Abstract
The last phase of the proton transfer cycle of bacteriorhodopsin calls for a passage of a proton from D38 to D96. This reaction utilizes a narrow shaft ∼10-Å long that connects the two carboxylates that cross through a very hydrophobic domain. As the shaft is too narrow to be permanently hydrated, there are two alternatives for the proton propagation into the channel. The proton may propagate through the shaft without solvation at the expense of a high electrostatic barrier; alternatively, the shaft will expand to accommodate some water molecules, thus lowering the Born energy for the insertion of the charge into the protein (B. Schätzler, N. A. Dencher, J. Tittor, D. Oesterhelt, S. Yaniv-Checover, E. Nachliel, and G. Gutman, 2003, Biophys. J. 84:671–686). A comparative study of nine published crystal-structures of bacteriorhodopsin identified, next to the shaft, microcavities in the protein whose position and surrounding atoms are common to the reported structures. Some of the cavities either shrink or expand during the photocycle. It is argued that the plasticity of the cavities provides a working space needed for the transient solvation of the shaft, thus reducing the activation energy necessary for the solvation of the shaft. This suggestion is corroborated by the recent observations of Klink et al. (B. U. Klink, R. Winter, M. Engelhard, and I. Chizhov, 2002, Biophys. J. 83:3490–3498) that the late phases of the photocycle (τ ≥ 1 ms) are strongly inhibited by external pressure.
INTRODUCTION
Bacteriorhodopsin (Br) is the best-studied membranal protein. The protein utilizes the energy of a photon for an active translocation of protons from the cytoplasmic space of the bacterium to the extracellular space. The sequence of events, initiated by the absorption of a photon by the ground state (Br state) protein, had been extensively investigated and characterized by the spectral properties of the protein's retinal chromophore. The various intermediates of this photocycle have been characterized by their spectral properties and labeled by the letters from H to O.
The structure of the Br state had been determined by various groups, using electron and x-ray diffraction methods of either the native two-dimensional crystal, as found in the bacterial membrane (Grigorieff et al., 1996; Henderson et al., 1990), epitaxis crystallization on benzamidine (Essen et al., 1998), or cubic lipid matrix (Landau and Rosenbusch, 1996; Sass et al., 1997). At present, the resolution is as high as 1.55 Å (Luecke et al., 1999a). The structures of some photo intermediates were also determined.
The most initial steps of the photocycle (H, I, and J) are extremely fast, falling in the range from 10−15 to 10−9 s (Wang et al., 2002; Kobayashi et al., 2001). The free proton is ejected to the extracellular space within ∼50–100 μs (referred to as the M formation). In the later phase of the photocycle (steps N, O, and the final reformation of the initial ground state), a proton is obtained from the cytoplasmic space and delivered to the Schiff-base (for reviews see Lanyi and Pohorille, 2001; and Subramaniam et al., 1999). These steps involve charge redistribution inside the protein and require tens to hundreds of milliseconds to accomplish.
The deprotonated Schiff-base regains its proton from the carboxylate of the nearby aspartate D96 that, due to its very hydrophobic environment, has a high pK and is not a proton donor with respect to the Schiff-base (Brown et al., 1999; Luecke et al., 2000; Schätzler et al., 2003). Only after a conformational change, caused by the ejection of the proton from the extracellular section of the protein, the immediate vicinity of D96 becomes more hydrated and a pK shift causes a proton transfer from its carboxylate to the Schiff-base. The reprotonation of the Schiff-base is coupled with a large shift in its absorbance maximum and is readily recorded. On the other hand, the reprotonation of D96 modulates the electrostatic potential too far from the Schiff-base to affect its spectrum. The protonation kinetics of D96 was inferred by a proton bookkeeping procedure (Schätzler et al., 2003). The protons released to the bulk by the photocycle are recorded by following the state of protonation of a pH indicator, which is not adsorbed to the membrane, whereas the reprotonation of the Schiff-base is recorded, in parallel, at its specific wavelength. The comparison between the two indices accounts for all available protons stored in the protein. These measurements indicated that reprotonation of D96 lags behind the protonation of the Schiff-base, indicating that the rate-limiting step of the late phase of the photocycle is the propagation of the proton from the bulk into the inner section of the protein where D96 is located. This process can take 10–100 ms, depending on the ionic strength of the solution. Minor modification of the charged residues located on the cytoplasmic surface of the protein can cause large modulation in the dynamics of the last phase of the photocycle (Schätzler et al., 2003).
The uptake of a proton from the cytoplasmic space of the bacterium necessitates picking up a single proton from a solution having pH ≥ 7 and keeping it ready to be transferred into the protein, for a period of more than 10 ms (Subramaniam et al., 1999). This is a mechanistic problem concerning the high abundance of small buffer molecules in the bacterium cytoplasmic matrix. The total of all carboxylates containing substrate molecules (as of the citric acid cycle), plus the phosphorylated compounds (ATP, ADP, AMP, and free phosphate), will amount to 20–50 mM of buffering molecules. Accordingly, delaying a proton for a 10-ms period at the orifice of the cytoplasmic channel necessitates its storage in a site that is secluded from the bulk; otherwise it will be carried off by encounter with a buffer molecule within a few nanoseconds. The temporary proton storage, used for the protonation of D96, is the carboxylate of D38 (Riesle et al., 1996), a site that is partially accessible to bulk protons but which is practically protected from anions dissolved in the bulk (Checover et al., 2001). This residue is located ∼10 Å from the carboxylate moiety of D96 and serves as its preferential proton donor (Nachliel et al., 2002).
The passage of proton from D38 to D96 proceeds through a narrow shaft that extends along a chain of hydrophobic residues with F42 at its center (Dencher et al., 1992; Dioumaev et al., 1999; Schätzler et al., 2003). Examination of the molecular surface, at the orifice of the shaft (see Fig. 1) reveals that heavy atoms are so close to each other that a water molecule cannot squeeze in unless the protein exercises some local reorganization leading to a transient widening of the shaft. Unless the conducting space is solvated, the Born energy associated with the insertion of the protonic charge into the shaft will be Ea ∼33 kcal/mol (calculated according to Gilson et al., 1985). Such a value is so high that the reaction is unlikely to take place. On the other hand, expansion of the shaft will allow penetration of water molecules, a process that will lower the energy penalty for inserting a charge into the low dielectric matrix. Proteins are characterized by a compressibility coefficient of 10–25 Mbar−1 (Kharakoz, 2000), very close to the value determined to Purple Membrane preparation (Marque et al., 1984). According to this value, the expansion of the shaft to the extent that water molecule can penetrate inside, will require an energy investment of E ∼10–20 kcal/mol (the value is based on an energy increment of ∼0.3 kcal/mol/Å2 estimated from the calculations of Kharakoz, 2000; and Kocher et al., 1996). This energy investment is less prohibitive than the insertion of a dry proton and yet will lower the probability of the event by a factor of ≤ 4.10−10. Accordingly, there should be a mechanism that allows the expansion of the shaft with a smaller energy barrier. As will be discussed below, this process can be accomplished through the dynamics of the void space of intraprotein cavities that can expand or shrink one at the expense of the other.
FIGURE 1.
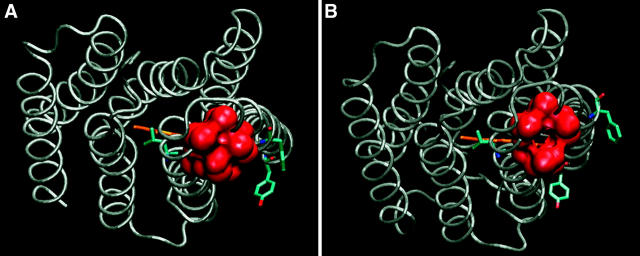
Expansion of the cytoplasmic surface of bacteriorhodopsin (1CWQ) near the vicinity of D38. Please note the shaft, which leads from the carboxylate of D38 (red), running in parallel with the benzene ring of F42 (white), toward the oxygen atoms of the carboxylate of D96 (yellow). The shaft is too narrow to accommodate a water molecule, as evident by comparison with water molecule HOH863 (presented by a transparent surface) located at the orifice of the shaft. Residue K41 is presented in blue.
A delay in the proton passage through the hydrophobic shaft was recorded for the WT protein (in the presence of 150 mM screening electrolyte) and for some mutations (D104C, E161C, and E164C) where some of the cytoplasmic surface charges were replaced. Yet, in two mutants, D102C and E166Q, it was observed that the proton passage through the shaft was faster than in the WT. Apparently, the local changes caused by the replacement of surface charges can affect the flexibility of the protein at the vicinity of the shaft. Other kinds of mutations, where the large hydrophobic residues that surrounded the shaft were replaced by smaller ones (Dioumaev et al., 2001), resulted in even smaller rates of proton transfer to D96. These peculiar observations imply that the free space vacated by the replacement of a large residue by a smaller one can collapse spontaneously under the pressure applied by surface tension ascribed to a cavity in a protein (Kharakoz, 2000). This can be taken as an indication that the internal motion of the heavy atoms inside the protein can translate the void from one site to the other, in reminiscence of propagation of fractures in a solid crystal.
In conclusion, the crucial step needed for the completion of the photocycle requires a transient, short time solvation of the hydrophobic domain that plugs the passage of proton between D38 and D96. Considering the energy expenditure needed for the enlargement of the shaft to be wide enough to contain few water molecules, the protein can balance its energy budget by withdrawing free space from nearby existing cavities. Thus, the intraprotein cavities can function as a lubricant that facilitates the relative motion of side chains or helices, as well as available free space that can be redistributed inside the protein. To evaluate whether such a mechanism is applicable for the conformation changes in the cytoplasmic section of the protein, we searched the structures of the protein, looking for small cavities located in the immediate vicinity of the shaft.
The role of permanent cavities in protein was first discussed by Rashin and Honig (Rashin et al., 1986). Due to the van der Waals repulsion, even in densely packed proteins there is always some free space between the atoms. Thus, a hypothetical particle with radius of ∼0.4 Å can permeate freely between the atoms of a protein (Rashin et al., 1986). In practice, the packing of the residues in a protein is looser and the cavities vary in size, so that some are large enough to contain one or more solvent molecules. Besides the permanent cavities whose location is derived from the crystal structure, cavities can be generated by the thermal fluctuations of a protein. Molecular dynamics simulations, carried out with soluble proteins (Kocher et al., 1996), indicated that the apolar domains near the surface of a protein have the highest tendency to form small cavities as part of the thermal motion. Accordingly, we might expect that the hydrophobic domain surrounding the shaft would exhibit high flexibility where the shaft can expand at the expense of nearby cavities. For this reason we have carried out a comparative study of the cavities located in the vicinity of the shaft, searching for the presence of cavities whose location is persistent in the resolved structures of bacteriorhodopsin.
The definition of intraprotein cavities is attained through programs that search for spaces large enough to accommodate a probing sphere of a selected radius. A cavity is defined when the probe sphere cannot access the void from the bulk. This definition is somewhat inappropriate in the present case, as an intraprotein free space that can function as a lubricant does not have to be disconnected from the bulk. Accordingly, when in some structures a cavity is missing, it should be examined whether the cavity had shrunk in size or, due to a minor motion of a few atoms, the cavity was opened to the bulk (in this text we shall use the terminology of cavity versus cave). For this reason we preferred to depict the cavities by the atoms that wrap the free space. This formalism also allowed us to present the boundary also in cases where the cavity was transformed into a cave, which was undetectable as a cavity as it had become accessible to the bulk, although it still functioned as a void in the protein.
METHODS
Model structures
The bacteriorhodopsin model structures used in this work are the PDB files detailed in Table 1. All models were taken from the Protein Data Bank (Berman et al., 2000). Six of the structures are of the Br state. Two are of the M state: 1CWQ (chain B; Sass et al., 2000) is in a late M configuration, whereas 1F4Z (Luecke, 2000) is an early M state protein generated by illumination of the E204Q bacteriorhodopsin mutant. Four out of the nine structures were of the WT protein in its Br state. Each of these four structures was determined by a different research group.
TABLE 1.
The protein structures that were used in this study
| Structure (PDB code) | Chain (monomeric unit) | State | Mutation | Reference | Resolution (Å) | Missing residues | Residues with missing atoms | Space group | Crystal cell (Å) |
|---|---|---|---|---|---|---|---|---|---|
| 1BRR* | C | Br | Essen et al., 1998 | 2.9 | 233–249 | 227, 229, 230, 232 | C 1 2 1 | 120.520 105.960 80.190 | |
| 1C3W | Br | Luecke et al., 1999a | 1.55 | 1–4, 157–161, 232–249 | P 6 3 | 60.631 60.631 108.156 | |||
| 1CWQ† | A | Br | Sass et al., 2000 | 2.2 | 0, 1, 240–249 | P 6 3 | 61.080 61.080 110.400 | ||
| 1CWQ‡ | B | M | Sass et al., 2000 | 2.2 | 0, 1, 240–249 | P 6 3 | 61.080 61.080 110.400 | ||
| 1C8R | Br | D96M | Luecke et al., 1999b | 2 | 1–4, 157–161, 232–249 | P 6 3 | 60.631 60.631 108.156 | ||
| 1F4Z | M | E204Q | Luecke et al., 2000 | 1.8 | 1–4, 157–161, 232–249 | P 6 3 | 60.631 60.631 108.156 | ||
| 1F50 | Br | E204Q | Luecke et al., 2000 | 1.7 | 1–4, 157–161, 232–249 | P 6 3 | 60.631 60.631 108.156 | ||
| 1JV7 | O | D85S | Rouhani et al., 2001 | 2.25 | 1–8, 64–77, 233–249 | C 2 2 21 | 51.800 121.300 85.700 | ||
| 1QHJ | Br | Belrhali et al., 1999 | 1.9 | 1–4, 233–249 | 163, 227, 232 | P 6 3 | 60.800 60.800 110.520 |
Noted throughout this article as 1BRRC.
Noted throughout this article as 1CWQA.
Noted throughout this article as 1CWQB.
Detection of cavities, volume, and surface calculations
The volumes and molecular surface areas of the nine protein structures listed in Table 1 and the locations, sizes, and surface areas of the cavities were calculated by the VOLBL program (Liang et al., 1998a,b). For the purpose of the analysis, only the heavy atoms were included. The heteroatoms, including the retinal, were removed. Atomic radii parameters were taken from the default set of the VOLBL program. The calculations were carried out using a probe sphere with a radius of 1 Å.
Addition of hydrogen atoms to the protein
The effect of addition of hydrogen atoms on the number, size, and shape of cavities was investigated by a comparison of these parameters as calculated for the 1BRR (subunit C) structure (Essen et al., 1998). First, all missing atoms were added to the incomplete residues (for the missing atoms and residues see Table 1) using the Biopolymer program (Accelrys, San Diego, CA). Hydrogen atoms were then added by the HBUILD program in CHARMM (Brooks et al., 1983). The van der Waals collisions were removed by 500 steps of energy minimization using the steepest-descents algorithm implemented in the CHARMM program. During these calculations, the shape and the size of the retinal-binding cavity was maintained constant by restraining the motion of the atoms lining the cavity.
The illustrations presented in this manuscript were generated by the VMD program (Humphrey et al., 1996; http://www.ks.uiuc.edu/Research/vmd/).
RESULTS AND DISCUSSION
Comparison of volume between different BR models
Table 2 lists the cavity inventory of the nine protein structures detailed in Table 1. The data refer to the volume of the protein, the number of cavities, and the cavities' total volume. The selected structures were determined and refined by leading research groups and the coordinates were taken as is, without considering the coordinate error. As is detailed in Table 2, the crystals were grown under two main conditions: epitaxis on benzamidine (Essen et al., 1998), yielding a crystal that preserves the trimer arrangement as in the Purple Membrane, or in the cubic phase of phospholipid (Landau and Rosenbusch, 1996; Nollert et al., 2001). The crystals also differ in their space group and in the unit cell dimension. Accordingly we can expect that the forces operating on the protein molecules during the crystal growth differed between the samples.
TABLE 2.
Comparison of total volume of the bacteriorhodopsin molecule, number of cavities, and sum of their volumes as calculated for nine crystalline structures of the protein
| PDB | Mutation | State | Mode of crystallization | Total volume | Number of voids | Total volume of voids |
|---|---|---|---|---|---|---|
| 1BRRC | WT | Br | Epitaxis on benzamidine | 29,084 | 51 | 1,617 |
| 1C3W | WT | Br | Cubic phase lipids | 27,940 | 41 | 1,408 |
| 1CWQA | WT | Br | Cubic phase lipids | 29,806 | 46 | 1,587 |
| 1QHJ | WT | Br | Cubic phase lipids | 28,691 | 42 | 1,492 |
| 1C8R | D96M | Br | Cubic phase lipids | 27,918 | 38 | 1,386 |
| 1F4Z | E204Q | Br | Cubic phase lipids | 27,812 | 32 | 1,214 |
| 1CWQB | WT | M | Cubic phase lipids | 29,005 | 30 | 1,323 |
| 1F50 | E204Q | M | Cubic phase lipids | 27,679 | 36 | 1,121 |
| 1JV7 | D85S | O | Cubic phase lipids | 26,094 | 39 | 796 |
The volume and surface of the cavities are given in Å3 and Å2 units, respectively.
Comparison of the total protein volumes reveals small variations in the total volume parameter (28,492 ± 719 Å3). One should note that although the 1BRR structure was crystallized under crystallization conditions that differ from all others, its volume is very similar to that of the proteins from the other structures. In contrast to the uniformity of the protein's volume, the numbers of cavities that are large enough to accommodate a probe particle with a radius of 1 Å vary dramatically between the structures, as does the total space of the cavities. This difference means that one should be very cautious with the assumption that cavities are reproducible structural elements. For this reason, we have decided not to treat the distribution of the cavities in a structure as a statistical population but to examine to what extent the cavities occupy analogous positions in the different protein models. Upon examination of the individual cavities, it was found that the largest cavity is the retinal binding site, and the smallest ones have a volume of ∼4.5 Å3.
The cytoplasmic cavities of the trimeric structure
Evaluation of the persistency of the cavities in the various structures can be achieved by screening for their presence in the reported structures. In this study we limited the search for cavities located next to the shaft connecting D96 and D38. Accordingly, of the many cavities detected in the structures, we concentrate on those that are detailed in Table 3. Table 3 features cavities from four structures, all of which are in the ground state form (Br). We have used structures that were determined by four different research groups in our search for the reproducibility of their presence.
TABLE 3.
The volume, surface area, and volume/area ratios are given for the cavities on the cytoplasmic part of the protein (as presented in Fig. 2) of the structures 1BRRC, 1CWQA, 1QHJ, and 1C3W
| 1BRRC | 1CWQA | 1QHJ | 1C3W | ||
|---|---|---|---|---|---|
| 1 blue | Volume | 89 | 81 | 82 | 74 |
| Area | 131 | 125 | 129 | 112 | |
| Ratio | 0.68 | 0.65 | 0.64 | 0.66 | |
| 2 white | Volume | 72 | 0* | 0* | 60 |
| Area | 119 | 0* | 0* | 95 | |
| Ratio | 0.61 | N/A* | N/A* | 0.63 | |
| 3 green† | Volume | 50 | 58 | 16 | 11 |
| Area | 83 | 96 | 33 | 25 | |
| Ratio | 0.60 | 0.60 | 0.48 | 0.44 | |
| 4 red | Volume | 33 | 0* | 0* | N/A‡ |
| Area | 58 | 0* | 0* | N/A‡ | |
| Ratio | 0.57 | N/A* | N/A* | N/A‡ | |
| 5 orange | Volume | 31 | 28 | 32 | 25 |
| Area | 56 | 62 | 54 | 46 | |
| Ratio | 0.55 | 0.45 | 0.59 | 0.54 | |
| 6 purple | Volume | 25 | 4.3 | 0§ | 0§ |
| Area | 47 | 13 | 0§ | 0§ | |
| Ratio | 0.53 | 0.33 | N/A§ | N/A§ | |
| 7 yellow† | Volume | 24 | 58 | 29 | 41 |
| Area | 46 | 96 | 52 | 73 | |
| Ratio | 0.52 | 0.60 | 0.56 | 0.56 | |
| 8 lilac | Volume | 14 | 12 | 12 | 8.5 |
| Area | 30 | 26 | 27 | 21 | |
| Ratio | 0.47 | 0.46 | 0.44 | 0.40 | |
| 9 tan¶ | Volume | 19 | 0* | 0* | 27 |
| Area | 41 | 0* | 0* | 48 | |
| Ratio | 0.46 | N/A* | N/A* | 0.56 | |
| 10 silver¶ | Volume | 14 | 0* | 7 | 27 |
| Area | 33 | 0* | 18 | 48 | |
| Ratio | 0.42 | N/A* | 0.39 | 0.56 |
The volume and surface of the cavities are given in Å3 and Å2 units, respectively.
In this structure, the cavity is open to the bulk (N/A, not applicable).
The yellow cavity and the green cavity are joined into a single void in 1CWQA.
The atoms that create the cavity are missing from the structure.
In this structure, the cavity is too small to be detected using a probe radius of 1Å.
The tan cavity and the silver cavity are joined in 1C3W.
The location of the cavities in the cytoplasmic section of the trimeric structure 1BRR is presented in Fig. 2. The cavities are defined by the heavy atoms that enclose the void as determined for subunit C (subunit C was selected because the average temperature factor of its atoms was lower than corresponding values obtained for subunits A and B). Each of the cavities is presented in a different color and the volume, surface area, and ratio between the two parameters (reflecting its shape) are all listed in Table 3.
FIGURE 2.
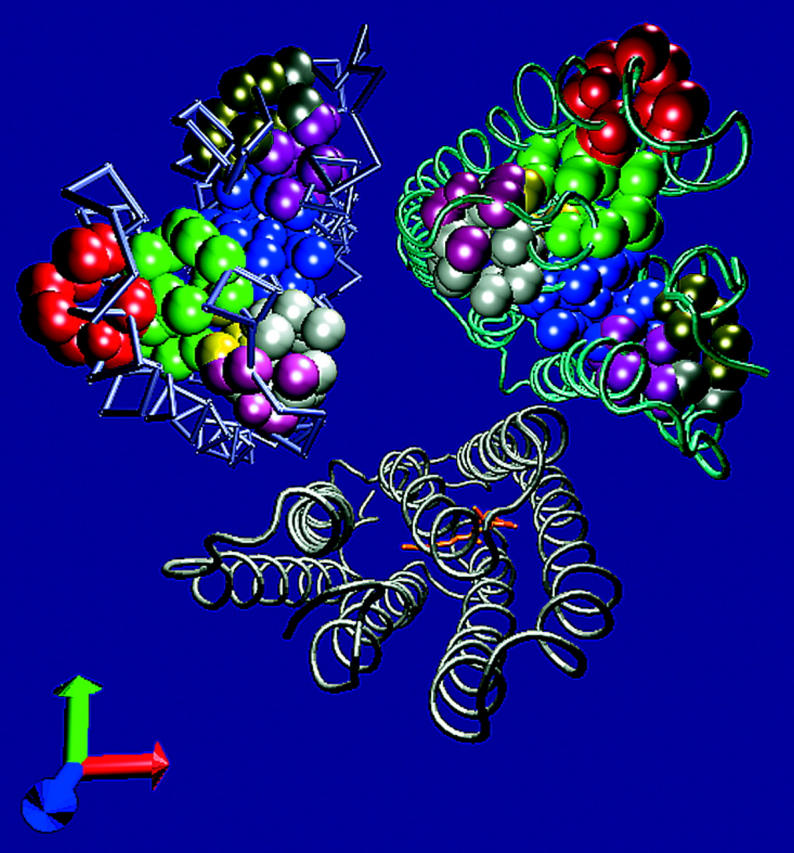
The cavities on the cytoplasmic side of the BR. The atoms, which border the cavities of the cytoplasmic side of the BR, are presented as VDW spheres on the helical structure of the protein trimer structure 1BRR. The cavities, which were found on subunit C, are presented on subunits B and C, whereas only the retinal residue is presented on subunit A for orientation. See Table 3 for the properties of the cavities.
The effect of adding hydrogen atoms to the cavity inventory
The absence of hydrogen atoms in the refined x-ray diffraction structures of protein reflects the inability to determine their precise locations. Thus, the PDB files do not specify the location of the hydrogen atoms. Addition of hydrogen atoms will naturally reduce the size of the intraprotein cavities, and some of the smaller ones are expected to vanish. Thus, before evaluating whether cavity deformation participates in conformation transitions, we have tested to what extent the addition of the hydrogen atoms affect the cavity inventory. The structure represented by the 1BRRC model was selected for this purpose as it has the minimal number of missing atoms and residues, and the results are presented in Table 4.
TABLE 4.
A comparison between the 1BRRC structure before and after the addition of hydrogens to the model
| 1BRRC (no H) | 1BRRC (with H) | ||
|---|---|---|---|
| Total number of cavities | 51 | 39 | |
| Total volume of cavities | 1,617 | 1,243 | |
| Total surface area of cavities | 2,475 | 1,961 | |
| 1 blue | Volume | 89 | 67 |
| Area | 131 | 113 | |
| Ratio | 0.68 | 0.59 | |
| 2 white | Volume | 72 | 43 |
| Area | 119 | 79 | |
| Ratio | 0.61 | 0.54 | |
| 3 green | Volume | 50 | 26 |
| Area | 83 | 53 | |
| Ratio | 0.60 | 0.49 | |
| 4 orange | Volume | 31 | 21 |
| Area | 56 | 39 | |
| Ratio | 0.55 | 0.54 | |
| 5 red | Volume | 33 | 6.8 |
| Area | 58 | 19 | |
| Ratio | 0.57 | 0.36 | |
| 6 purple | Volume | 25 | 30 |
| Area | 47 | 53 | |
| Ratio | 0.53 | 0.57 | |
| 7 yellow | Volume | 24 | 7.1 |
| Area | 46 | 18 | |
| Ratio | 0.52 | 0.39 | |
| 8 lilac | Volume | 14 | 4.4 |
| Area | 30 | 13 | |
| Ratio | 0.47 | 0.34 | |
| 9 tan | Volume | 19 | 13 |
| Area | 41 | 29 | |
| Ratio | 0.46 | 0.45 | |
| 10 silver | Volume | 14 | 30 |
| Area | 33 | 53 | |
| Ratio | 0.42 | 0.57 | |
The volume and surface of the cavities are given in Å3 and Å2 units, respectively.
The purple and the silver cavities are joined into a single cavity in the 1BRRC structure, where the hydrogens are present, due to the energy minimization procedure.
After the hydrogen addition, the number of cavities dropped from 51 to 39 and their total volume was reduced by ∼22%. Thus, the fully hydrogenated protein still contains a large number of small cavities. A detailed inspection of the cavities surrounding the shaft at the cytoplasmic side reveals an inconsistent variation of size and surface. The volumes of cavities 1, 2, 3, 4, and 9 were reduced by ∼30–40%, as expected from the introduction of more nuclei into the cavities space. Cavities 5, 7, and 8 were almost eliminated, indicating that the insertion of hydrogen atoms caused local conformation rearrangements. The distortion introduced by the addition of the hydrogen atoms made cavities 6 and 10 merge into one unified cavity. Ideally, we would assume that the addition of hydrogen atoms, after a short relaxation to release the van der Waals interaction, would have no effect on the location of the heavy atoms. Apparently, the coordinates of the heavy atoms derived by the refinement are affected by the stress introduced through the addition of the hydrogen atoms to the structure. Even the short minimization process we had carried out already caused a shift of the heavy atoms from the initial coordinated with a total RMSD of 0.33 Å (the RMSD of the Cα atoms was only 0.1 Å). It should be mentioned that the cavities listed in Table 3 were detected both in the original structure and after the addition of the hydrogen atoms. Still, for the sake of adherence to the original information, we shall limit the present discussion to the heavy atom model of the proteins, with no attempt to improve the presentation by addition of hydrogen atoms.
Comparison between the cavities on the cytoplasmic side of the bacteriorhodopsin
Fig. 3 depicts the four ground state structures of bacteriorhodopsin. Each structure is presented from two sides with the cytoplasmic section on the top of the frame. The orientation can be deduced from the retinal molecule where the ring points either to the left or to the right. The cavities are presented with the same color code as in Fig. 2. As clearly seen in the figure, and detailed in Table 3, most of the cavities are persistent in the four structures but vary in their volumes and surface areas. Sometimes a specific cavity appears to be missing from the structure. The fate of the missing cavities can be resolved by tracing the heavy atoms that define the cavities. Thus, using the atoms which border the cavities in the 1BRRC structure, we could deduce that cavities 2, 4, 9, and 10 are missing from some structures because the heavy atoms that define their borders are slightly ajar and the program failed to recognize them as closed cavities. This is demonstrated in Fig. 4 where an ∼1-Å shift in the relative positions of the heavy atoms leads to an exposure of the inner space of the cavity to the bulk. The fact that the program was not able to define this void as a cavity has no effect on its projected function as an available free space needed to support conformational changes. For this reason, we regard this void as a possible participant in the mechanism leading to the shaft expansion procedure, needed for the reprotonation of D96.
FIGURE 3.
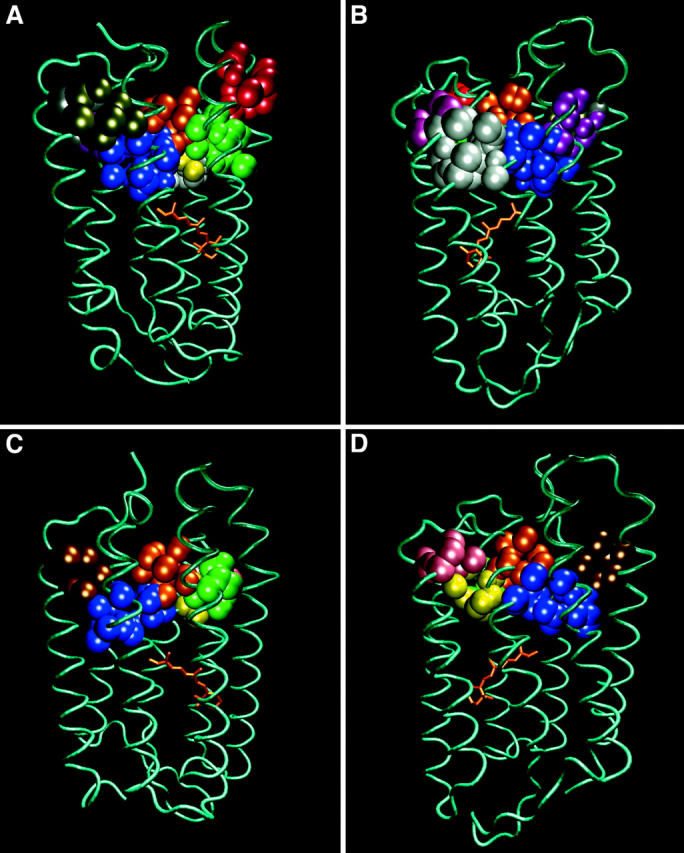
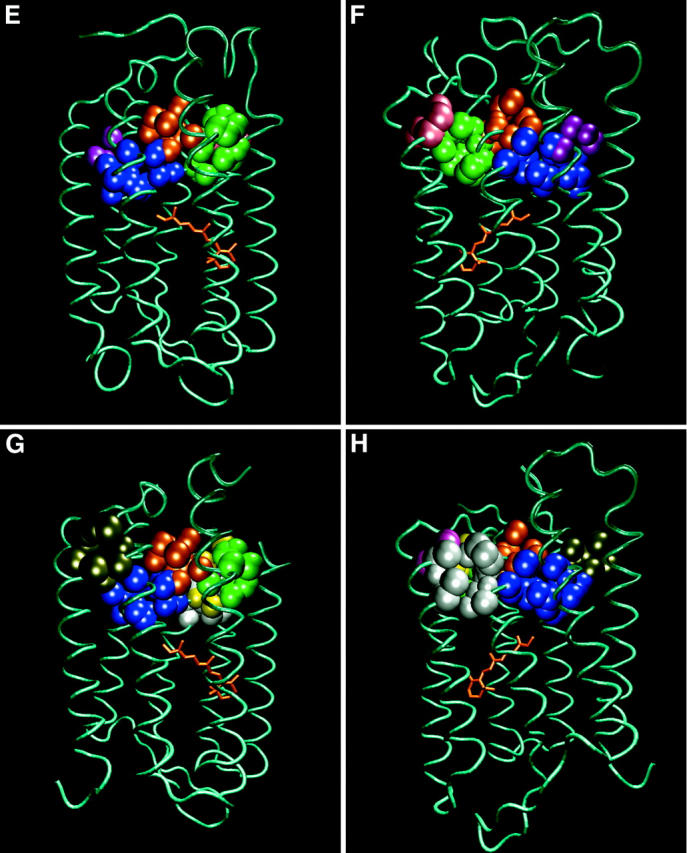
A comparison of the shapes and sizes of the 10 cavities located in the cytoplasmic section of bacteriorhodopsin, as calculated for four ground state structures of the protein. Each pair of frames depicts the position of the cavities, colored as defined in Table 2. The orientation of the protein can be deduced by the location of the retinal molecule. Frames A and B: 1BRRC. Frames C and D: 1QHJ. Frames E and F: 1CWQA. Frames G and H: 1C3W.
FIGURE 4.
A detailed presentation of cavity 4 in the 1JV7 (frame A) and 1QHJ (frame B) structures. Please note that the relative motion of a few atoms on the surface of the cavity exposes its inner space to the bulk. Residues L174 (of helix F, located to the left of the cavity in the figure), F154 (of helix E, located to the right of the cavity in the figure), and Y131 (found at the extracellular origin of helix E) are presented on the model. The retinal cofactor is shown in orange.
The flexibility of the cavities and their varying sizes and shapes with the crystallization conditions can be demonstrated on inspection of cavity 6, which shrinks from 25 Å3 in 1BRRC to 4.3 Å3 in 1CWQA. In the other Br structures (1QHJ and 1C3W), the heavy atoms are too close together for the accommodation of a 1-Å probe. The reshaping of the cavities due to the mild forces operating during the crystal formation can lead to merging of cavities. For example, cavities 3 and 7 are well-separated voids in three crystal structures although they are joined into a single space in 1CWQA. Similarly, cavities 9 and 10 are merged in the 1C3W structure.
On the basis of these observations we can deduce that the cavities are persistent features of the structure, as they appear to be located in comparable locations. However, the cavities have nonidentical properties. These variations probably reflect the yielding of the cavity's shape under the shear forces applied to the protein during the crystallization. To summarize, by adopting the terminology borrowed from computer science we can state that the cavities are “features” of the protein rather than “bugs” introduced into the structure during the crystallization process. In the following sections, we shall discuss how these cavities contribute to the functionality of the protein.
Modulation of the cavities during the photocycle
The presence of flexible elements in the structure, as ascribed to the cavities, implies that their sizes and shapes vary in parallel to the structural modulations associated with the photocycle. This projection was tested first for the WT protein, as revealed by the structure of the two states reported by Sass and co-workers (1CWQ; see Sass et al., 2000, and the supplement of the article).
The transition of the protein from the Br to the M state is associated with an enhanced solvation of the cavity where the carboxylate of D96 is located. In the expanded cavity the carboxylate is solvated and its pK is reduced, thus becoming a suitable proton donor to the Schiff-base (Brown et al., 1995; Cao et al., 1991; Weik et al., 1998). Besides the modulation of this cavity, other cavities are also altered during the Br → M transition. Of the six shaft-surrounding cavities present in the Br state 1CWQA (see Table 3), only four vary during the photocycle (see Table 5). Two of the cavities (3 and 8, located between helices C, D, E, and F) diminished in size: cavity 8 had vanished whereas cavity 3 lost ∼50% of its volume and surface area. In parallel, cavities 5 and 6 (located between helices B, G, and F and A, B, and G, respectively) expanded significantly, increasing their volume by ∼4-fold. It should be pointed out that cavity 5 became more hydrophilic.
TABLE 5.
The volume and surface area of cavities that have changed during the Br → M transition in the 1CWQ model
| Br (1CWQA) | M (1CWQB) | |||||
|---|---|---|---|---|---|---|
| Cavity | Volume | Surface area | Boundary helices | Volume | Surface area | Boundary helices |
| 8 lilac* | 12 | 26 | C, D, F | 0 | 0 | |
| 3 green† | 58 | 96 | C, D, E, F | 23 | 44 | C, D, E |
| 5 orange‡ | 28 | 62 | B, G, F | 121 | 179 | B, C, F, G |
| 6 purple§ | 4.3 | 13 | A, B, G | 19 | 37 | B, G, |
The volume and surface of the cavities are given in Å3 and Å2 units, respectively.
Contributing atoms: 1CWQA: Helix C:A98:CB; Loop CD:A103:CB; Helix D: T107: C, CG2; T108:N, CA, CG1; L111:CG1; Helix E:F156:CE1. In 1CWQB this cavity becomes too small to be detected with a probe radius of 1 Å.
Contributing atoms: 1CWQA: Helix C: L94:CA, O, CB, CG, CD1, CB; L97:CD1; A98:N, CB; Helix D: L111:CD1; L152:CB, CG, CD1, CD2; Helix E: F156: CE2, CZ; F171:CE1, CZ; Helix F: R175:CB, CG, CD.
1CWQB: Helix C; L97:CD2; Helix D: L152:CD2; Helix E: F171:CE1, CZ; L174: C, CB; R175: CA, CB; T178:OG1.
Contributing atoms: 1CWQA: Helix B: F42:CE1; Helix C: D96:C, O, CB; L97:CA, CD2; L100:CG, CD; Helix F: L174:CD1; F219:CZ; L223:CD1, CD2.
1CWQB: Helix B: F42:CD1, CE1; Helix C: D96:C, O, CB, CG; L97:CA, CD2; L100:CB, CD1; Helix F: T170:CG2; F171:CD1; L174:CD1, CD2; Helix G:F219:CE2; L223:CA, O, CB, CD1, CD2; S226:O, CB, OG; A228:N, CB; I229:N, CG2.
Contributing atoms: 1CWQA: Helix A: F27:CB; Helix B: T46:C; T47:OG1; Helix G: L224:CD1. 1CWQB: Helix A: F27:CB, CD1; Helix B: Y43: CA, O, CD1; T46:CB, CG2; T47:OG1 Helix G: L224:CD1.
The other set of successive photocycle structures is presented in Table 6. In this case, the proteins under study are mutants whose structures were specifically modulated to introduce a rate-limiting step in the photocycle so that, upon illumination, a specific photointermediate will accumulate. Naturally, such mutations alter the structure of the protein even in the Br state. These results therefore are separately discussed.
TABLE 6.
The volumes of six cavities in the cytoplasmic side of BR structures 1F50 (Br state), 1F4Z (early M state), and 1JV7 (O-like state)
| BR (1F50) | M (1F4Z) | O (1JV7) | |||||
|---|---|---|---|---|---|---|---|
| Color* | Volume | Boundary helices | Volume | Boundary helices | Volume | Boundary helices | |
| Blue† | 1 | 71 | A, B, C, G | 93 | A, B, C, G | 95 | A, B, C, G |
| Green‡ | 3 | 18 | C, E, F | 66 | C, E, F | 53 | C, E, F |
| Orange§ | 4 | 23 | B, C, F, G | 27 | C, F, G | 34 | B, C, F, G |
| Purple¶ | 6 | Too small | – | 14 | A, B, G | – | – |
| Yellow‖ | 7 | 21 | C, D, E | Fractured | – | 19 | C, D, E |
| Lilac** | 8 | 9.7 | C, D | 7.5 | D, E | Fractured | – |
Bold numbers correspond with cavities that expanded significantly during the Br → Early M transition. The volume is given in Å3 units.
Color of the corresponding cavities in Fig. 2
Contributing atoms: 1F50: Helix A: F27:CE2; Helix B: I45:CG2; T46:CA, O, CG2; V49:CB, CG1, CG2; P50:CD; Helix C: L93:CA, CD2; D96:OD2; Helix G:K216:CA, C, O; F219:CD1; G220:CA. 1F4Z: Helix A:F27:CE2; Helix B: I45:CG2; T46:CA, O, OG1, CG2; V49:CB, CG1, CG2; P50:CD; Helix C: L93:CD1; D96:OD2; Helix G:A215:O; K216:CA, C, O; F219:CB, CD1;G220:N, C; L223:CB, CD2. 1JV7: Helix A: F27:CE1; Helix B: I45:CG2; T46:CA, O, CG2; V49:CB, CG1, CG2; P50:CB, CG, CD; Helix C: L92:C, O, CA, CD2, CB, OD1; Helix G: K216:CA, C, O, CB; V217:CA, CD2, CE2; G220:CA; L223:CD2.
Contributing atoms: 1F50: Helix C: L97:CD1; Helix E: L152:CB, CD1, CD2; Helix F: F171:CE1; R175:CA, CB, CG. 1F4Z: Helix C: L97:CD1, CD2; Helix E: L149:CA, O, CD2; L152:CB, CD1, CD2 F153:N, CA, CB; Helix F: F171:CE1, CZ, CA; R175:O, CB, CG; T178:CB, CG2; V179:CG2. 1JV7: Helix C: L97:CG, CD1, CD2; Helix E: L152:CB, CD1, CD2; F171:CD1, CE1, CZ; L174:C, CB; Helix F: R175:N, CA, O, CB, CG; T178:CB, CG2; V179:CG2.
Contributing atoms: 1F50: Helix B: F42:CD1, CE1; Helix C:D96:C, O, CB; L97:CD2;L100:CD1, CD2; Helix F: L174:CD1; Helix G: F219:CZ; L223:CD1, CD2. 1F4Z: Helix C: D96:C, O, CB; L97: CD2; L100:CD2; Helix F: L174:CD1; Helix G: F219:CE1, CZ, CD1; L223:CD2. 1JV7: Helix B: F42:CD2, CE2; Helix C: D96:C, O, CB; L97:CA, CD1; L100:CG, CD1; Helix F: L174:CD1; Helix G: F219:CE2, CZ; L223:CD1, CD2.
Contributing atoms: 1F4Z: Helix A: F27: CB; Helix B: Y43:CA, O, CD1; T46:C, CB, OG1; Helix G: L224:CD1. In 1F50 and 1JV7, this cavity becomes too small to be detected with a probe radius of 1 Å.
Contributing atoms: 1F50: Helix C: L94:CA, O, CB, CD2; L97:CB; A98:N, CA, CB; Helix D: L111: CD1; Helix E: L152:CD2; F156:CE2, CZ. 1JV7: Helix C: L94:CA, O, CB, CD2; L97:CB; A98:CB; Helix D: L111:CD1; Helix E: L152:CD2; F156:CE2. In 1F4Z, this cavity is accessible to the bulk, and hence is not detected by our screening.
Contributing atoms: 1F50: Helix C: A98:CB; Loop: A103:CB; Helix D: T107:C, CG2; I108:N, CA, CG1; L111:CD1. 1F4Z: Loop: A103:CB; Helix D: T107:C, CG2; I108:N, CA, CG1; L111: CD1; Helix E: F156:CZ. In 1JV7, this cavity is accessible to the bulk, and hence is not detected by our screening.
The formation of the early M state is associated with a 3-fold expansion of cavity 3, located between helices C, E, and F. In the same way cavity 6, which is too small to accommodate the 1-Å probe in the Br state of the E204Q mutant, expanded to 14 Å3 in the early M state. In the O-like state structure, 1JV7, cavity 3 seems to vanish again. However, we cannot state for sure that it is not a contribution of the D85S mutation to the structure of the protein.
The other two cavities, 7 and 8, seem to retain a constant size during the photocycle, but as the integrity of their surface was lost, they are better defined as caves.
The results presented in Table 5 confirm that during the photocycle the protein undergoes conformational changes that affect the size and integrity of the cavities.
Energetic considerations involved in the passage of charge through the shaft
The passage of a positive charge through the shaft is associated with energy barriers that vary with the mechanism. Insertion of a unit charge 5–10 Å below the surface of a low dielectric matrix sphere (r ∼20 Å, comparable with the general size of bacteriorhodopsin) necessitates, as calculated according to Gilson et al. (1985), an investment of 33 kcal/mol. This is an extremely high energy barrier, implying that the mechanism of the reaction must involve a lower barrier.
The temporal solvation of the shaft will reduce the electrostatic barrier by the interaction of the water molecules with the proton, but this calls for investment of energy required for the expansion of the shaft. In the following section we assume that the dimension of the shaft, as evaluated from the crystal structures, is a fair approximation of its cross section in solution. To allow water molecules to enter the shaft, its radius should increase from 1 Å up to R ≥ ∼1.6 Å. For a shaft 10-Å long that expansion will generate ∼30 Å2 of new surface, necessitating a work against the surface tension and the rigid structure of the protein. Using the values published by Kharakoz (2000) and Kocher et al. (1996), we can estimate that the needed work is 10–20 kcal/mol. This mechanism appears as a better alternative than the insertion of a dry proton into the protein's low dielectric matrix, but it is still an appreciable barrier. Both mechanisms seem to call for a very high investment of energy, making the reaction almost impossible. Naturally, if the protein structure would allow the expansion of the shaft at the expense of constriction of nearby cavities, the mechanism will be much more favorable. Accordingly, we can consider the cavities near the shaft as transferable free space, which can be shifted inside the protein, thus facilitating the expansion of the shaft. This hypothesis implies that upon application of external pressure the intraprotein cavities will collapse and the propagation of the proton in the shaft will be delayed.
The packing of the atoms in proteins avoids van der Waals collisions. Thus, there is always some free space between the atoms. Precise calculations carried out by Eilers et al. (2000) indicated that the packing value, a parameter that quantitates the leeway between the nearby residues, depends on the general features of the residues and on the nature of the protein (globular versus transmembranal). In transmembranal helices, the packing value of hydrophobic residues is smaller than that of polar ones. A similar tendency of hydrophobic residues to be loosely packed was determined by molecular dynamics simulations (Kocher et al., 1996). It was noted that the energy barrier for the generation of a spherical cavity in a protein varies with the general features of its local domain. Hydrophobic domains located close to the protein's surface require a lower energy investment compared to polar domains located in the interior of the structure. The hydrophobic nature of the region of the protein that surrounds the shaft, and its location close to the protein-water interface, render this region suitable for modulation of existing cavities during the dynamic cycle of the protein.
The above conjuncture had been corroborated by the recent observations of Klink and co-workers (Klink et al., 2002) who monitored the effect of external pressure on the dynamics of the photocycle. In their measurements the authors recorded the dynamics of the photocycle and reconstructed it as a sum of nine exponents that vary in their half-life times from a microsecond up to tens of milliseconds. This mode of analysis is capable of reconstructing the intermediate spectrum of the sample at any given time after excitation, yet these terms are macroscopic descriptors and cannot be associated with a defined chemical step. Thus, we shall refer to the time constant as representation of a process.
The kinetic analysis indicated that the dynamics of the process with the shortest time constant (τ1 = 0.7 μs measured at 1 bar 25°C) varied monotonically with the applied pressure (ln(τ) vs. pressure), yielding an activation volume ΔV* = RT(∂ln(τ)/∂p)T = 19 Å3/molecule. The following five macroscopic time constants (τ2 = 7 μs, τ3 = 37 μs, τ4 = 100 μs, τ5 = 370 μs, and τ6 = 1.3 ms) are practically unaffected by external pressure. The last three processes (τ7 = 3.9 ms, τ8 = 7.3 ms, and τ9 = 40 ms) exhibited a sharp dependence on pressure; the first two (τ7 and τ8) deviated from a linear function, whereas τ9 exhibited a monotonic dependence with ΔV* = 14 Å3/molecule.
Of special interest are the two nonlinear processes corresponding with the time constants τ7 and τ8. Up to ∼1,500 bar the two processes were slowed down by the pressure, with apparent activation volumes of ∼40 Å3/molecule each. However, at higher pressure, the processes are pressure insensitive. The time frames in which two processes are detected correspond to the time window where the proton transfer through the shaft is expected to take place. This pressure dependence can be explained as a gradual collapse of the intraprotein cavities, leading to a diminishment of the free space that can be available for the expansion of the shaft. At pressure exceeding 1,500 bar, the collapse had consumed the free space reservoir needed for the shaft expansion so that higher pressure has no effect on the process. It should be mentioned that the last macroscopic process (τ9 = 40 ms 1 bar, 25°C) is also pressure sensitive, indicating that the last phases in the relaxation of the photocycle involve some more free space shuffling besides the shaft expansion. According to Klink and co-workers (Klink et al., 2002), the dependence of the macroscopic dynamics descriptors on the pressure is milder at 40°C than at 25°C, except for τ7 and τ8. For these two processes, the saturating pressure (the value where the rate becomes pressure insensitive) has increased to 2,000 bar (at 40°) and the activation volumes changed to 80 Å3 and 50 Å3 respectively. It is of interest to point out that, even at high pH values (pH = 10; Varo and Lanyi, 1995), where the rate-limiting step of the photocycle is the protonation of D38 (the kinetics were measured ∼4-pH units above the pK of D38) rather than the proton transfer through the shaft, the kinetics exhibit some dependence on the activation volume: the N(−1) → N(0) transition in which the N state gains a proton from the bulk exhibits an activation volume of 15.6 ml/mol (26 Å3/molecule).
The compressibility coefficient of the Purple Membrane was determined to be β = 26.7 Mbar−1 (Marque et al., 1984). Considering that the protein is the dominant component of the Purple Membrane, and that β for phospholipid is almost twice as large (Marque et al., 1984), we can assume that the reported values well represent the properties of the protein. This value serves as in the upper limit of the compressibility coefficients of proteins as compiled by Kharakoz (β = 10–25 Mbar−1; Kharakoz, 2000).
Accordingly, under external pressure of ∼1,500 bar the protein will lose ∼2% of its total volume, which amounts to ∼550 Å3. This value is ∼30–40% of the total volume taken by the cavities in the protein (see Table 2). Thus the collapse of the free space in the protein, as estimated for the whole protein, exceeds the volume of the cavities near the shaft. The contribution of the void shuffling to the overall energy barrier can be estimated from the extent of the slowdown of τ7 and τ8. According to the reconstructing procedure (Klink et al., 2002), the overall process is a set of sequential reactions; thus the pressure effect on the photocycle at its saturating level (1,500 bar at 25°) delays the photocycle by ∼65-fold, which is equivalent to an energy barrier of 2.5 kcal/mol. We suggest that a collapse of transferable free space in the vicinity of the shaft below a threshold level eliminates an accessory mechanism that lowers the activation energy by at least 2.5 kcal/ mol.
We have demonstrated in this study that the immediate vicinity of the shaft is surrounded by small cavities, and few of them indeed change size and shape during the photocycle. The shaft expansion is most likely to be a very brief event; generated at low frequency during the thermal fluctuation of the protein the lifetime of the expanded shaft is probably very short. Yet, the efficiency of proton transfer along the short stretch of the shaft is very high, and the time needed for the reaction is in the order of a nanosecond or less. Thus, any opportunity of shaft expansion can yield a productive step, given that D38 is loaded with a proton.
The proposed mechanism of the shaft expansion is most probably driven by mobilization of free space from the nearby cavities, but an alternative mechanism may be operating based on a coherent (cooperative) motion of the protein within the trimeric packing in the Purple Membrane. The cooperativity within the trimers had been noticed on measuring the optical unsiothropy of Purple Membrane preparations; it is manifested up to the late phases of the photocycle (Bauer et al., 1976; Tokaji and Dancshazy, 1997; Varo et al., 1996) and can be recorded over the full length of the photocycle. Thus, it may be envisioned that some coordinated motion of the proteins may facilitate the shaft expansion mechanism. Although we have no evidence to support or negate this process, the higher compressibility of the lipid matter between the protein in the trimer is likely to absorb fast, short free space translation between the constituent of the trimer so that the intraprotein source of free space is a more likely mechanism.
Acknowledgments
This research is supported by the German-Israeli Foundation for Scientific Research and Development (GIF grant I-140-207.98) and the Israeli Science Foundation (427/01-1).
References
- Bauer, P. J., N. A. Dencher, and M. P. Heyn. 1976. Evidence for chromophore-chromophore interactions in the purple membrane from reconstitution experiments of the chromophore-free membrane. Biophys. Struct. Mech. 2:79–92. [DOI] [PubMed] [Google Scholar]
- Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187–217. [Google Scholar]
- Brown, L. S., R. Needleman, and J. K. Lanyi. 1999. Functional roles of aspartic acid residues at the cytoplasmic surface of bacteriorhodopsin. Biochemistry. 38:6855–6861. [DOI] [PubMed] [Google Scholar]
- Brown, L. S., G. Varo, R. Needleman, and J. K. Lanyi. 1995. Functional significance of a protein conformation change at the cytoplasmic end of helix F during the bacteriorhodopsin photocycle. Biophys. J. 69:2103–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y., G. Varo, M. Chang, B. F. Ni, R. Needleman, and J. K. Lanyi. 1991. Water is required for proton transfer from aspartate-96 to the bacteriorhodopsin Schiff base. Biochemistry. 30:10972–10979. [DOI] [PubMed] [Google Scholar]
- Checover, S., Y. Marantz, E. Nachliel, M. Gutman, M. Pfeiffer, J. Tittor, D. Oesterhelt, and N. A. Dencher. 2001. Dynamics of the proton transfer reaction on the cytoplasmic surface of bacteriorhodopsin. Biochemistry. 40:4281–4292. [DOI] [PubMed] [Google Scholar]
- Dencher, N. A., J. Heberle, G. Buldt, H.-D. Holtje, and M. Holtje. 1992. Active and passive proton transfer steps through bacteriorhodopsin are controlled by a light-triggered hydrophobic gate. In Structures and Functions of Retinal Proteins. J. L. Rigaud, editor. John Libbey Eurotext Ltd., Montrouge, France. 213–216.
- Dioumaev, A. K., L. S. Brown, R. Needleman, and J. K. Lanyi. 1999. Fourier transform infrared spectra of a late intermediate of the bacteriorhodopsin photocycle suggest transient protonation of Asp-212. Biochemistry. 38:10070–10078. [DOI] [PubMed] [Google Scholar]
- Dioumaev, A. K., L. S. Brown, R. Needleman, and J. K. Lanyi. 2001. Coupling of the reisomerization of the retinal, proton uptake, and reprotonation of Asp-96 in the N photointermediate of bacteriorhodopsin. Biochemistry. 40:11308–11317. [DOI] [PubMed] [Google Scholar]
- Eilers, M., S. C. Shekar, T. Shieh, S. O. Smith, and P. J. Fleming. 2000. Internal packing of helical membrane proteins. Proc. Natl. Acad. Sci. USA. 97:5796–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen, L., R. Siegert, W. D. Lehmann, and D. Oesterhelt. 1998. Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl. Acad. Sci. USA. 95:11673–11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson, M. K., A. Rashin, R. Fine, and B. Honig. 1985. On the calculation of electrostatic interactions in proteins. J. Mol. Biol. 184:503–516. [DOI] [PubMed] [Google Scholar]
- Grigorieff, N., T. A. Ceska, K. H. Downing, J. M. Baldwin, and R. Henderson. 1996. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259:393–421. [DOI] [PubMed] [Google Scholar]
- Henderson, R., J. M. Baldwin, T. A. Ceska, F. Zemlin, E. Beckmann, and K. H. Downing. 1990. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 213:899–929. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—Visual Molecular Dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- Kharakoz, D. P. 2000. Protein compressibility, dynamics, and pressure. Biophys. J. 79:511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink, B. U., R. Winter, M. Engelhard, and I. Chizhov. 2002. Pressure dependence of the photocycle kinetics of bacteriorhodopsin. Biophys. J. 83:3490–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., T. Saito, and H. Ohtani. 2001. Real-time spectroscopy of transition states in bacteriorhodopsin during retinal isomerization. Nature. 414:531–534. [DOI] [PubMed] [Google Scholar]
- Kocher, J.-P., M. Prevost, S. J. Wodak, and B. Lee. 1996. Properties of the protein matrix revealed by the free energy of cavity formation. Structure (London). 4:1517–1529. [DOI] [PubMed] [Google Scholar]
- Landau, E. M., and J. P. Rosenbusch. 1996. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA. 93:14532–14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi, J. K., and A. Pohorille. 2001. Proton pumps: mechanism of action and applications. Trends Biotechnol. 19:140–144. [DOI] [PubMed] [Google Scholar]
- Liang, J., H. Edelsbrunner, P. Fu, P. V. Sudhakar, and S. Subramaniam. 1998a. Analytical shape computation of macromolecules: II. Inaccessible cavities in proteins. Proteins. 33:18–29. [PubMed] [Google Scholar]
- Liang, J., H. Edelsbrunner, and C. Woodward. 1998b. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 7:1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke, H. 2000. Atomic resolution structures of bacteriorhodopsin photocycle intermediates: the role of discrete water molecules in the function of this light-driven ion pump. Biochim. Biophys. Acta. 30:133–156. [DOI] [PubMed] [Google Scholar]
- Luecke, H., B. Schobert, J. P. Cartailler, H. T. Richter, A. Rosengarth, R. Needleman, and J. K. Lanyi. 2000. Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin. J. Mol. Biol. 300:1237–1255. [DOI] [PubMed] [Google Scholar]
- Luecke, H., B. Schobert, H. T. Richter, J. P. Cartailler, and J. K. Lanyi. 1999a. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 291:899–911. [DOI] [PubMed] [Google Scholar]
- Luecke, H., B. Schobert, H.-T. Richter, J.-P. Cartailler, and J. K. Lanyi. 1999b. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 286:255–260. [DOI] [PubMed] [Google Scholar]
- Marque, J., L. Eisenstein, E. Gratton, J. M. Sturtevant, and C. J. Hardy. 1984. Thermodynamic properties of purple membrane. Biophys. J. 46:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachliel, E., M. Gutman, J. Tittor, and D. Oesterhelt. 2002. Proton transfer dynamics on the surface of the late M state of bacteriorhodopsin. Biophys. J. 83:416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollert, P., H. Qiu, M. Caffrey, J. P. Rosenbusch, and E. M. Landau. 2001. Molecular mechanism for the crystallization of bacteriorhodopsin in lipidic cubic phases. FEBS Lett. 504:179–186. [DOI] [PubMed] [Google Scholar]
- Rashin, A. A., M. Iofin, and B. Honig. 1986. Internal cavities and buried waters in globular proteins. Biochemistry. 25:3619–3625. [DOI] [PubMed] [Google Scholar]
- Riesle, J., D. Oesterhelt, N. A. Dencher, and J. Heberle. 1996. D38 is an essential part of the proton translocation pathway in bacteriorhodopsin. Biochemistry. 35:6635–6643. [DOI] [PubMed] [Google Scholar]
- Sass, H. J., G. Buldt, R. Gessenich, D. Hehn, D. Neff, R. Schlesinger, J. Berendzen, and P. Ormos. 2000. Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin. Nature. 406:649–653. [DOI] [PubMed] [Google Scholar]
- Sass, H. J., I. W. Schachowa, G. Rapp, M. H. Koch, D. Oesterhelt, N. A. Dencher, and G. Buldt. 1997. The tertiary structural changes in bacteriorhodopsin occur between M states: x-ray diffraction and Fourier transform infrared spectroscopy. EMBO J. 16:1484–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schätzler, B., N. A. Dencher, J. Tittor, D. Oesterhelt, S. Yaniv-Checover, E. Nachliel, and G. Gutman. 2003. Sub-second proton-hole propagation in bacteriorhodopsin. Biophys. J. 84:671–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, S., M. Lindahl, P. Bullough, A. R. Faruqi, J. Tittor, D. Oesterhelt, L. Brown, J. Lanyi, and R. Henderson. 1999. Protein conformational changes in the bacteriorhodopsin photocycle. J. Mol. Biol. 287:145–161. [DOI] [PubMed] [Google Scholar]
- Tokaji, Z., and Z. Dancshazy. 1997. Cooperativity-induced optical anisotropy changes during the photocycle of bacteriorhodopsin. Biochem. Biophys. Res. Commun. 233:532–536. [DOI] [PubMed] [Google Scholar]
- Varo, G., and J. K. Lanyi. 1995. Effects of hydrostatic pressure on the kinetics reveal a volume increase during the bacteriorhodopsin photocycle. Biochemistry. 34:12161–12169. [DOI] [PubMed] [Google Scholar]
- Varo, G., R. Needleman, and J. K. Lanyi. 1996. Protein structural change at the cytoplasmic surface as the cause of cooperativity in the bacteriorhodopsin photocycle. Biophys. J. 70:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., S. Link, C. D. Heyes, and M. A. El-Sayed. 2002. Comparison of the dynamics of the primary events of bacteriorhodopsin in its trimeric and monomeric states. Biophys. J. 83:1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weik, M., G. Zaccai, N. A. Dencher, D. Oesterhelt, and T. Hauss. 1998. Structure and hydration of the M-state of the bacteriorhodopsin mutant D96N studied by neutron diffraction. J. Mol. Biol. 275:625–634. [DOI] [PubMed] [Google Scholar]