Abstract
Complex conformational changes influence and regulate the dynamics of ion channels. Such conformational changes are stochastic and often inhomogeneous, which makes it extremely difficult, if not impossible, to characterize them by ensemble-averaged experiments or by single-channel recordings of the electric current that report the open-closed events but do not specifically probe the associated conformational changes. Here, we report our studies on ion channel conformational changes using a new approach, patch-clamp fluorescence microscopy, which simultaneously combines single-molecule fluorescence spectroscopy and single-channel current recordings to probe the open-closed transitions and the conformational dynamics of individual ion channels. We demonstrate patch-clamp fluorescence microscopy by measuring gramicidin ion channel conformational changes in a lipid bilayer formed at a patch-clamp micropipette tip under a buffer solution. By measuring single-pair fluorescence resonance energy transfer and fluorescence self-quenching from dye-labeled gramicidin channels, we observed that the efficiency of single-pair fluorescence resonance energy transfer and self-quenching is widely distributed, which reflects a broad distribution of conformations. Our results strongly suggest a hitherto undetectable correlation between the multiple conformational states of the gramicidin channel and its closed and open states in a lipid bilayer.
Subtle conformational changes of ion channels play an important role in regulating the function and dynamics of ion channels (Karlin, 2002; Madden, 2002; Choe, 2002). Channel conductance recording analysis and kinetic modeling (Cross et al., 1999; Andersen et al., 1999), together with site-directed mutagenesis, protein modifications, and static structural analysis provide extensive insights into the conformations and the conformational rearrangements associated with channel activity (Karlin, 2002; Madden, 2002; Choe, 2002; Glauner et al., 1999; Cha et al., 1999). However, the conformational dynamics underlying the mechanisms of ion channel action are still largely unknown due to lack of direct measurements. Mechanistic characterization of the conformational changes has been primarily based on kinetic model analyses of the electric current trajectories obtained by single-channel and whole-cell patch-clamp recordings that are not sensitive to subtle conformational intermediate states, including electrically undetectable “silent” conformational states. Protein dynamics may involve significant static and dynamic inhomogeneities (Lu et al., 1998; Xie and Trautman, 1998). For instance, ion channel dynamics can be modulated by inhomogeneous local membranes and a fluctuating environment. These static and dynamic inhomogeneous conformational dynamics are extremely difficult to study by conventional experimental approaches.
Recently, our group (Orr et al., 2001, 2002; Harms et al., 2002, 2003) and others have made significant progress in developing new approaches to studying ion channels using both fluorescence imaging and electric patch recording. Utilizing a micro-pinhole patch technique, Yanagida and coworkers (Ide and Yanagida, 1999; Ide et al., 2002) first reported fluorescence images acquired before or after a single-channel current measurement on the same sample lipid bilayer. Woolley, Schuetz, and their co-workers (Lougheed et al., 2001; Borisenko et al., 2003) have elegantly demonstrated the feasibility of combining single-molecule fluorescence imaging and electric recording on gramicidin ion channels. Isacoff and co-workers (Sonnleitner et al., 2002) have reported a single-molecule imaging and whole-cell patch recording study of structural rearrangement of voltage-gated Shaker K+ channels in the plasma membrane of living cells. A related experimental effort by Schmidt and his co-workers (Harms et al., 2001) has demonstrated wide-field fluorescence microscopy imaging of the diffusion and aggregation of yellow-fluorescent protein-labeled L-type Ca2+ channels in living cells. Another related experiment by Zagotta and his co-workers (Zheng and Zagotta, 2000) has studied gating rearrangements of cyclic nucleotide-gated channel proteins by combined fluorescence imaging and patch-clamp recording at multiple-channel-averaged measurements. These research efforts have begun to provide a new paradigm for studying ion channel conformational dynamics and mechanisms and for obtaining information not obtainable by the conventional patch current recording measurements, a primary experimental approach for decades.
Here, we demonstrate a new approach, patch-clamp fluorescence microscopy (PCFM), by combining single-molecule fluorescence imaging of fluorescence resonant energy transfer (FRET) and fluorescence self-quenching with a single-channel electric current patch recording. Using PCFM, we were able to correlate in real-time the single-channel open-closed kinetics with simultaneous changes in optical signals associated with conformational changes in a gramicidin ion channel in a lipid bilayer. By utilizing a typical patch-clamp technique in this new approach, the application of PCFM for studying ion channel dynamics in living cells (Orr et al., 2002) is now feasible. A two-state kinetic model has been widely referenced to illustrate the gramicidin ion channel kinetic behavior, i.e., the association and dissociation of the gramicidin dimer corresponding to the channel's open and closed states. However, the nature of the conformational changes and the dynamics that regulate channel activity are still largely unclear, since there are no methods of providing a direct real-time single-molecule measurement of both ion channel electric current and conformational changes. Our new experimental results, not obtainable by using ensemble-averaged methods or single-channel patch electric recording alone, suggest the occurrence of multiple intermediate conformational states underlying the gramicidin channel dynamics.
MATERIALS AND METHODS
Linear gramicidin produced naturally from the bacterium Bacillus brevis (Hladky and Haydon, 1970) is a typical “model” ion channel (Koeppe and Andersen, 1996; Cross et al., 1999; Andersen et al., 1999), and its conductive open state is presumably a dimer (Veatch et al., 1975; Koeppe and Andersen, 1996; Cross et al., 1999; Andersen et al., 1999) of two 15-amino acid peptides forming β-helices that meet head-to-head at the N-termini in the interior of a lipid bilayer (Koeppe and Andersen, 1996; Cross et al., 1999; Andersen et al., 1999; Roux and Karplus, 1994). Because of the available extensive knowledge on the gramicidin ion channels in lipid bilayers, we chose this system to demonstrate our combined single-molecule approach of PCFM.
Fig. 1 shows the experimental setup for the PCFM single-channel electric current recording and fluorescence imaging. We recorded the single-channel current of dye-labeled gramicidin dimers that were incorporated in lipid bilayers formed at the tip of a patch pipette (Montal and Mueller, 1972; see Fig. 1, inset) as we simultaneously imaged the pipette tip using wide-field single-molecule fluorescence microscopy (Schmidt et al., 1996; Ma et al., 2000; Han et al., 2001; Bartko et al., 2002; Seisenberger et al., 2001; Deschenes and Vanden Bout, 2001; Cognet et al., 2000).
FIGURE 1.
Experimental apparatus and measurement configuration of PCFM. Lipid bilayers were formed at the tip of a patch pipette by apposition of two monolayers of mixed lipids (4:1 dPhPE:DPhPC). Dye-labeled gramicidin monomers are introduced from both sides of the bilayer, and single-channel electric currents are recorded by using typical patch-clamp instrumentation. The laser excitation was focused through a microscope objective on the pipette tip, and the fluorescence was imaged on a CCD camera.
Single-molecule fluorescence microscope
A microscope (Nikon Diaphot 300, Nikon, Kanagawa, Japan) equipped with a 60× objective (N.A. = 1.4) and 4× relay lens was used in this work. In all experiments, the illumination intensity was set to 5 kW/cm2 for tetramethylrhodamine (TMR)-gramicidin at 514 nm (Ar-ion laser, Molecular Probes, Eugene, OR; MWK Laser, Riverside, CA) or 532 nm (Verdi 10 W diode laser, Coherent, Palo Alto, CA), and 1.5 kW/cm2 for Cy5-gramicidin at 632 nm (HeNe laser, Uniphase or dye laser, CR-5999, Coherent). The polarization of the excitation light was controlled by using a λ/4 waveplate (Meadowlark Optics, Frederick, CO). Filter combinations for TMR consisted of DCLP555, HQ625/100 (Chroma Technology Corp., Brattleboro, VT) and OG550 (Schott, Duryea, PA), and for Cy-5 of DCLP645, HQ690/90, and RG645 (Schott). The single-pair fluorescence resonant energy transfer (spFRET) measurements were done using a custom-built dual-dichroic beam-splitter (a combination of a DCLP555 and DCLP645, Chroma Technology Corp.) and band-pass filter in addition to either OG550 or RG645, permitting the detection of single fluorophores by a nitrogen-cooled CCD camera (Spec-10 1340 × 400B or 700B, Roper Scientific, Trenton, NJ). The microscope system total detection efficiency was 10 ± 2% for TMR and 8 ± 2% for Cy5. We estimated the total detection efficiency from the collection efficiency of the objective, the spectral throughput of the filter set, and the spectral detection efficiency of the CCD camera. We also subtracted the estimated reflections for each glass surface in the optical beam path. This device ensured the toggling of the excitation wavelength between 532 nm and 632 nm, and the detection of the emission at 8 ± 2% efficiency for TMR and 5 ± 2% efficiency for Cy5 with the FRET filters. The images were taken using 5-ms exposures at 1–10 Hz.
Lipid bilayer formation at the tip of a patch-pipette
Lipid bilayers were formed at the tip of patch pipettes by the apposition of two monolayers (Montal and Mueller, 1972), using 4:1 diphytanoylphosphatidylethanolamine (dPhPE):diphytanoylphosphatidylcholine (dPhPC) (Avanti, Alabaster, AL). Gramicidin dissolved in ethanol (10−9 M) was added to the aqueous subphase (1 M KCl, 1 mM CaCl2, and 5 mM Hepes of pH 7.5) after bilayer formation (Fig. 1, inset). Gramicidin monomers were labeled with either TMR or Cy5. Homodimers of TMR-labeled gramicidin were assembled by adding gramicidin-TMR monomers to both sides of the membrane; gramicidin-TMR-gramicidin-Cy5 heterodimers were generated by introducing TMR-gramicidin and Cy5-gramicidin from opposite sides of the bilayer (O'Connell et al., 1990). The latency period to form an active channel is typically a few minutes after gramicidin insertion into the bilayer and presumably arises from the slow diffusion of individual gramicidin monomers to form a dimer.
Single-channel electric current patch recording
Single-channel electric currents elicited at constant applied voltage (−100–+100 mV) were recorded using an EPC-9 amplifier (HEKA Electronics, Lambrecht, Germany). The recorded channel electric signal was sampled at 4 kHz and filtered at 3 kHz. Control experiments using unlabeled gramicidin A revealed distributions of current amplitude and open/closed dwell-time similar to those of the dye-labeled gramicidin. The dye-labeling of the gramicidin C-terminal did not significantly alter the gramicidin ion channel activities (Lougheed et al., 2001; Borisenko et al., 2003). The presence of a single channel at the patch was confirmed by recorded channel-current trajectories showing typical single-channel open-closed behavior (O'Connell et al., 1990; Sawyer et al., 1989; Veatch et al., 1975). The following criteria were applied to ensure that the recorded channel activity was from a single ion channel and not due to contamination or membrane instability: 1), no channel activity was detected before adding gramicidin; 2), only bursts of channel activity were considered; 3), a high membrane resistance (at the giga-ohm range) leading to a high signal-to-noise ratio (S/N) was achieved; and 4), membrane breakdown occurred when 300–500 mV polarization voltage was applied to the patch. These criteria are also practiced commonly in other labs (O'Connell et al., 1990; Sawyer et al., 1989; Veatch et al., 1975; Sigworth et al., 1987) to ensure a single gramicidin channel patch recording. About 10% of all the attempts were successful in incorporating a single gramicidin channel at the patch; membranes without channel activity or with more than one channel were not pursued.
Dye labeling of gramicidin monomers
Gramicidin C with Gly and Lys residues attached to the C-terminal (synthesized by Genemed Synthesis, Inc.) was used in this work. TMR or Cy5 HNS esters were covalently bonded by acylation to the ɛ-amino group of the Lys residue. The full sequence of the gramicidin used for the fluorescence labeling was Formyl - V - G - A - (d)L - A - (d)V - V - (d)V - W - (d)L - Y - (d)L - W - (d)L - W - G – K-NHCH2OH. Multiple HPLC purifications (C8 column with 80% methanol solvent) ensured the purity of the dye-labeled gramicidin samples, which was further confirmed by MALDI-TOF mass spectroscopy (molecular weight of gramicidin C-TMR, calculated = 2412 and measured = 2415; molecular weight of gramicidin C-Cy5, calculated = 2637 and measured = 2641).
Fluorescence imaging data analyses
The fluorescence background was subtracted by averaging photon counts for each pixel or by fitting the background to a wide 2-D Gaussian. The individual fluorescent “hot spots” were identified by the maxima of a 2-D Gaussian-correlation filter (Schmidt et al., 1996). The region around these maxima was fitted to a 2-D Gaussian, reporting the intensity, width, position, and errors of the least square fit (Schmidt et al., 1995, 1996; Press et al., 1990).
Single-molecule fluorescence control experiments
Single-molecule imaging was conducted of dye-labeled gramicidin in Langmuir-Blodgett (LB) bilayers and spin-coated over polymethyl methyacrylate (PMMA) polymer-coated cover slips. The single-molecule fluorescence imaging signal was confirmed by several typical verification measurements, including observations of single-step photobleaching events (Fig. 2 a, inset) and of linearly polarized dipoles (data not shown). The dPhPE:dPhPC 4:1 was doped with 10−8 mol TMR-gramicidin C or Cy5-gramicidin C per mol lipid and were deposited at 22°C and 28 dynes/cm on glass slides using LB deposition. Single-molecule fluorescence imaging measurements, using essentially the same experimental parameters and configurations that were used to image the bilayers containing individual dye-labeled gramicidin molecules at the tip, provided a quantitative reference for the single-molecule photon count rates under the wide-field imaging configuration (Fig. 2 a) in our experiments, which is a typical calibration commonly practiced in single-molecule imaging experiments (Schmidt et al., 1995, 1996; Seisenberger et al., 2001; Harms et al., 2001). The single-molecule images were analyzed by the least-square 2-D Gaussian fitting, as discussed above.
FIGURE 2.
(a) Histogram of single hot-spot fluorescence intensities deduced from 2-D Gaussian fitting to each spot imaged at the patch-clamp tip. Inset: intensity trajectory from a “hot spot” image of a single TMR-gramicidin molecule. (b) The probability density function (normalized) deduced from the histogram in a, and from the histogram of hot-spot intensities on glass-supported lipid bilayers (dotted line) as a control. Our control experiments and those of others (Lougheed et al., 2001) show no observable perturbation by dye labeling on the gramicidin channel activity.
The probability density function (PDF) of the photon counts,
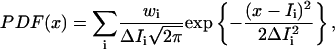 |
is constructed using parameters from the single-molecule image photon counts, where Ii, ΔIi, and wi represent the photon counts, its standard deviation, and its relative weight. The PDF statistically evaluates the imaging photon counts with their standard deviations and occurrences and provides a more reliable distribution than an occurrence histogram does (Schmidt et al., 1995). Under our set-up and experimental conditions, the average photon count detection rate for the TMR-gramicidin C monomers was 68 ± 5 counts/ms with a PDF peak of 45 ± 5 counts/ms and PDF width of 32 counts/ms. For Cy5-gramicidin C, the average photon count was 46 ± 5 counts/ms with a PDF peak of 36 ± 5 counts/ms and width of 30 counts/ms. The width of the PDF distributions indicates that the level of signal fluctuations is due primarily to shot noise and to the fluorescence quantum yield distribution of the individual dye molecules in a spatially inhomogeneous local environment (Lu and Xie, 1997; Xie and Trautman, 1998).
There are inevitable differences in the fluorescence photon detection efficiency and the signal-to-noise ratio of imaging single molecules on the LB layer or the PMMA polymer, as in a control experiment, and in imaging single molecules at a patch-clamp pipette tip, as in a real PCFM measurement (Fig. 2 b). There are at least three reasons for these differences: 1), There are different configurations for the pipette tip under a buffer solution in the PCFM measurement and the cover slip with individual dye-labeled gramicidin molecules in LB layers under water or spin-coated individual dye-labeled gramicidin molecules on PMMA polymer films. These intrinsic differences in the experimental configuration apparently cause different signals and noise levels associated with a change in the refractive index and light-scattering conditions. 2), There are different fluorescence quantum efficiencies under different conditions for the LB bilayer, a polymer, and the lipid bilayer at a pipette tip under a buffer. 3), There are different focal planes of the wide-field laser excitation and fluorescence collection, which are at the upper surface of the cover slip for the control experiment but at ∼70 μm above the cover slip in the buffer for imaging at the pipette tip.
The single-molecule fluorescence signal was distinguished from the background at the pipette tip based on characteristic single-molecule spectroscopic signatures: fluorescence blinking, single-step photobleaching, specific polarization orientation, and, most importantly, simultaneous single-channel ion current recording. The last simultaneous verification was that only a single-channel ion current associated with a single ion channel in the patch was observed during fluorescence imaging. The signal-to-noise ratio was typically within a range of 5–10 in the control LB bilayer imaging at the LB bilayer or PMMA polymer film, and 3–6 in the PCFM imaging at the pipette tips.
RESULTS AND DISCUSSION
Single-molecule spectroscopy is powerful in resolving complex and inhomogeneous dynamics and kinetics of protein systems. However, the fluctuation of most of the measured parameters is intrinsically pertinent to single-molecule measurements (Zwanzig, 1990; Lu and Xie, 1997; Xie and Trautman, 1998; Moerner, 2002). For example, single-molecule fluorescence has spectral and intensity fluctuations, FRET efficiency fluctuations, and lifetime fluctuations. These fluctuations are not observable in a typical ensemble-averaged experiment, and in most cases they are associated with single-molecule conformations and local environments. A reliable conclusion should typically be drawn from sound statistical analyses of a sufficient number of measurements of single-molecule events. Although the fluctuations give a larger standard deviation in the data of a single-molecule measurement compared to that of an ensemble-averaged measurement, the information obtained from single-molecule experiments has not been obtainable from ensemble-averaged experiments.
Heterodimers of gramicidin statistically correlate with maximum spFRET at an open state
Using PCFM, we conducted colocalization and spFRET imaging experiments simultaneously with single-channel patch recording. Asymmetric incorporation of Cy5-gramicidin C and TMR-gramicidin C led to the formation of single channels tagged with two different dyes, the donor and acceptor of the fluorescence energy transfer. Colocalization and spFRET within a single gramicidin heterodimer were probed by dual-color excitation and donor-acceptor two-channel fluorescence imaging, using an instrument configuration similar to one reported previously (Cognet et al., 2000). Fig. 3 a shows two fluorescence images taken at the tip with 514-nm (image 1) and 632-nm (image 2) excitation wavelengths, using a filter that allows imaging of both Cy5 and TMR emissions, both of which are clearly discernable in Fig. 3 a. From the 2-D Gaussian fitting of the colocalized signals, the two dye molecules were found to be within the fluorescence imaging diffraction-limited spot (∼300-nm width), which suggests that a pair of colocalized TMR-gramicidin and Cy5-gramicidin monomers was present at the tip when the active single-channel current signal was observed (Fig. 3 a). The colocalization imaging measurement is important and was used periodically to check the coexistence of the donor and acceptor molecules and to differentiate the situations of non-FRET from acceptor photobleaching events. To further characterize the heterodimerization states of TMR-gramicidin and Cy5-gramicidin, spFRET images were collected using a 645-nm long-pass filter, allowing only the fluorescence of acceptor Cy5 to pass. Toggling between 514-nm and 632-nm excitation wavelengths enabled the measurement of spFRET and verification of the coexistence of the single acceptor (Fig. 3 b). The spFRET that occurred during the open states of the channel is shown in Fig. 3 b. By counting images correlated with the electric signal that reflected the open-closed activity of single channels at the patch, we found that ∼70% of channel-opening events were correlated with spFRET. Estimating that ∼25% of the molecules undergo blinking events, we attributed the small fraction of non-FRET to acceptor photobleaching. Interestingly, we also observed that spFRET occurred in >∼10% of the cases where the channels were closed. We have further verified the FRET anticorrelation of the donor-acceptor fluorescence intensities by photon time-stamping, time-correlated single-photon counting (Hu and Lu, 2003) and proved the presence of FRET (Harms et al., 2003; G. S. Harms, G. Orr and H. P. Lu, unpublished results).
FIGURE 3.
PCFM measurement of single TMR/Cy5-gramicidin heterodimer ion channels. Green and red arrows mark the timing of the toggled excitation at 514 nm and 632 nm, respectively. (a) Colocalization of two gramicidin monomers labeled with two different dyes, TMR and Cy5. Single-channel current trajectory recorded at 100-mV polarization potential, with arrows indicating the point of exposure of two consecutively recorded frames. The first frame was taken at 514-nm excitation (with the OG550 long-pass filter), indicating single-molecule fluorescence of TMR. The second frame was taken at 632-nm excitation, indicating single-molecule fluorescence of Cy5. From a 2-D Gaussian fitting, the molecules were found to be within 30 nm of each other (the minimal positional accuracy allowed by the measurement system). “C” and “O” denote the current corresponding to closed and open states, respectively. (b) spFRET within a pair of gramicidin monomers labeled with two different dyes, TMR and Cy5. Single-channel current trajectory (upper panel) with four arrows indicating the exposure of four consecutively acquired frames (lower panel). Frames 1 and 3 were taken at 514-nm excitation with a red-emission long-pass filter (RG645). Frames 2 and 4 were taken at 632-nm excitation with a red-emission long-pass filter (RG645). Frame 1 indicates that FRET occurred between the TMR/Cy5 spFRET pair when the gramicidin channel was at an open state. Frame 2 verifies that a single Cy5 molecule was present. Frame 3 indicates that no spFRET occurred when the channel was at a closed state. Frame 4 verifies that the Cy5 was still present. Subsequent images taken without the red long-pass filter (RG645) did show colocalization as shown in a. (c) FRET efficiency distribution of “channel open” TMR/Cy5 heterodimers (shaded bars) with mean of 0.59, width of 0.21, and “channel closed” (open bars) with a maximum value of 0.57. An asterisk is inserted to indicate the 70% of channels showing no appreciable FRET (below 0.3) in the channel-closed state (or 30% of channels in the channel-open state).
A broad distribution of FRET efficiency (Ha et al., 1999; Brasselet et al., 2000; Cognet et al., 2000),
 |
was found at gramicidin heterodimers, and the efficiency was evaluated by the activity of the channels at the time that the images were taken (Fig. 3 c). ITMR and ICy5 are the total photon counts for single-molecule images of TMR and Cy5, η is detection efficiency, and Φ is the quantum yield. We used the values of ηTMR and ηCy5 discussed above as well as ΦTMR = ΦCy5 = 0.28 (Cognet et al., 2000). When the channels were open, the Gaussian-like distribution of the efficiency showed a mean value of 0.59 ± 0.05 with a full-width half-maximum (FWHM) of 0.21. The standard deviation of the F was estimated to be ±0.05 based on shot-noise error propagation analysis (Press et al., 1990). Our control experiment of polarization-dependent imaging (Harms et al., 2003) indicated that the rotational diffusion of the TMR and Cy5 dye molecules had a much faster rate than the imaging rate, so that the orientation effect of the dye dipoles on the FRET efficiency was averaged out during the 5-ms imaging collection time for each frame. The mean spFRET efficiency and the FWHM of 0.21 correspond to a mean donor-acceptor distance (d) of ∼56 Å and a range of ∼52 Å ≤ d ≤ ∼63 Å, respectively. However, ∼30% of the images obtained from the channel open states showed no detectable FRET efficiency, mostly because of photobleaching and spectral fluctuation. We cannot differentiate the data between no FRET and low-efficiency FRET below 0.3, due to the experimental limitation of the signal-to-noise ratio of the single-molecule PCFM imaging. The distribution of FRET efficiency of the closed state of single channels had a mean of 0.42 ± 0.05 (excluding the low signal data) and extended to a maximum efficiency of 0.57. Therefore, the difference in the FRET between open and closed states was statistically significant and beyond the measurement noise. The broad FRET distributions of both open and closed states of the single gramicidin channels suggest that the channels could be in closed states even when the two gramicidin monomers are still in an intermediate dimerized state (Fig. 4, a and b). We postulate that the partially dissociated dimer may have had a distorted ion channel pore, creating a significant change in the channel electric current, though the physical distance between the two monomers did not significantly change (Fig. 4 b). It has been suggested that there are six hydrogen bonds at the N-terminal head-to-head cross-linking the dimer (Szabo and Urry, 1978; Koeppe and Andersen, 1996). It is possible that one or more broken hydrogen bonds can give different conformations of the pore structure of a gramicidin dimer. In Fig. 4, we show that the energy-minimized structure of gramicidin heterodimer FRET pair (represented by using TMR molecular structures) has the minimum donor-acceptor distance of ∼50–52 Å. The relative dihedral angle between the two monomers at different conformations of the conducting dimer (Fig. 4) can give a distance change up to 6 Å, which can result in a 0.15 change in FRET efficiency. We note that the calculated spFRET efficiency is only reliable in the range of 0.3 < F < 0.7 due to the measurement noise associated with the effects of shot noise and photophysical dark states. Nevertheless, spFRET imaging results (Fig. 3 c) provide unambiguous evidence that the gramicidin channel dynamics involve multiple open and closed states associated with different conformations of the gramicidin dimers.
FIGURE 4.

Molecular structures (top and side views) of the possible open (a) and closed (b) conformational states of a gramicidin dimer in a lipid bilayer. The molecular structures were calculated by molecular dynamics simulation using Discovery and Insight II software. It is evident that slight rotational distortion along the dihedral angle can significantly narrow the channel pore size and separate the dye probe pair.
The spFRET imaging measurement using toggled dual-color excitation and simultaneous correlation with the single gramicidin ion channel patch electric recording is a novel approach, and the results of the correlation between the spFRET and the open-closed states of the heterodimer gramicidin strongly suggest the existence of more than one conformational state at each open and closed state of the gramicidin ion channels in the lipid bilayer. These hidden multiple conformational states are not observable from ensemble-averaged measurements, and also are “silent” and not resolvable in a conventional patch recording analysis.
Homodimers of dye-labeled gramicidin statistically coincide with maximal fluorescence self-quenching at an open state
We have obtained fluorescence images of single TMR-gramicidin homodimers at bilayer patches simultaneously with measurements of their single-channel electric currents using PCFM. A segment of a single-channel electric current trajectory (Fig. 5 a) with synchronous fluorescence images (Fig. 5 b) illustrates the correlation between the transitions of the channel from the closed to the open state and fluorescence hot spots (Fig. 5). Single hot spots were analyzed for width (Fig. 6 a) and intensities (Fig. 6, b and c). These mean values are from single homodimers of the dye-labeled gramicidin, an interpretation based on the coincidence of the recorded single-channel electric currents with the fluorescence images and on the various control experimental analyses discussed above.
FIGURE 5.
PCFM measurement of single TMR-gramicidin homodimer ion channels. (a) Upper trace: single-channel electric current trajectory recorded at 50-mV polarization potential from a single TMR-gramicidin channel at a patch-clamp pipette tip. Lower trace: time flags indicating the 5 ms of laser excitation and CCD camera exposure simultaneously acquired with the single-channel electric recording. “C” and “O” denote the electric current corresponding to closed and open states, respectively, of the single channel. (b) Top panel: Consecutive fluorescence images of the pipette tip taken simultaneously with the single-channel electric current recording shown in a. Bottom panel: plot of the 2-D Gaussian fitting for the above images. Right inset: 3-D plot of the first image (top) and 3-D plot of the 2-D Gaussian fitting (bottom) with subtracted background intensity added to the base shown in gray.
FIGURE 6.
(a) Fluorescence intensities of single hot spots versus fluorescence imaging width of the spots as derived from 2-D Gaussian fit. (b, c) Distributions of the single-channel fluorescence intensity for closed and open states of single channels with PDF and fitting.
We further observed that the width of the hot spot fluorescence increased with fluorescence intensity (Fig. 6 a). Theoretically, the spread functions of the one-point-source and two-point-source images are Gaussian (G(x,y)) = A exp{[−(x−x0)2 + (y−y0)2]/B} and a convolution of two Gaussian functions (G × G), respectively. Assuming that the imaging spatial resolution is optical diffraction-limited, the FWHM is a constant, ∼2B0.5, for G(x, y) at different signal intensity and the FWHM of G × G is dependent only on the two-point-source separation. The error bars for data points in Fig. 6 a are contributed from the shot noise, finite signal-to-noise ratio, digitized photon counting, and digitized pixels. The widths of the hot spots were essentially diffraction-limited but wider when the channels were fully closed (Figs. 5 b and 6 a). This observed behavior is beyond the standard deviation (Fig. 6 a). These observations are consistent with the expectation that the channel is fully open when the dimer is fully associated, leading to maximal fluorescence self-quenching because the two labeled dye molecules have the closest distance across the lipid bilayer at the patch (Fig. 4 a). Rarely (<5% of images) were two separate and spatially resolved imaging spots observed when a single channel was closed, i.e., a dissociated dimer. In most cases, the two dissociated monomers do not separate beyond the imaging spatial resolution, ∼300 nm, or even beyond the diffraction limit. The imaging signal-to-noise ratio is not high enough for obtaining a clear image of two separate spots, as we observe that the widths of the spots are larger than the diffraction limit at higher fluorescence intensity (Fig. 6 a).
We found that the diffusional motion of single gramicidin channels at the dPhPE:DPhPC bilayers was confined within the imaging spatial resolution. According to viscosity studies, dPhPE is likely to be in a gel-phase at room temperature (Janko and Benz, 1977). Based on our control measurements in a large, supported LB-membrane bilayer, the diffusion coefficient of TMR-gramicidin was estimated to be <10−10 cm2/s in a dPhPE LB layer, yielding ∼0.2-μm average displacement in 1 s. This is consistent with the fact that we rarely observed diffusion of a dissociated dimer into two resolved monomers. Although the diffusion coefficient of the TMR-gramicidin can be as high as 10−8 cm2/s in DPhPC at room temperature (Borisenko et al., 2003), the 4:1 dPhPE:DPhPC bilayers may have small domains (<500 nm) of the DPhPC formed (Heller et al., 1998), confining the diffusional motions of the TMR-gramicidin molecules within the DPhPC. Such “corralled” motion may be accountable for the rare observation of an optically resolved separation of a pair of TMR-gramicidin monomers dissociated from a dimer. It has been known (Heller et al., 1998) that domains of DPhPC can form in DPhPE bilayers and that gramicidin channels typically prefer to stay in the DPhPC regions.
The diffraction-limited single hot-spot images of TMR-gramicidin dimers also reflected different degrees of fluorescence self-quenching (Zhuang et al., 2000). The self-quenching critical radius of the rhodamine 6G dyes is ∼46 Å (Penzkofer and Lu, 1986). The fluorescence self-quenching between the two dye molecules causes an intensity decrease as the two monomers become closer and form a conductive channel. The average distance between the transition dipoles of the TMR dyes was calculated to be ∼50 Å, and the transition dipoles were nearly aligned in the fully dimerized state (Fig. 4 a).
The mechanism of fluorescence self-quenching is still largely unknown and system-dependent (Zhuang et al., 2000; Penzkofer and Lu, 1986). In our gramicidin ion channel system, because the two TMR dye molecules are not in contact to form a dimer or excimer, the possible mechanism of the observed weak self-quenching effect is likely associated with reabsorption, and secondary fluorescence depends on the fluctuating orientation of the two TMR dye molecules across the lipid bilayer.
Fig. 6, b and c, shows the distributions of the fluorescence imaging intensity in correlation with the open and closed states of the gramicidin channels, measured by PCFM. The intensity of the hot spots was statistically higher when the channel was closed (Fig. 6 b), and lower when the channel was open (Fig. 6 c). The means of the distribution were found to be 67 ± 8 counts/ms at closed states and 52 ± 7 counts/ms at open states. Furthermore, the difference in fluorescence intensities of the single-channel images taken for the open and closed states was also found to be statistically significant, based on the paired t-test (t = 2.58, P = 0.0297). We also noted that the broadness of the distribution is significant and beyond the measurement noise and that both quenched and unquenched gramicidin channel dimers probably existed in the channel closed state. Because the efficiency of self-quenching depends on the distance and the relative orientation between the two molecules, the broader distributions of the fluorescence intensity at the closed (Fig. 6 b) and open (Fig. 6 c) states also suggests that the two monomers form “closed” or “open” dimers of different conformations (Fig. 4). This result is consistent with the spFRET imaging result (Fig. 3), also suggesting that there are multiple intermediate conformation states corresponding to different distances between the homo-pair of TMR molecules in a single gramicidin channel based on the wide distribution of the fluorescence self-quenching efficiency among the single gramicidin channels in the lipid bilayer.
Autocorrelation analysis of single-channel electric current trajectories characterizes the dynamics of conformational changes of the multiple closed and open states
To further explore the possible existence of the multiple conformational states involved in gramicidin ion channel dynamics, we applied autocorrelation function analysis (Zwanzig, 1990; Neher and Stevens, 1977; Labarca et al., 1985; Lu et al., 1998) of the single-channel electric current trajectories. Detailed single-channel current analyses support our attribution that multiple conformational states are involved in the spatially confined gramicidin dimer channel dynamics. Fig. 7 a shows a typical single gramicidin channel electric current trajectory. The intermediate conductive states, in addition to the fully open and closed states, are evident in the trajectory and are further revealed in an electric current amplitude distribution shown in Fig. 7 b. There is a significant population beyond the measurement noise and within the time resolution in the distribution between the typical open-closed electric current amplitude bimodal distribution (Fig. 7 b). The autocorrelation function deduced from the single-channel electric current trajectory (Fig. 7 a) shows nonexponential decay (Fig. 7 c), and the nonexponential autocorrelation functions were commonly observed for both dye-labeled and non-dye-labeled gramicidin channels (D. J. Panther, G. S. Harms, G. Orr, and H. P. Lu, unpublished results). The nonexponential autocorrelation functions suggest that the transitions between multiple open-closed states of a single channel either occur at a fluctuating rate (Zwanzig, 1990; Xie, 2002; Yang and Xie, 2002; Jung et al., 2002) or involve a complex kinetic mechanism associated with multiple intermediate states (Schenter et al., 1999; Jung et al., 2002; Reilly and Skinner, 1993). We have analyzed the dynamics using an autocorrelation function of sequential open dwell-time (index number, m) {ton(m)}, calculated by 〈Δton(0)Δton(m)〉, where Δton(m) = ton(m) − 〈ton(m)〉 (Fig. 7 d). The single-exponential decays that were fitted to the autocorrelation functions of ton(m) (Fig. 7 d) and toff(m) (not shown) were 2.6(±1.0) × 10−2 m−1 and 0.5 ± 0.2 m−1, respectively. The open and closed dwell-time correlations suggest that the channel open and closed states are dependent within a short period of “memory” time (Lu et al., 1998; Zwanzig, 1990; Yang and Cao, 2001; Vlad et al., 2002; Jung et al., 2002; Xie, 2002; Yang and Xie, 2002). The memory time for the open dwell-time was determined to be ∼8 s (38 cycles), but only ∼0.4 s (8 cycles) for the closed dwell-time. We postulate that the “memory” effect originates from the channel open-closed conformational motions that are trapped in metastable processes under different rates of toggling among different conformational states. This dynamically inhomogeneous behavior, also defined as dynamic disorder (Zwanzig, 1990), has been observed in other ion channel systems (Sakmann and Neher, 1995; Neher and Stevens, 1977). Recently, single-molecule spectroscopy has made progress in understanding this dynamic disorder behavior, which is beyond the scope of the conventional kinetics (Lu et al., 1998; Zwanzig, 1990; Yang and Cao, 2001; Vlad et al., 2002; Jung et al., 2002; Xie, 2002; Yang and Xie, 2002). Typically, the mechanism of the dynamic disorder can be analyzed by the “Agmon-Hopfield” diffusive model (Agmon and Hopfield, 1983; Agmon, 2000) or by the multiple-state Markovian models (Schenter et al., 1999; Sakmann and Neher, 1995; Neher and Stevens, 1977). The Fig. 7 d inset shows the distribution of the “open” dwell-time. At least two subgroups of “open” dwell times contributed to this broad distribution. We postulate that the dynamic disorder of this gramicidin system in dPhPE:DPhPC bilayers can be characterized by a simple coupled Markovian kinetic model, for example, the 2 × 2 model (Schenter et al., 1999), which describes two slowly interconverting open-closed kinetic processes that have different kinetic rates.
FIGURE 7.
(a) Single-channel electric current trajectory, {i(t)}, recorded at 100-mV polarization potential from a single gramicidin channel in a lipid bilayer patch (4:1 dPhPE:DPhPC) formed at a patch-clamp pipette tip. The single-channel electric current trajectory indicates that there are only single-channel events (O'Connell et al., 1990). (b) Electric current amplitude histogram calculated from the single-channel trajectory in a. Intermediate electric current amplitude reflecting the inhomogeneous conductivity is evident although the channel shows essentially two-state kinetics. “C” and “O” denote the current corresponding to closed and open states, respectively. (c) Autocorrelation function, C(t) = 〈Δi(0)Δi(t)〉, calculated from the single-channel trajectory in a. Biexponential fitting gives k1 = 4.0 ± 1.0 s−1, A1 = 0.48, and k2 = 53 ± 5 s−1, A2 = 0.52; using A1 exp(−k1t) + A2 exp(−k2t). (d) Autocorrelation function of “open” dwell times, r(m) = 〈Δt(0)Δt(m)〉, where m is the index of the “open” dwell-time trajectory for the single TMR-gramicidin ion channel recorded in a. The single-exponential decay rate constant is 2.6 (±1.0) × 10−2 m−1, and the first point (not shown) of the autocorrelation contains the uncorrelated noise and fast fluctuation beyond the instrument resolution. Error bars are shown for the first ten data points. The calculated autocorrelation times are in the timescale of seconds and subseconds (c and d). The channel open-closed dwell times are also at subsecond timescale (a). Using a 4-kHz filter instead of a 6-kHz filter does not affect the results, even if the possible aliasing effect exists at millisecond timescale. Inset: The distribution of “open” dwell time deduced from the same trajectory in a. The solid line shows a distribution envelope fitting with a bi-Gaussian function with a relative weight of 0.808 and 0.192 at averaged dwell time of 1.13 × 10−1 and 8.67 × 10−3 s, respectively. Inhomogeneous distribution is evident.
The open and closed dwell times are found to be dependent over periods of hundreds of milliseconds, which suggests a trapped or not fully dissociated subset of channel conformers. We note that the gramicidin channel dynamics can be significantly different using different lipid bilayers because the channel open and closed activities are associated with the gramicidin diffusion motion in a particular bilayer. Nevertheless, the gramicidin ion channel dynamics at the dPhPE:DPhPC bilayers are apparently associated with more than two states: 1), The open states of the gramicidin dimer involve slow, strongly correlated conformational fluctuations that lead to dynamic disorder of the dissociation rate (Lu et al., 1998; Zwanzig, 1990; Yang and Cao, 2001; Vlad et al., 2002). 2), Gramicidin dimerization involves multiple, weakly correlated or noncorrelated association kinetics at different rates. Although the association kinetics of gramicidin have been studied and discussed in terms of fast fluctuations (Sigworth et al., 1987) and the existence of possible “mini-channels” (Busath and Szabo, 1981; Sawyer et al., 1989), our observations of the “memory” effect in the gramicidin channel dynamics in dPhPE:DPhPC bilayers provide a new insight into the molecular level. However, we are not sure, at this stage, if the complex mechanism we observe is generally true for the gramicidin ion channels in other lipid bilayer systems.
A multistate model for gramicidin channel dynamics
Based on the information derived from the PCFM imaging, we propose a working model of multi-state gramicidin channel dynamics at dPhPE:DPhPC bilayers (Fig. 8). The open (left), closed (right), and intermediate (middle) states can be presumably the outcome of intra- and intermolecular conformational changes, geminate recombination, and nongeminate recombination. Although the valine (Townsley et al., 2001; Sigworth et al., 1987) and tryptophan residues (Townsley et al., 2001) may undergo large structural fluctuations and alter channel conductance by perturbing the structure of the dimeric channel (Koeppe and Andersen, 1996; Townsley et al., 2001; Koeppe et al., 2000; Salom et al., 1995; Woolf and Roux, 1994), we postulate that the intermediate conformers result predominately from the six intermolecular H-bond fluctuations that disrupt the pore structure of the gramicidin dimer. Conformational changes at the N-terminal linkage of the dimer may significantly disrupt the column of structured water, the channel cavity structure, and the electrostatic field, thereby perturbing channel conductance (Eisenberg, 1998). Drastic conductance modifications were reported for gramicidin channels with mutated N-terminal head-to-head connections (Sigworth et al., 1987; Szabo and Urry, 1978), indicating that the six intermolecular H-bonds formed at the contact region of the dimer stabilize the fully conductive gramicidin channel (Szabo and Urry, 1978; Koeppe and Andersen, 1996). Partial disruption of these H-bonds results in incomplete association or dissociation of the dimers (Szabo and Urry, 1978). Given that each H-bond has ∼3–5 kcal/mol dissociation activation energy, the dissociation of these bonds plausibly occurs in the millisecond timescale at room temperature. These estimated H-bond dissociation times are in the same timescale as the gramicidin dimer changes between open and closed states. The conformational changes that could originate from intramolecular changes or lipid-channel interactions are expected to occur at a faster timescale and, therefore, should not result in the observed self-quenching and FRET effects at the millisecond timescale. Therefore, we hypothesize that the conformational changes that we measured arise from the N-terminal H-bond dynamic changes. We are not able to provide direct structural evidence of the H-bond fluctuation using our current PCFM. The structural and dynamical information may be obtained by integrating a Raman spectroscopic probe with the PCFM, which is beyond the scope of this article.
FIGURE 8.
Proposed multistate model of gramicidin dynamics indicating a third “multistate” of nonconducting or partially conducting channels with larger separation between the fluorophore probes. In this model, the third “multistate” represents more than one intermediate conformation state. Alternative rotationally distorted conformations are possible for the fully conductive state of the gramicidin channels (Roux and Karplus, 1994).
Diffusion of individual monomers that dissociate from the dimerized state (Tank et al., 1982) is confined within the optical diffraction-limited spot size, based on our experimental observation using wide-field imaging. This observation has been further confirmed by time-dependent and static anisotropy analysis of gramicidin C-TMR in the LB bilayer at the patch-clamp tips. The detailed results and discussion will be reported elsewhere. Briefly, the decay time of the anisotropy is ∼5 ns, and the static anisotropy is 0.25 ± 0.05 (Harms et al., 2003; G. S. Harms, G. Orr, and H. P. Lu, unpublished data). Based on our observation, in most of the events, the pairs of gramicidin monomers did not diffuse away from each other beyond the imaging diffraction-limited scale (≤300 nm), and we postulate that the geminate recombination may be associated with single-channel conformational changes. It is possible that the diffusion of gramicidin monomers was confined to domains (≤300 nm) likely associated with the inhomogeneity (Nielsen and Andersen, 2000) of the dPhPE:DPhPC bilayers, as discussed above.
CONCLUSIONS
We have developed a new single-molecule approach, PCFM, combining single-molecule fluorescence imaging microscopy and single-channel patch recording to simultaneously record the single-channel trajectories of fluorescence imaging and channel electric current recording. We have demonstrated the application of PCFM in studying the ion channel dynamics of single gramicidin dimers in a lipid bilayer at the patch-clamp tip. By correlating single-channel fluorescence quenching and spFRET imaging with single-channel electric current recording simultaneously acquired, our results strongly suggest the existence of multiple conformational states of a single gramicidin channel associated with the channel open-closed activity at a dPhPE:DPhPC bilayer. Although the inhomogeneous dynamics of the conformational change were probed in a lipid bilayer, the exact structures of the conformational states require further studies, for example, by correlated Raman spectroscopy. The new single-molecule experimental approach for studying ion channel conformational dynamics and mechanisms should be applicable for direct study of ion channel proteins in living cells (Orr et al., 2002).
Acknowledgments
This work was supported by the Laboratory Directed Research and Development Program of Pacific Northwest National Laboratory, operated for the U.S. Department of Energy by Battelle Memorial Institute. Research (MM) at University of California, San Diego, was supported by grants from the National Institutes of Health (GM-49711) and the Department of the U.S. Army Medical Research and Materiel Command (DAMD 17-02-C-0106).
G. S. Harms and G. Orr contributed equally to this work.
References
- Agmon, N. 2000. Conformational cycle of a single working enzyme. J. Phys. Chem. B. 104:7830–7834. [Google Scholar]
- Agmon, N., and J. J. Hopfield. 1983. Transient kinetics of chemical reactions with bounded diffusion perpendicular to the reaction coordinate: intramolecular processes with slow conformational changes. J. Chem. Phys. 78:6947. [Google Scholar]
- Andersen, O. S., H. J. Apell, E. Bamberg, D. D. Busath, R. E. Koeppe, F. J. Sigworth, G. Szabo, D. W. Urry, and A. Woolley. 1999. Gramicidin channel controversy–the structure in a lipid environment. Nat. Struct. Biol. 6:609. [DOI] [PubMed] [Google Scholar]
- Bartko, A. P., K. Xu, and R. M. Dickson. 2002. Three-dimensional single molecule rotational diffusion in glassy state polymer films. Phys. Rev. Lett. 89:026101. [DOI] [PubMed] [Google Scholar]
- Borisenko, V., T. Lougheed, J. Hesse, E. Fureder-Kitzmuller, N. Fertig, J. C. Behrends, G. A. Woolley, and G. J. Schutz. 2003. Simultaneous optical and electrical recording of single gramicidin channels. Biophys. J. 84:612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasselet, E. J. G., A. Miyawaki, and W. E. Moerner. 2000. Single-molecule fluorescence resonant energy transfer in calcium concentration dependent cameleon. J. Phys. Chem. B. 104:3676–3682. [Google Scholar]
- Busath, D., and G. Szabo. 1981. Gramicidin forms multi-state rectifying channels. Nature. 294:371–373. [DOI] [PubMed] [Google Scholar]
- Cha, A., G. E. Snyder, P. R. Selvin, and F. Bezanilla. 1999. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 402:809–817. [DOI] [PubMed] [Google Scholar]
- Choe, S. 2002. Ion channel structure: potassium channel structures. Nat. Rev. Neurosci. 3:115–121. [DOI] [PubMed] [Google Scholar]
- Cognet, L., G. S. Harms, G. Blab, P. H. M. Lommerse, and Th. Schmidt. 2000. Simultaneous dual-color and dual-polarization imaging of single molecules. Appl. Phys. Lett. 77:1–3. [Google Scholar]
- Cross, T. A., A. Arseniev, B. A. Cornell, J. H. Davis, J. A. Killian, R. E. Koeppe, L. K. Nicholson, F. Separovic, and B. A. Wallace. 1999. Gramicidin channel controversy–revisited. Nat. Struct. Biol. 6:610–611. [DOI] [PubMed] [Google Scholar]
- Deschenes, L. A., and D. A. Vanden Bout. 2001. Molecular motions in polymer films near the glass transition: a single molecule study of rotational dynamics. J Phys. Chem. B. 105:11978–11985. [DOI] [PubMed] [Google Scholar]
- Eisenberg, B. 1998. Ionic channels in biological membranes. Natural nanotubes. Acc. Chem. Res. 31:117–125. [Google Scholar]
- Glauner, K. S., L. M. Mannuzzu, C. S. Gandhi, and E. Y. Isacoff. 1999. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 402:813–817. [DOI] [PubMed] [Google Scholar]
- Ha, T., A. Y. Ting, J. Liang, W. B. Caldwell, A. A. Deniz, D. S. Chemla, P. G. Schultz, and S. Weiss. 1999. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. USA. 96:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M., X. Gao, J. Z. Su, and S. Nie. 2001. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 19:631–635. [DOI] [PubMed] [Google Scholar]
- Harms, G. S., L. Cognet, P. H. M. Lommerse, G. A. Blab, H. Kahr, R. Gamsjager, H. P. Spaink, N. M. Soldator, C. Romanin, and Th. Schmidt. 2001. Single-molecule imaging of L-type Ca2+ channels in live cells. Biophys. J. 81:2639–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, G. S., G. Orr, M. Montal, B. Thrall, S. Colson, and H. P. Lu. 2002. Combined patch-clamp recording and single-molecule imaging microscopy study of single molecule ion channel dynamics. Biophys. J. 82:193a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, G. S., G. Orr, and H. P. Lu. 2003. Probing single-molecule ion channel conformational dynamics using combined ultrafast spectroscopy, and patch-clamp recording. Biophys. J. 84:123a. [Google Scholar]
- Heller, W. T., A. J. Waring, R. I. Lehrer, and H. W. Huang. 1998. Multiple states of beta-sheet peptide protegrin in lipid bilayers. Biochemistry. 37:17331–17338. [DOI] [PubMed] [Google Scholar]
- Hladky, S. B., and D. A. Haydon. 1970. Discreteness of conductance change in bimolecular lipid membranes in the presence of certain antibiotics. Nature. 5231:451–453. [DOI] [PubMed] [Google Scholar]
- Hu, D., and H. P. Lu. 2003. Single-molecule nanosecond anisotropy dynamics of tethered proteins. J. Phys. Chem. B. 107:618–626. [Google Scholar]
- Ide, T., Y. Takeuchi, and T. Yanagida. 2002. Development of an experimental apparatus for simultaneous observation of optical and electrical signals from single ion channels. Single Molecules. 3:33–42. [Google Scholar]
- Ide, T., and T. Yanagida. 1999. An artifical bilayer formed on an agarose-coated glass for simultaneoous electrical and optical measurement of single ion channels. Biochem. Biophys. Res. Commun. 265:595–599. [DOI] [PubMed] [Google Scholar]
- Janko, K., and R. Benz. 1977. Properties of lipid bilayer membranes made from lipids containing phytanic acid. Biochim. Biophys. Acta. 470:8–16. [DOI] [PubMed] [Google Scholar]
- Jung, Y., E. Barkai, and R. J. Silbey. 2002. Current status of single-molecule spectroscopy: theoretical aspects. J. Chem. Phys. 117:10980–10995. [Google Scholar]
- Karlin, A. 2002. Ion channel structure: emerging structure of the nicotinic acetylcholine receptors. Nat. Rev. Neurosci. 3:102–114. [DOI] [PubMed] [Google Scholar]
- Koeppe, R. E., and O. S. Andersen. 1996. Engineering the gramicidin channel. Annu. Rev. Biophys. Biomol. Struct. 25:231–258. [DOI] [PubMed] [Google Scholar]
- Koeppe, R. E., J. Hatchett, A. R. Jude, L. L. Providence, O. S. Andersen, and D. V. Greathouse. 2000. Neighboring aliphatic/aromatic side chain interactions between residues 9 and 10 in gramicidin channels. Biochemistry. 39:2235–2242. [DOI] [PubMed] [Google Scholar]
- Labarca, P., J. A. Rice, D. R. Fredkin, and M. Montal. 1985. Kinetic analysis of channel gating. Application to the cholinergic receptor channel and the chloride channel from Torpedo californica. Biophys. J. 47:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougheed, T., V. Borisenko, C. E. Hand, and G. A. Woolley. 2001. Fluorescent gramicidin derivatives for single-molecule fluorescence and ion channel measurements. Bioconjug. Chem. 12:594–602. [DOI] [PubMed] [Google Scholar]
- Lu, H. P., and X. S. Xie. 1997. Single-molecule spectral fluctuations at room temperature. Nature. 385:143–146. [Google Scholar]
- Lu, H. P., L. Xun, and X. S. Xie. 1998. Single-molecule enzymatic dynamics. Science. 282:1877–1882. [DOI] [PubMed] [Google Scholar]
- Ma, Y., M. R. Shortreed, and E. S. Yeung. 2000. High-throughput single-molecule spectroscopy in free solution. Anal. Chem. 72:4640–4645. [DOI] [PubMed] [Google Scholar]
- Madden, D. R. 2002. The structure and function of glutamate receptor ion channels. Nat. Rev. Neurosci. 3:91–101. [DOI] [PubMed] [Google Scholar]
- Moerner, W. E. 2002. A dozen years of single-molecule spectroscopy in physics, chemistry, and biophysics. J. Phys. Chem. B. 106:910–927. [Google Scholar]
- Montal, M., and P. Mueller. 1972. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA. 69:3561–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher, E., and C. F. Stevens. 1977. Conductance fluctuations and ionic pores in membranes. Annu. Rev. Biophys. Bioeng. 6:345–381. [DOI] [PubMed] [Google Scholar]
- Nielsen, C., and O. S. Andersen. 2000. Inclusion-induced bilayer deformations: effects of monolayer equilibrium curvature. Biophys. J. 79:2583–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, A. M., R. E. Koeppe, and O. S. Andersen. 1990. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 250:1256–1259. [DOI] [PubMed] [Google Scholar]
- Orr, G., M. Montal, B. D. Thrall, S. Colson, and H. P. Lu. 2001. Single-channel patch-clamp recording coupled with linear and non-linear confocal scanning fluorescence spectroscopy: towards the simultaneous probing of single-ion channel conformational changes and channel kinetics. Biophys. J. 80:151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, G., G. S. Harms, B. D. Thrall, M. Montal, S. Colson, and H. P. Lu. 2002. Probing single-molecule ligand-channel interaction dynamics in a living cell. Biophys. J. 82:255a. [Google Scholar]
- Penzkofer, A., and Y. Lu. 1986. Fluorescence quenching of rhodamine 6G in ethanol at high concentration. Chem. Phys. 103:399–405. [Google Scholar]
- Press, W. H., B. P. Flannery, S. A. Tenkolsky, and W. T. Vetterling. 1990. Numerical Recipes. Cambridge University Press, Cambridge, UK
- Reilly, P. D., and J. L. Skinner. 1993. Spectral diffusion of single molecule fluorescence: a probe of low-frequency localized excitations in disordered crystals. Phys. Rev. Lett. 71:4257–4260. [DOI] [PubMed] [Google Scholar]
- Roux, B., and M. Karplus. 1994. Molecular dynamics simulations of the gramicidin channel. Annu. Rev. Biophys. Biomol. Struct. 23:731–761. [DOI] [PubMed] [Google Scholar]
- Sakmann, B., and E. Neher. 1995. Single-Channel Recordings. Plenum Press, New York.
- Salom, D., M. C. Bano, L. Braco, and C. Abad. 1995. HPLC demonstration that an all Trp→Phe replacement in gramicidin A results in a conformational rearrangement from beta-helical monomer to double-stranded dimer in model membranes. Biochem. Biophys. Res. Commun. 209:466–473. [DOI] [PubMed] [Google Scholar]
- Sawyer, D. B., R. E. Koeppe, and O. S. Andersen. 1989. Induction of conductance heterogeneity in gramicidin channels. Biochemistry. 28:6571–6583. [DOI] [PubMed] [Google Scholar]
- Schenter, G. K., H. P. Lu, and X. S. Xie. 1999. Statistical analyses and theoretical models of single-molecule enzymatic dynamics. J. Phys. Chem. A. 103:10477–10488. [Google Scholar]
- Schmidt, Th., G. J. Schuetz, W. Baumgartner, H. J. Gruber, and H. Schindler. 1995. Characterization of photophysics and mobility of single molecule in a fluid lipid membrane. J. Phys. Chem. 99:17662–17668. [Google Scholar]
- Schmidt, Th., G. J. Schuetz, W. Baumgartner, H. J. Gruber, and H. Schindler. 1996. Imaging of single molecule diffusion. Proc. Natl. Acad. Sci. USA. 93:2926–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger, G., M. U. Ried, T. Endress, H. Buening, M. Hallek, and C. Braeuchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 294:1929–1932. [DOI] [PubMed] [Google Scholar]
- Sigworth, F. J., D. W. Urry, and K. U. Prasad. 1987. Open channel noise. III. High-resolution recordings show rapid current fluctuations in gramicidin A and four chemical analogues. Biophys. J. 52:1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner, A., L. M. Mannuzzu, S. Terakawa, and E. Y. Isacoff. 2002. Structural rearrangements in single ion channels detected optically in living cells. Proc. Natl. Acad. Sci. USA. 99:12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, G., and D. W. Urry. 1978. N-acetyl gramicidin: single-channel properties and implications for channel structure. Science. 203:55–57. [DOI] [PubMed] [Google Scholar]
- Tank, D. W., E. S. Wu, P. R. Meers, and W. W. Webb. 1982. Lateral diffusion of gramicidin C in phospholipid multibilayers. Effects of cholesterol and high gramicidin concentration. Biophys. J. 40:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley, L. E., W. A. Tucker, S. Sham, and J. F. Hinton. 2001. Structures of gramicidins A, B, and C incorporated into sodium dodecyl sulfate micelles. Biochemistry. 40:11676–11686. [DOI] [PubMed] [Google Scholar]
- Veatch, W. R., R. Mathies, M. Eisenberg, and L. Stryer. 1975. Simultaneous fluorescence and conductance studies of planar bilayer membranes containing a highly active fluorescent analog of gramicidin A. J. Mol. Biol. 99:75–92. [DOI] [PubMed] [Google Scholar]
- Vlad, M. O., F. Moran, F. W. Schneider, and J. Ross. 2002. Memory effects and oscillations in single-molecule kinetics. Proc. Natl. Acad. Sci. USA. 99:12548–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf, T. B., and B. Roux. 1994. Molecular dynamics simulation of the gramicidin channel in a phospholipid bilayer. Proc. Natl. Acad. Sci. USA. 91:11631–11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. S. 2002. Single-molecule approach to dispersed kinetics and dynamic disorder: probing conformational fluctuation and enzymatic dynamics. J. Chem. Phys. 117:11024–11032. [Google Scholar]
- Xie, X. S., and J. K. Trautman. 1998. Optical studies of single molecules at room temperature. Annu. Rev. Phys. Chem. 49:441–480. [DOI] [PubMed] [Google Scholar]
- Yang, S., and J. Cao. 2001. Two-event echoes in single-molecule kinetics: a signature of conformational fluctuations. J. Phys. Chem. B. 105:6536–6549. [Google Scholar]
- Yang, H., and X. S. Xie. 2002. Probing single-molecule dynamics photon by photon. J. Chem. Phys. 117:10965–10979. [Google Scholar]
- Zheng, J., and W. N. Zagotta. 2000. Gating rearrangements in cyclic neurotechnique nucleotide-gated channels revealed by patch-clamp fluorometry. Neuron. 28:369–374. [DOI] [PubMed] [Google Scholar]
- Zhuang, X., T. Ha, H. D. Kim, T. Centner, S. Labeit, and S. Chu. 2000. Fluorescence quenching: a tool for single-molecule protein-folding study. Proc. Natl. Acad. Sci. USA. 97:14241–14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwanzig, R. 1990. Rate-processes with dynamic disorder. Acc. Chem. Res. 23:148–152. [Google Scholar]












