Abstract
Histonelike nucleoid structuring protein (H-NS) is an abundant prokaryotic protein participating in nucleoid structure, gene regulation, and silencing. It plays a key role in cell response to changes in temperature and osmolarity. Force-extension measurements of single, twist-relaxed λ-DNA-H-NS complexes show that these adopt more extended configurations compared to the naked DNA substrates. Crosslinking indicates that H-NS can decorate DNA molecules at one H-NS dimer per 15–20 bp. These results suggest that H-NS polymerizes along DNA, forming a complex of higher bending rigidity. These effects are not observed above 32°C or at high osmolarity, supporting the hypothesis that a direct H-NS-DNA interaction plays a key role in gene silencing. Thus, we propose that H-NS plays a unique structural role, different from that of HU and IHF, and functions as one of the environmental sensors of the cell.
INTRODUCTION
The bacterial chromosome or nucleoid is a dynamic entity that is continuously replicating and repaired, and it allows for rapid gene expression in response to specific cellular demands (Pettijohn, 1996; Trun and Marko, 1998). A dozen DNA-binding proteins known as histonelike proteins serve as a dynamic scaffold for nucleoid organization (Drlica and Rouviere-Yaniv, 1987). In Escherichia coli, it was shown that the relative concentration of the various nucleoid-associated proteins changes as the cells enter the stationary phase of growth (Azam et al., 1999). One of the major nucleoid proteins, the histonelike nucleoid structuring protein (H-NS; see Ussery et al., 1994; Williams and Rimsky, 1997), is an abundant protein, present at ∼20,000 copies in exponential phase. The H-NS protein is structurally unrelated to histones. It does not exhibit high affinity for specific sequences but binds preferentially to intrinsically curved DNA and A+T-rich sequences (Dame et al., 2001; Rimsky et al., 2001). Cyclization experiments suggest that H-NS can bend DNA upon binding (Spurio et al., 1997). It was claimed that H-NS compacts DNA strongly (Spassky et al., 1984). Recent atomic force microscopy (AFM) observations have suggested a mechanism of lateral compaction based on H-NS-induced bridging of distal DNA segments along the same molecule, in a hairpin configuration (Dame et al., 2000, 2001). In contrast, when H-NS is present at high concentration, electron microscopy suggested that H-NS coats long DNA substrates end-to-end with no appreciable compaction (Tupper et al., 1994). At low concentration, H-NS was shown to affect nucleoid structure by constraining negative supercoils of DNA in vivo and in vitro (Tupper et al., 1994). Thus the precise way by which H-NS participates in shaping nucleoid structure remains uncertain.
In addition to its role in nucleoid architecture, H-NS plays a pleiotropic role in bacterial response to environmental stimuli such as starvation, changes in pH, temperature, and osmolarity (Atlung and Ingmer, 1997; McLeod and Johnson, 2001; Williams and Rimsky, 1997). H-NS was found to affect gene expression in a number of different ways, as the expression of over 5% of the E. coli genes are affected in an hns mutant (Hommais et al., 2001). A remarkable property of H-NS is its ability to act in gene silencing, a mechanism requiring, in contrast to direct repression, a high affinity H-NS binding site located at a distance from the promoter. Silencing by H-NS was recently demonstrated for gene clusters in pathogenicity islands (Bustamante et al., 2001; Falconi et al., 1998; Friedberg et al., 1999). Although the molecular basis for gene silencing is not known, it was suggested that H-NS could polymerize along DNA from the silencer region to the promoter (Caramel and Schnetz, 1998; Rimsky et al., 2001).
To shed more light on H-NS function and on its role in shaping nucleoid structure, we studied the elastic response of single twist-free DNA-H-NS complexes. Single λ-phage DNA molecules (16.5 μm long) tethering a magnetic bead to glass surface were stretched, in a solution containing H-NS, by an external magnetic field. The setup allows the direct measurement of the end-to-end extension of the nucleoprotein complex as a function of the stretching force. This experimental approach offers a number of advantages. The DNA molecules, attached both to bead and glass substrate by single bonds, are free to fluctuate and swivel, supporting no torsion. Furthermore, the DNA-H-NS complexes are in thermodynamic equilibrium with the protein solution, allowing H-NS to bind and dissociate from DNA unhindered. These features are particularly important for studying weak nonspecific protein-DNA interactions that are difficult to study by most molecular biology techniques.
We find that at physiological concentrations, H-NS-DNA complexes assume extended configurations that on average are ∼twofold larger than bare DNA. This is in stark contrast to the effects of two other important nucleoid-associated proteins, IHF (Jaffar Ali et al., 2001) and HU (Schnurr et al., unpublished results). In addition, crosslinking experiments indicate that H-NS is associated in very large numbers with DNA. These findings suggest a picture in which massive polymerization of H-NS on DNA, covering extended tracts, increases DNA stiffness. This picture is consistent with H-NS participation in gene silencing as has been recently proposed (Rimsky et al., 2001). Interestingly, we find that in this purified system one can reproduce the temperature and osmolarity response of H-NS in vivo. Thus it appears that H-NS can act directly as a high-resolution temperature and osmolarity sensor in the control of gene expression.
MATERIALS AND METHODS
DNA-bead constructs
Lambda-phage DNA-bead constructs were prepared by mixing λ-phage DNA labeled at the right and left ends with biotin and digoxigenin, Dig (Roche Molecular Biochemicals, Indianapolis, IN), with 2.8 μm M280 magnetic tosyl-activated beads (Dynal, Lake Success, NY) coated with anti-digoxigenin and passivated with α-casein, respectively. Labeling was achieved by hybridizing and ligating bare λ-phage DNA (Roche Molecular Biochemicals, Indianapolis, IN) with labeled complementary DNA oligomers. H-NS purification was carried out with minor modifications of existing protocols (Dersch et al., 1993).
Sample cells
Cells for DNA elasticity measurements have already been described in detail (Jaffar Ali et al., 2001). In brief, samples were prepared in capillaries of square cross section (Vitro Dynamics, Rockway, NJ), which were washed first in a NH4:H2O2:H2O solution (1:1:5), rinsed in H2O for 10 min, and dried. The inner surface of each capillary was then made hydrophobic by soaking in Sigmacote (Sigma-Aldrich, St. Louis, MO), and rinsed in both ethanol and H2O. Next, the capillary was coated with BSA-biotin (Sigma-Aldrich) (incubation for 2 h at 37°C), followed by incubation with streptavidin for 2 h at 37°C. DNA-bead constructs in casein buffer (CB: 10 mM Tris-HCl, 200 mM KCl, 5% DMSO, 0.1 mM EDTA, 0.2 mg/ml α-casein, pH 8.0) were then introduced and incubated at room temperature, to allow for enough constructs to bind to the lower inner side of the capillary.
Force measurements and optical setup
Samples containing λ-DNA-tethered magnetic beads were observed by bright-field illumination using a home-built inverted microscope. Extension-force measurements were made by using a magnetic force technique (Jaffar Ali et al., 2001; Strick et al., 1996). In brief, a vertical magnetic force stretching the tethering DNA molecule was applied on a bead by means of a pair of magnets, whose height could be controlled accurately. The vertical force F was obtained from measurements of the bead's Brownian motion transverse to the direction of the force, using the equipartition theorem: F/L = kBT/〈δX2〉. Here, L is the extension of the tether, 〈δX2〉 representing an average over the square of the bead transverse displacements; T is the temperature; and kB is Boltzmann's constant. The extension L was measured by correlating the bead's image with a library of images taken of the same bead-DNA construct, when stretched by a large force (>10 pN) to prevent large fluctuations in the longitudinal z direction. The images in the library were obtained at different foci separated by a known amount over a range of 20 μm. The reproducibility of force-extension curves was checked by making measurements on five different λ-DNA molecules for each H-NS concentration.
Data processing and analysis
The extension of λ-DNA molecules was measured by the following procedure: the center of the first bead image in an experimental run and the center of all the bead images in the library were initially determined. The radial intensity profiles of the bead image and those in the library were then calculated. Next, the radial intensity profile of the bead image was correlated with the profiles corresponding to each image in the library. The bead height was determined by performing a parabolic fit around the maximum of the correlation, as a function of the height corresponding to each library image. The height value corresponding to the maximum of the fit was then defined as the bead height. This procedure was repeated for each image in an experimental run after the first, calculating the correlation only with images in the library corresponding to small differences in height relative to the previous image in the experimental run. We estimate the error in our measurements at ∼1% of the total length, or ∼150 nm.
DNA digestion
Reaction mixtures, 20 μl, in buffer (50 mM NaCl, 10 mM Tris-HCl pH 7.9, 10 mM MgCl2, 1 mM DTT) containing 0.5 μg of λ-DNA was incubated with the restriction enzyme Sau3A (New England Biolabs, Beverly, MA) in the absence or presence of H-NS at a concentration of 1 μM for 30 min. The DNA fragments were separated on 1% agarose gel and visualized with ethidium bromide.
Density gradient centrifugation of H-NS-DNA complexes
These experiments were carried out as described previously (Jaffar Ali et al., 2001) with the following modifications: formaldehyde and sarcosyl were omitted during the run. Centrifugation was carried out at 32,000 rpm for 64 h.
RESULTS
H-NS induces larger end-to-end extension of DNA
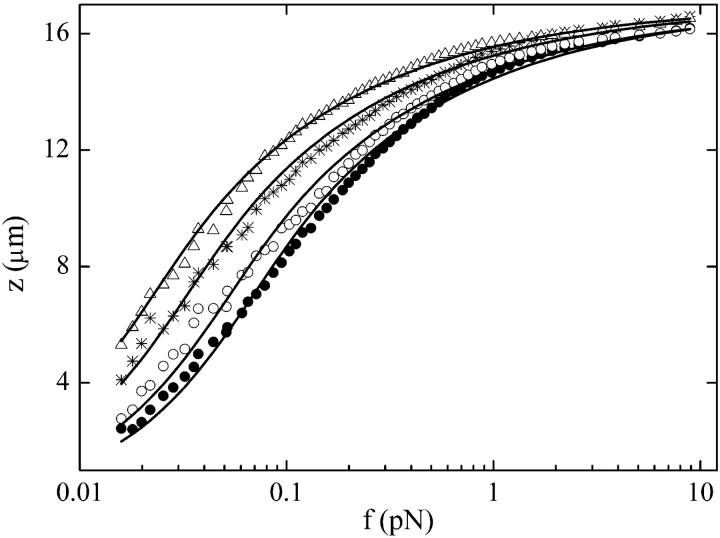
To determine the influence of H-NS on single DNA molecules in solution, we measured DNA extension (z) versus force (f) for different concentrations of wild-type H-NS at 50 mM KCl, and 25°C (Fig. 1). The H-NS-DNA complexes exhibit larger extension values than naked DNA in the low-force regime, whereas at large forces, the elastic behavior of the complexes and naked DNA tend to converge. The difference in behavior at low force increases with H-NS concentration, reaching a ∼twofold increase in end-to-end extension. This increase is not a result of an H-NS-induced change in the contour length of DNA as in the case of RecA (Hegner et al., 1999; Leger et al., 1998; Shivashankar et al., 1999). No hysteresis was observed upon repeated extension-retraction cycles. After elasticity measurements were completed, flushing with buffer without H-NS led to the recovery of the elastic behavior of naked DNA. Furthermore, addition of poly(dI-dC), a nonspecific competitor, eliminates all H-NS effects (data not shown). These results suggest that H-NS can bind to DNA with low specificity.
FIGURE 1.
Increased end-to-end distance of DNA by H-NS for low tension. Extension z of single H-NS-DNA complexes as a function of the force f pulling on their ends for four H-NS concentrations: 0 nM (solid circles), 250 nM (open circles), 2 μM (stars), and 4 μM (triangles). All the measurements were carried out at 25°C and 50 mM KCl in Tris buffer (Jaffar Ali et al., 2001). Lines are fits to the data using an extended wormlike chain model (Bouchiat et al., 1999).
A large number of H-NS molecules associate with single DNA molecules
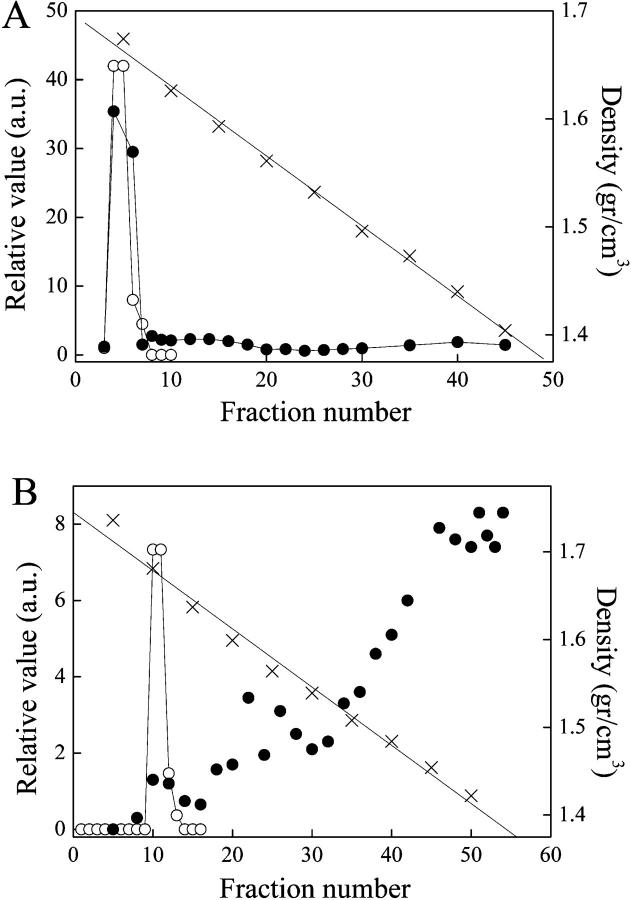
What is the mechanism by which H-NS induces an end-to-end extension upon binding to DNA? One possibility is that a large number of H-NS molecules polymerize along DNA, forming a complex of higher rigidity than the bare DNA substrate. To estimate the number of H-NS molecules associated with λ-DNA, we measured the shift in buoyant density of the H-NS-DNA complexes. Radioactively labeled λ-DNA was incubated with H-NS, crosslinked with formaldehyde to prevent dissociation, and resolved by equilibrium density centrifugation in CsCl gradients. Without H-NS, the radioactively labeled DNA appears as a sharp band overlapping with marker unlabeled λ-DNA (Fig. 2 A). In contrast, nucleoprotein complexes formed in the presence of H-NS (Fig. 2 B) appear in a wide distribution of densities, indicating the predominance of complexes with a large number of H-NS molecules. We estimate the number of H-NS dimers per λ-DNA to be 2500–3200, equivalent to one H-NS dimer per every 15–20 bp. To obtain an indication whether H-NS is uniformly distributed along the DNA, the DNA was subjected to cleavage by the restriction enzyme Sau3A that recognizes 116 GATC sites along λ-DNA. The results show that in the absence of H-NS, discrete bands are easily discerned in the range 1–3 kbp. The addition of H-NS greatly inhibits digestion by the enzyme and most of the discrete bands disappear (Fig. 3) (the same pattern was seen under low UV exposure).
FIGURE 2.
(A, Left axis) Relative value as a function of fraction number of marker DNA (open circles), radioactively-labeled DNA (solid circles). (A, Right axis) Density along the CsCl gradient where the fractions appear (crosses). (B) Same as in A but with the radioactively-labeled DNA crosslinked with H-NS in a 1 μM solution.
FIGURE 3.
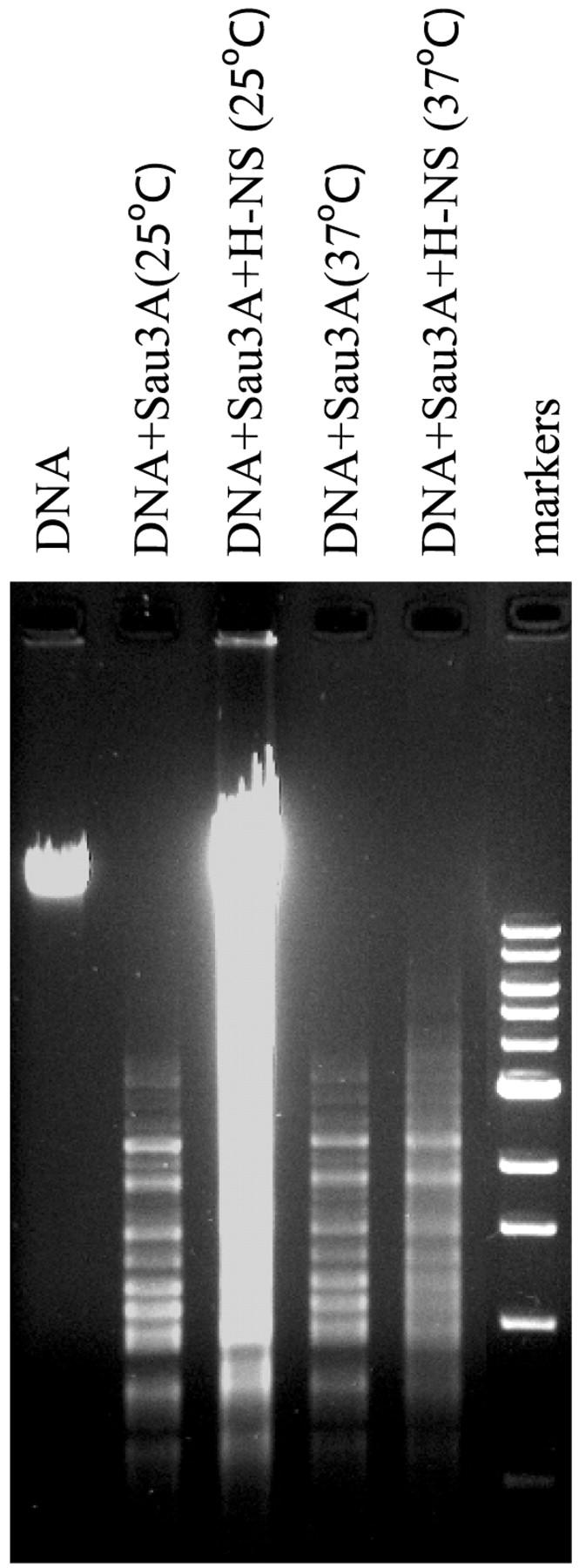
Electrophoretic analysis of enzymatic digestion of H-NS-DNA complexes with Sau3A, at 25 and 37°C. Reaction mixtures, 20 μl, in buffer (50 mM NaCl, 10 mM Tris-HCl pH 7.9, 10 mM MgCl2, 1 mM DTT) containing 0.5 μg of λ DNA was incubated with the restriction enzyme Sau3A in the absence of presence of H-NS at a concentration of 1 μM for 30 min. The DNA fragments were separated on 1% agarose gel and visualized with ethidium bromide.
Complexes formed by the polymerization of H-NS on DNA have a higher rigidity than bare DNA
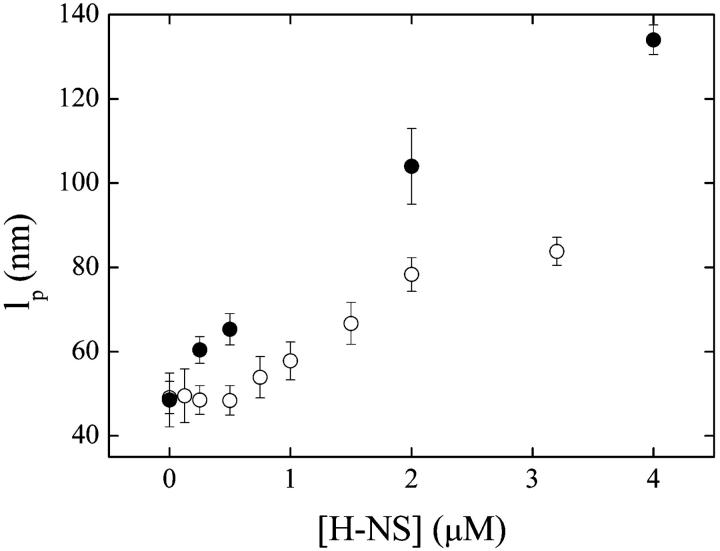
The results described above are consistent with a model in which H-NS polymerizes along the DNA substrate, covering extensive tracts. Provided that these tracts are large in number, and their typical size is much smaller than the length of the nucleoprotein complex, the elastic behavior of nucleoprotein complexes may be adequately described by the worm-like chain model (Bouchiat et al., 1999; Bustamante et al., 1994), with an associated effective persistence length lp. This quantity is an unknown function of the (small-scale) persistence lengths of naked DNA tracts, tracts fully covered by H-NS, their number and spatial distribution. We show fits to the elasticity data using the worm-like chain model in Fig. 1. We determined from the fits values of lp characterizing complexes at different H-NS concentrations. The persistence length increases, as a function of H-NS concentration, reaching a value of ∼130 nm at the highest concentration measured, nearly threefold that of naked DNA (Fig. 4). In comparison, this ratio lies between 4 and 15 for RecA-DNA filaments (Hegner et al., 1999; Leger et al., 1998; Shivashankar et al., 1999).
FIGURE 4.
The effect of osmolarity on H-NS-DNA complexes. Persistence length lp as a function of H-NS concentration for 50 mM KCl (solid circles) and 200 mM KCl (open circles). All measurements were carried out at 25°C.
H-NS alone can act as a temperature and osmolarity sensor in vitro
H-NS has been shown to be a key regulator in the response to changes in osmolarity, pH, and temperature. However, since in vivo such stimuli induce a number of concomitant changes such as supercoling, the specific mechanisms by which H-NS acts have proved elusive. To test for possible direct effect of high osmolarity on H-NS-DNA interaction, the effect of H-NS on DNA elasticity as a function of increasing salt concentration was determined. We found that the H-NS effect is reduced at high ionic strength, independent of H-NS concentration (Fig. 4). Note, that at higher osmolarity, H-NS increases the persistence length measurably only above ∼0.5 μM. Replacement of KCl by NaCl resulted in the same behavior shown in Fig. 2 A (data not shown), demonstrating that H-NS responds to both ions. Since our DNA substrates support no torsion, the attenuation of H-NS effects we observe at high osmolarity is not a result of changes in the supercoiling level of the substrate, but to H-NS-DNA interactions directly. These results are consistent with the inhibitory effect of H-NS observed on transcription of the proV gene at 50 mM KCl, the relief of inhibition at 200 mM KCl (Ueguchi and Mizuno, 1993), and with gel retardation and fluorescence analysis of H-NS-DNA complexes (Tippner and Wagner, 1995).
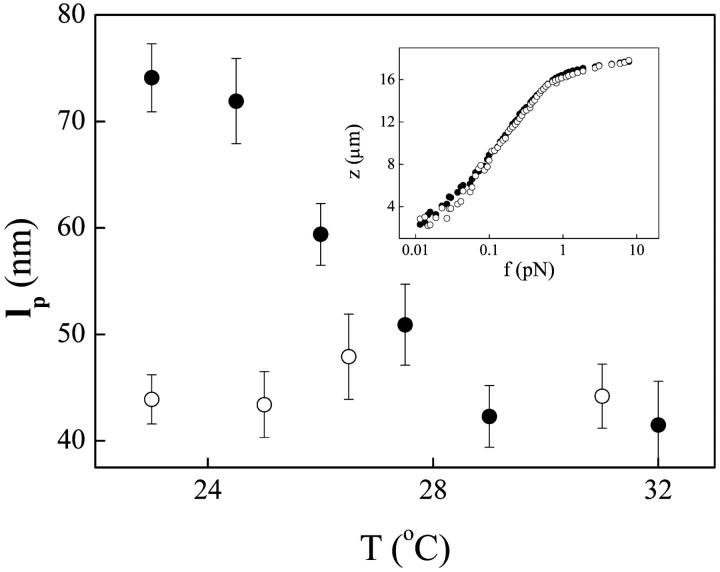
The effects of temperature on H-NS-DNA complex formation was investigated by elasticity measurements in the range 23°C to 37°C at low osmolarity (Fig. 5). Remarkably, lp decreases monotonically with increasing temperature. A change of 5–7°C is sufficient to abolish most H-NS function. At 37°C, the full force-extension curves of naked DNA and DNA in the presence of a 2 μM solution of H-NS coincide within experimental error (inset, Fig. 5). Thus H-NS does not exert any effects on DNA conformation at high temperatures. Consistent with these data, H-NS was found to be ineffective in protecting DNA from digestion with Sau3A at 37°C (Fig. 3, compare lanes 4 and 5). Taken together, our experimental system appears to display similar signal response to that observed in vivo.
FIGURE 5.
The effect of temperature on H-NS-DNA complexes. Persistence length lp of H-NS-DNA complexes (solid circles) and naked DNA (open circles) as a function of temperature. All measurements were carried out in buffer with 50 mM KCl. Complexes were formed in a 0.8 μM H-NS solution. (Inset) Extension z of single H-NS-DNA complexes as a function of force f measured at 37°C, for 0 nM (solid circles) and 2 μM (open circles) H-NS.
DISCUSSION
Our study of the H-NS/DNA interaction reveals the following features: H-NS covers extensive tracts along the DNA molecule, thereby increasing its bending rigidity. As a result, the end-to-end extension of the complex is larger than that of bare DNA. This increase in end-to-end extension, in contrast to the case of RecA, is not ATP-dependent and is not due to an increase in the contour length of the DNA substrate. Furthermore, temperature and osmolarity effects on the activity of H-NS in vivo were mimicked in our minimal system. These results suggest the model shown schematically in Fig. 6. Based on genetic, biochemical and AFM observations (Dame et al., 2001), H-NS binds at AT-rich regions of high intrinsic curvature along the DNA backbone. These regions may serve as initiation sites for the polymerization of H-NS along the double-stranded tracts. Polymerization of H-NS on DNA has already been suggested by electron microscopy studies (Tupper et al., 1994). Without polymerization it would be impossible to account for the larger end-to-end extension of the nucleoprotein complex observed in our experiments. The overall length of double-stranded DNA covered by H-NS must be large enough to compensate for hairpin formation at curved sites. Furthermore, our measurements suggesting the polymerization of H-NS were obtained from experiments in which the H-NS-DNA complex is minimally perturbed. In contrast, direct methods of visualization such as EM (Tupper et al., 1994) and AFM techniques (Dame et al., 2000, 2001) involve larger perturbations of the H-NS-DNA complexes and are not mutually consistent. The large number of H-NS molecules interacting with DNA helps to explain the increased sedimentation behavior of H-NS-DNA complexes that was a major factor in assuming that H-NS compacts DNA (Spassky et al., 1984).
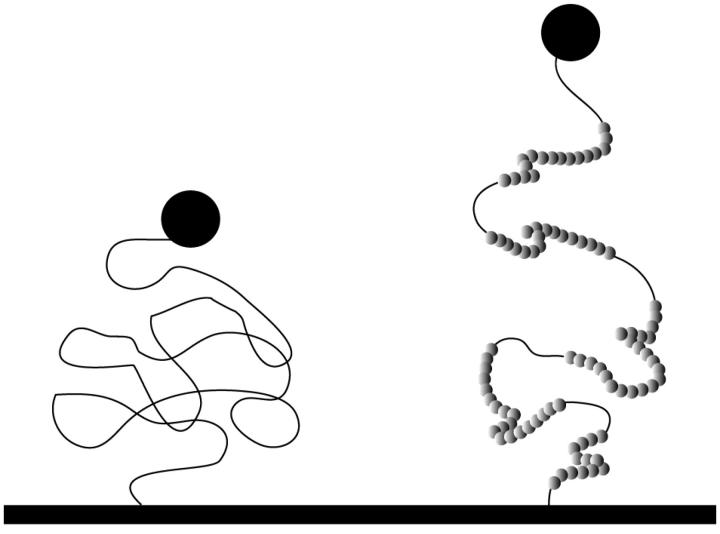
FIGURE 6.
Schematic model of configurations of naked DNA and H-NS-DNA complexes in the low tension regime. The random coil configuration of the complex is more open due to its larger effective persistence length.
The H-NS molecule is composed of two distinct domains joined by a flexible linker region. The C-terminal domain, determined by NMR, contains a unique DNA binding motif. The N-terminal domain is involved in H-NS oligomerization. It is made of a short coiled-coil motif (Dorman et al., 1999; Renzoni et al., 2001; Smyth et al., 2000). It has been previously shown that a coiled-coil motif in proteins induces oligomer dissociation in response to increased osmolarity or temperature, suggesting that this region participates in the response of H-NS to high temperature and osmolarity. We propose that the changes in H-NS structure under these conditions prevents polymerization of H-NS along the DNA.
Our model is consistent with the proposed role of H-NS as a transcriptional silencer (Goransson et al., 1990). Curved sequences far away from a promoter serve as nucleation sites for H-NS polymerization, which extend up to the promoter, thereby inhibiting transcription. This situation may occur naturally in the downward regulating element (DRE) of the proU operon in Salmonella typhimurium. The DRE is 200 bp away from the site of transcription initiation. Sequences involved in H-NS-mediated silencing of the bgl promoter are also ∼200 bp away from the latter (Caramel and Schnetz, 1998). Only long insertions (≥800 bp) between the nucleation site and the promoter disrupt silencing (Fletcher and Csonka, 1995). Thus the extent of polymerization around each nucleation site may be taken to be of the order of 200–500 bp, equivalent to ∼100–250 polymerization regions along the λ-DNA substrate. In agreement with this estimate, an analysis of high intrinsic curvature sites along the λ-DNA molecule which we have performed using the CURVATURE software, revealed 90–200 high curvature loci (Shpigelman et al., 1993).
Our studies and previous investigations provide a new perspective for the role played by H-NS in organizing nucleoid structure and its function. H-NS can nucleate on sites of intrinsic curvature, but can also polymerize along short stretches of DNA, increasing DNA rigidity, and leading to gene silencing. These stretches, located within regulatory regions are probably limited on one hand by nucleation sites from which polymerization is initiated, and on the other by roadblocks generated by DNA binding proteins. Our results demonstrate that H-NS alone does not lead to DNA compaction. In contrast to IHF, Fis, and HU, H-NS contributes more to the increased rigidity of small segments of the chromosome. One can only speculate about the implications this may have in nucleoid organization. A unique property of the bacterial nucleoid is its ability to respond rapidly to external input signals. Interestingly, the structure of the H-NS-DNA complex alone permits the nucleoid to respond, with high sensitivity, to changes in temperature and osmolarity, independent of additional host proteins and changes in DNA supercoiling.
Acknowledgments
We thank Dr. Ilan Rosenshine and Dr. Erhard Bremer for providing us with plasmid pPD3, Dinah Teff and Simi Koby for technical assistance, and John Marko for very useful correspondence.
This research was supported by The Israel Science Foundation (grant 489/01-1), and the Minerva Foundation.
References
- Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7–17. [DOI] [PubMed] [Google Scholar]
- Azam, T. A., A. Iwata, A. Nishimura, U. Susumu, and A. Ishihama. 1999. Growth phase dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchiat, C., M. D. Wang, J.-F. Allemand, T. R. Strick, S. M. Block, and V. Croquette. 1999. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 76:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664–678. [DOI] [PubMed] [Google Scholar]
- Caramel, A., and K. Schnetz. 1998. Lac and lambda repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J. Mol. Biol. 284:875–883. [DOI] [PubMed] [Google Scholar]
- Dame, R. T., C. Wyman, and N. Goosen. 2000. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 28:3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame, R. T., C. Wyman, and N. Goosen. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie. 83:231–234. [DOI] [PubMed] [Google Scholar]
- Dersch, P., K. Schmidt, and E. Bremer. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8:875–889. [DOI] [PubMed] [Google Scholar]
- Dorman, C. J., J. C. Hinton, and A. Free. 1999. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 7:124–128. [DOI] [PubMed] [Google Scholar]
- Drlica, K., and J. Rouviere-Yaniv. 1987. Histone-like proteins of bacteria. Microbiol. Rev. 51:301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17:7033–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, S. A., and L. N. Csonka. 1995. Fine-structure deletion analysis of the transcriptional silencer of the proU operon of Salmonella typhimurium. J. Bacteriol. 177:4508–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941–952. [DOI] [PubMed] [Google Scholar]
- Goransson, M., B. Sonden, P. Nilsson, B. Dagberg, K. Forsman, K. Emanuelsson, and B. E. Uhlin. 1990. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 344:682–685. [DOI] [PubMed] [Google Scholar]
- Hegner, M., S. B. Smith, and C. Bustamante. 1999. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl. Acad. Sci. USA. 96:10109–10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J.-P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20–36. [DOI] [PubMed] [Google Scholar]
- Jaffar Ali, B. M., R. Amit, I. Braslavsky, A. Oppenheim, B. O. Gileadi, and J. Stavans. 2001. Compaction of single DNA molecules induced by binding of integration host factor (IHF). Proc. Natl. Acad. Sci. USA. 98:10658–10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger, J. F., J. Robert, L. Bourdieu, D. Chatenay, and J. F. Marko. 1998. RecA binding to a single double-stranded DNA molecule: a possible role of DNA conformational fluctuations. Proc. Natl. Acad. Sci. USA. 95:12295–12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod, S. M., and R. C. Johnson. 2001. Control of transcription by nucleoid proteins. Curr. Opin. Microbiol. 4:152–159. [DOI] [PubMed] [Google Scholar]
- Pettijohn, D. E. 1996. The nucleoid. In Escherichia coli and Salmonella. F. C. Neidhardt, R. Curtis, III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger, editors. ASM Press, Washington, D.C. pp. 158–166.
- Renzoni, D., D. Esposito, M. Pfuhl, J. C. Hinton, C. F. Higgins, P. C. Driscoll, and J. E. Ladbury. 2001. Structural characterization of the N-terminal oligomerization domain of the bacterial chromatin-structuring protein, H-NS. J. Mol. Biol. 306:1127–1137. [DOI] [PubMed] [Google Scholar]
- Rimsky, S., F. Zuber, M. Buckle, and H. Buc. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 45:1311–1323. [DOI] [PubMed] [Google Scholar]
- Shivashankar, G. V., M. Feingold, O. Krichevsky, and A. Libchaber. 1999. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc. Natl. Acad. Sci. USA. 96:7916–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpigelman, E. S., E. N. Trifonov, and A. Bolshoy. 1993. CURVATURE: software for the analysis of curved DNA. Comput. Appl. Biosci. 9:435–440. [DOI] [PubMed] [Google Scholar]
- Smyth, C. P., T. Lundback, D. Renzoni, G. Siligardi, R. Beavil, M. Layton, J. M. Sidebotham, J. C. Hinton, P. C. Driscoll, C. F. Higgins, and J. E. Ladbury. 2000. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 36:962–972. [DOI] [PubMed] [Google Scholar]
- Spassky, A., S. Rimsky, H. Garreau, and H. Buc. 1984. H1a, an Escherichia coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 12:5321–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16:1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick, T. R., J.-F. Allemand, D. Bensimon, A. Bensimon, and V. Croquette. 1996. The elasticity of a single supercoiled DNA molecule. Science. 271:1835–1837. [DOI] [PubMed] [Google Scholar]
- Tippner, D., and R. Wagner. 1995. Fluorescence analysis of the Escherichia coli transcription regulator H-NS reveals two distinguishable complexes dependent on binding to specific or nonspecific DNA sites. J. Biol. Chem. 270:22243–22247. [DOI] [PubMed] [Google Scholar]
- Trun, N. J., and J. F. Marko. 1998. Architecture of a bacterial chromosome. Am. Soc. Microbiol. Rev. 64:276–283. [Google Scholar]
- Tupper, A., E., T. A. Owen-Hughes, D. W. Ussery, D. S. Santos, D. J. Ferguson, J. M. Sidebotham, J. C. Hinton, and C. F. Higgins. 1994. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 13:258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi, C., and T. Mizuno. 1993. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 12:1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussery, D., W., J. C. Hinton, B. J. Jordi, P. E. Granum, A. Seirafi, R. J. Stephen, A. E. Tupper, G. Berridge, J. M. Sidebotham, and C. F. Higgins. 1994. The chromatin-associated protein H-NS. Biochimie. 76:968–980. [DOI] [PubMed] [Google Scholar]
- Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the Escherichia coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175–185. [DOI] [PubMed] [Google Scholar]