Arlequin 1.1 [41].
Nucleotide expected heterozygosity and haplotype expected heterozygosity calculated from allele and haplotype frequencies, respectively:
![]()
where n is the number of gene copies in the sample, pi is the frequency of the i-th allele or i-th haplotype [52]. The reported values of nucleotide expected heterozygosity are averages over all polymorphic nucleotide sites within the locus. Expected heterozygosity per nucleotide site π was calculated from nucleotide expected heterozygosity values:
![]()
Where Hi is nucleotide expected heterozygosity at a polymorphic site i, and L is the length of the sequence segment analysed, which contains n polymorphic sites. Insetion / deletion rates are likely to be different from single nucleotide mutation rates, and may not be caused by single molecular events, causing complications in the estimation of divergence times. Therefore, calculations involving genetic parameters were determined for single nucleotide polymorphisms only, and, in some cases, separately, for all polymorphic sites including insertions / deletions (indels). For the purpose of this calculation, each indel was treated as a single event.
Neighbor-joining trees were based on the haplotype sequences, using nucleotide number of differences as a distance measure and were calculated with Mega 2.0 (S. Kumar, K. Tamura, I.B. Jakobsen and M. Nei, http://www.bio.psu.edu/People/Faculty/Nei/Lab/Programs.html. For the purposes of tree calculation indels were treated as equivalent to single nucleotide differences. The support level for branching points in the trees was determined by 1000 bootstrap re-samplings of the data.
Authors' contributions
Author 1 initials AC carried out most of the experimental studies and data analysis. Author 2 initials KSC and author 3 initials MJ carried out some initial molecular studies and contributed to DNA sequencing. Author 4 initials MD was responsible for DNA sequencing Author 4 initials OSS contributed to selection of germplasm and discussion of the effects of germplasm subdivision. Author 5 initials ST contributed to the discussion of linkage disequilibrium and in the coordination of the study. Author 6 initials MM contributed to the study design and to data analysis. Author 7 initials JAR conceived the study, participated in its design and in data analysis.
Supplementary Material
List of genes and PCR primers
Frequency of polymorphisms and population parameters in coding and non-coding regions
Aligned sequence data in an interleaved text format, similar to NEXUS. Sequences from all loci examined are concatenated, the identity and location of sequences corresponding to each locus is in the file header.
Same data as file 1 but no locus identity information. DNAsp – compatible NEXUS format format. May be opened directly with DNAsp software (see Materials and Methods).
Same data as file 2 in MEGA format. May be opened directly with MEGA or DNAsp software (see Materials and Methods).
Excel file containing information about positions of individual loci in the concatenated sequence files Ching_data1, Ching_data2 and Ching_data3
Acknowledgments
Acknowledgements
We thank Jim Register, Pioneer Hi-Bred International, for helping us obtain the plant material and Phyllis Biddle for DNA sequencing support. We thank Mike Clegg, Mark Williams, Tim Helentjaris for helpful comments and Barbara Mazur for support.
Contributor Information
Ada Ching, Email: ada.s.ching@usa.dupont.com.
Katherine S Caldwell, Email: kcaldw@scri.sari.ac.uk.
Mark Jung, Email: mark.t.jung@usa.dupont.com.
Maurine Dolan, Email: maureen.dolan@usa.dupont.com.
Oscar S (Howie) Smith, Email: howie.smith@pioneer.com.
Scott Tingey, Email: scott.v.tingey@usa.dupont.com.
Michele Morgante, Email: michele.morgante@usa.dupont.com.
Antoni J Rafalski, Email: j-antoni.rafalski@usa.dupont.com.
References
- Lindblad-Toh K, Winchester E, Daly M, Wang D, Hirschhorn JN, Laviolette JP, Ardlie K, Reich DE, Robinson E, Sklar P, Shah N, Thomas D, Fan JB, Gingeras T, Warrington J, Patil N, Hudson TJ, Lander ES. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- Bhattramakki D, Rafalski A. Discovery and Application of Single Nucleotide Polymorphism Markers in Plants. In: Henry RJ, editor. Plant Genotyping: The DNA Fingerprinting of Plants. Wallingford Oxon, UK: CABI Publishing; 2001. [Google Scholar]
- Syvanen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- Jorde LB. Linkage Diseqilibrium as a Gene-Mapping Tool. Am J Hum Genet. 1995;56:11–14. [PMC free article] [PubMed] [Google Scholar]
- Risch NJ. Searching for genetic determinants for the new millenium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- Jorde LB. Linkage Disequilibrium and the Search for Complex Disease Genes. Genome Research. 2000;10:1435–1444. doi: 10.1101/gr.144500. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ESI. Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetics. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- Weber J, May PE. Abundant Class of Human DNA Polymorphisms Which Can Be Typed Using the Polymerase Chain Reaction. Am J Hum Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- Viard F, Franck P, Dubois MP, Estoup A, Jarne P. Variation of microsatellite size homoplasy across electromorphhs, loci, and populations in three invertebrate species. J Mol Evol. 1998;47:42–51. doi: 10.1007/pl00006361. [DOI] [PubMed] [Google Scholar]
- Estoup A, Tailliez C, Cornuet JM, Solignac M. Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae). Mol Biol Evol. 1995;12:1074–1084. doi: 10.1093/oxfordjournals.molbev.a040282. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Atshuler D. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Buckler 4th ES, Thornsberry JM. Plant Molecular Diversity and Applications to Genomics. Curr Opin Plant Biol. 2002;5:107–11. doi: 10.1016/S1369-5266(02)00238-8. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Le Thierry d'Ennequin M, Peek AS, Sawkins MC. Maize as a model for the evolution of plant nuclear genomes. Proc Natl Acad Sci USA. 2000;97:7008–7015. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck-Eidens DM, Bell RN, Neuhausen SL, Helentjaris T. DNA Sequence Variation Within Maize and Melon: Observations From Polymerase Chain Reaction Amplification and Direct Sequencing. Genetics. 1990;126:207–217. doi: 10.1093/genetics/126.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to the domestication of maize. Proc Natl Acad Sci USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Peek AS, Morton BR, Clegg MT. Patterns of genetic diversification within the Adh gene family in the grasses (Poaceae). Mol Biol Evol. 1999;16:1086–1097. doi: 10.1093/oxfordjournals.molbev.a026198. [DOI] [PubMed] [Google Scholar]
- Golubinoff P, Paabo S, Wilson AC. Evolution of maize inferred from sequence diversity of an Adh2 gene segment from archeological specimens. Proc Natl Acad Sci USA. 1993;90:1997–2001. doi: 10.1073/pnas.90.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A-M, Damerval C. High rates of polymorphism and recombination in the Opaque-2 locus in cultivated maize. Mol Gen Genet. 1997;256:147–157. doi: 10.1007/s004380050556. [DOI] [PubMed] [Google Scholar]
- Selinger DA, Chandler VL. Major recent and independent changes in levels and patterns of expression have occured at the b gene, a regulatory locus in maize. Proc Natl Acad Sci USA. 1999;96:15007–15012. doi: 10.1073/pnas.96.26.15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton H, Gaut BS. Speciation and domestication in the maize and its wild relatives: evidence from the globulin-1 gene. Genetics. 1998;150:863–872. doi: 10.1093/genetics/150.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R-L, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Gaut BS, Stec AO, Fuerstenberg SI, Goodman MM, Coe EH, Doebley JF. Evolution of anthocyanin biosynthesis in maize kernels: the role of regulatory and enzymatic loci. Genetics. 1996;143:1395–1407. doi: 10.1093/genetics/143.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SE, Doebley JF. The molecular evolution of terminal ear 1, a regulatory gene in the genus Zea. Genetics. 1999;153:1455–1462. doi: 10.1093/genetics/153.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin MT AC. DNA sequence variation and the recombinational landscape in Drosophila pseudoobscura: a study of the second chromosome. Genetics. 1999;153:859–869. doi: 10.1093/genetics/153.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattramakki D, Dolan M, Hanafey M, Wineland R, Vaske D, Register III JC, Tingey SV, Rafalski A. Insertion-Deletion Polymorphisms in 3' Regions of Maize Genes Occur Frequently and Can Be Used as Highly Informative Genetic Markers. Plant Mol Biol. 2002;48:539–547. doi: 10.1023/A:1014841612043. [DOI] [PubMed] [Google Scholar]
- Kruglyak S, Durrett RT, Schug MD, Aquadro CF. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc Natl Acad Sci USA. 1998;95:10774–8. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G, Tingey S. Simple sequence repeats for germplasm analysis and mapping in maize. Genome. 1996;39:277–287. doi: 10.1139/g96-038. [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR. A general approach to single-nucleotide polymorphism discovery. Nature Genetics. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- Sunyaev SR, Lathe 3rd WC, Ramensky VE, Bork P. SNP frequencies in human genes an excess of rare alleles and differing modes of selection. Trends Genet. 2000;16:335–337. doi: 10.1016/S0168-9525(00)02058-8. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BM, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh1 parallel rate differences at the plastid gene rblL. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Clegg MT. Molecular evolution of the Adh1 locus in the genus Zea. Proc Natl Acad Sci USA. 1993;90:5095–5099. doi: 10.1073/pnas.90.11.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith OS, Sullivan H, Hobart B, Wall SJ. Evaluation of a Divergent Set of SSR Markers to Predict F1 Grain Yield Performance and Grain Yield Heterosis in Maize. Maydica. 2000;45:235–241. [Google Scholar]
- Zhu ZF, Sun CQ, Jiang TB, Fu Q, Wang XK. The comparison of genetic divergences and its relationships to heterosisrevealed by SSR and RFLP markers in rice (Oryza sativa L.). Yi Chuan Xue Bao. 2001;28:738–745. [PubMed] [Google Scholar]
- Stuber CW, Lincoln SE, Wolff DW, Helentjaris T, Lander ES. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:823–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. DNA polymorphism in a subdivided population: the expected number of segregating sites in the two-subpopulation model. Genetics. 1989;123:229–240. doi: 10.1093/genetics/123.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Kueffer J-M, Roessli D, Excoffier L. Arlequin ver 1.1. A software for population genetic analysis. Software manual. 1997. http://anthropologie.unige.ch/arlequin
- Aris-Brosou S, Excoffier L. The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol Biol Evol. 1996;13:494–504. doi: 10.1093/oxfordjournals.molbev.a025610. [DOI] [PubMed] [Google Scholar]
- Tivang JG, Nienhuis J, Smith OS. Estimation of sampling variance of molecular-marker data using the bootstrap procedure. Theor App Genet. 1994;89:259–264. doi: 10.1007/BF00225151. [DOI] [PubMed] [Google Scholar]
- Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G, Motto M. Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs and AFLPs. Theor App Genet. 1998;97:1248–1255. doi: 10.1007/s001220051017. [DOI] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Nordborg M. Coalescent Theory. In: Balding DJ, Bishop M, Cannings C, editor. Handbook of Statistical Genetics. Chichester, England: John Wiley and Sons; 2001. pp. 179–212. [Google Scholar]
- Wilkes HG. Hybridization of maize and teosinte in Mexico and Guatemala and the improvement of maize. Economic Bot. 1977;31:254–293. [Google Scholar]
- Fu H, Dooner HK. Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Linkage Disequilibrium and Recombination. In: Balding DJ, Bishop M, Cannings C, editor. Handbook of Statistical Genetics. Chichester: John Wiley and Sons, Ltd; 2001. pp. 309–324. [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ESt. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II Sunderland, MA: Sinauer Associates, Inc. 1996.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes and PCR primers
Frequency of polymorphisms and population parameters in coding and non-coding regions
Aligned sequence data in an interleaved text format, similar to NEXUS. Sequences from all loci examined are concatenated, the identity and location of sequences corresponding to each locus is in the file header.
Same data as file 1 but no locus identity information. DNAsp – compatible NEXUS format format. May be opened directly with DNAsp software (see Materials and Methods).
Same data as file 2 in MEGA format. May be opened directly with MEGA or DNAsp software (see Materials and Methods).
Excel file containing information about positions of individual loci in the concatenated sequence files Ching_data1, Ching_data2 and Ching_data3