Abstract
Changes in the elastic properties of single deoxyribonucleic acid (DNA) molecules in the presence of different DNA-binding agents are identified using atomic force microscope single molecule force spectroscopy. We investigated the binding of poly(dG-dC) dsDNA with the minor groove binder distamycin A, two supposed major groove binders, an α-helical and a 310-helical peptide, the intercalants daunomycin, ethidium bromide and YO, and the bis-intercalant YOYO. Characteristic mechanical fingerprints in the overstretching behavior of the studied single DNA-ligand complexes were observed allowing the distinction between different binding modes. Docking of ligands to the minor or major groove of DNA has the effect that the intramolecular B-S transition remains visible as a distinct plateau in the force-extension trace. By contrast, intercalation of small molecules into the double helix is characterized by the vanishing of the B-S plateau. These findings lead to the conclusion that atomic force microscope force spectroscopy can be regarded as a single molecule biosensor and is a potent tool for the characterization of binding motives of small ligands to DNA.
INTRODUCTION
The investigation of interactions between double-stranded deoxyribonucleic acid (DNA) and DNA-binding agents is crucial to a deeper understanding of such important biochemical processes as replication, repair, recombination, and expression of genes. In principle, the possible binding mechanisms of ligands to double-stranded (ds) DNA can be divided into sequence-specific binding, and, on the other hand, binding modes that lack sequence specificity. Specific binding between ligand (protein) and receptor (dsDNA), often also termed “molecular recognition,” is the basis for the interaction of many transcription factors with DNA. Small agents that bind unspecifically or with lower sequence specificity to dsDNA are often capable of influencing or inhibiting these processes and intrinsically exhibit mutagenic properties. By consequence, these molecules find applications as pharmaceuticals, mainly in the treatment of cancer. Others are employed as DNA staining agents, for example in fluorescence assays.
We compared the effects exerted on the mechanical properties of dsDNA by unspecific binding of seven different ligands. When considering unspecific, noncovalent binding of molecules to dsDNA, at least three different modes are known. Binding of small, positively charged peptides may occur in the minor groove of DNA. This binding mode requires only slight conformational adaptions of the double helix. An example for this binding mechanism is the interaction of the peptide distamycin A with the minor groove. Distamycin A induces chromosome decondensation and is used as a lead structure for a whole class of nonintercalating mutagenic drugs (Baguley, 1982; Turner and Denny, 1996; Bailly and Chaires, 1998). As for minor groove binding, major groove binding is dominated by electrostatic interactions of helical ligands with the backbone assisted by hydrogen bonds. We investigated the synthetic amphipathic peptides Ac-(Leu-Ala-Arg-Leu)3-NH-linker (linker: 1,8-diamino-3,6-dioxaoctane), forming a 3.616-(α-)helix, and the 310-helix Ac-(Aib-Leu-Arg)4-NH-linker containing the β-loop-builder α-aminoisobutyric acid (Aib). Both peptides are supposed to be major groove binding agents (Niidome et al., 1996).
Intercalation is a different mode of interaction of small molecules (not necessarily peptides) with DNA. It is characterized by the sliding-in of flat, planar molecules into the base pair stack of dsDNA via interaction of their aromatic ring systems with the π-systems of the adjacent base pairs. The anthracycline-antibiotic daunomycin (also known as daunorubicin), which is a potent anticancer drug primarily used in the treatment of leukemia (Aubel-Sadron and Londos-Gagliardi, 1984; Hortobagyi, 1997), serves as an important example of an intercalant. The fluorescence dye ethidium bromide for DNA staining (Morgan et al., 1979) has a central phenanthridine ring system that intercalates into DNA. Other very stable fluorescent dyes are the rather complicated molecules YO and YOYO (Glazer and Rye, 1992). YO also has an extended aromatic system which enables the compound to intercalate into the double helix. YOYO is a bridged YO-dimer and a bis-intercalant: when sliding into the base sequence, the two ring systems enclose two base pairs. It has been proposed that at higher concentrations, both YO and YOYO are also able to interact with DNA by a major groove binding mode (Larsson et al., 1994).
The development and maturation of ultrasensitive force sensors during the past fifteen years has rendered experiments with single molecules or molecule complexes possible. In contrast to classical ensemble measurements, single molecule techniques focus on molecular individuals. In atomic force microscope (AFM) force spectroscopy, forces on the single molecule level are detected by measuring the deflection of an AFM cantilever, yielding a force versus distance plot. This technique has been applied to the investigation of intermolecular forces in receptor-ligand interactions like biotin-streptavidin/avidin (Florin et al., 1994; Moy et al., 1994; Lee et al., 1994b), antibody-antigen (Hinterdorfer et al., 1996; Dammer et al., 1996; Ros et al., 1998), or selectin-ligand (Fritz et al., 1998), interactions between complementary strands of DNA (Lee et al., 1994a; Florin et al., 1995; Strunz et al., 1999), and cell adhesion proteoglycans (Dammer et al., 1995). AFM force spectroscopy also proves a potent tool to examine intramolecular forces. In this setup, a single molecule is mechanically stretched between the tip and the surface. The plot of pulling force against molecular extension contains information about intramolecular structural transitions, which were observed in single dextran (Rief et al., 1997b), titin (Rief et al., 1997a), and DNA (Rief et al., 1999; Clausen-Schaumann et al., 2000) molecules. In addition, force spectroscopy experiments on DNA have also been performed using related techniques such as optical tweezers (Smith et al., 1996) and magnetic tweezers (Smith et al., 1992; Strick et al., 1998). Therein a highly cooperative transition to an overstretched conformation 1.7 times as long as the B-DNA contour length, which was termed S-DNA, was reported (Cluzel et al., 1996). An alternative explanation for this overstretching transition was given by Rouzina and Bloomfield (Rouzina and Bloomfield, 2001a,b), interpreting the phenomenon as an equilibrium force-induced melting process. (For simplicity, the overstretching transition, however interpreted, will be called “B-S transition” in the following text.)
This transition was reported by Clausen-Schaumann et al. (2000) to appear in the force-extension curve as a plateau at ∼65 pN for poly(dG-dC) dsDNA. Recent investigations (Wenner et al., 2002) examined the salt-dependence of this overstretching plateau, exhibiting an increase in the overstretching force with decreasing Na+ concentration. At higher pulling forces, a second structural transition is observed. This transition, as opposed to the B-S transition, is a nonequilibrium process on the time scale of the AFM experiment and interpreted as a rate-dependent melting of the double helix induced by the action of the external force (Clausen-Schaumann et al., 2000; Rouzina and Bloomfield, 2001a,b). Melting proceeds until the strands are fully separated, leaving only a single strand attached to the tip.
It has recently been shown that binding to DNA significantly affects the force response and allows for the differentiation between binding mechanisms of small molecules to DNA via their force-extension profiles (Anselmetti et al., 2000; Krautbauer et al., 2002a,b). These studies centered on the interaction of DNA with the mutagenic agents ethidium bromide, berenil, and cis-platin. Our study further corroborates these results by extending the measurements to a wider range of systems such as supposed major-groove binding helical peptides and bis-intercalants.
MATERIALS AND METHODS
Sample preparation
For all experiments poly(dG-dC) dsDNA (Amersham Bioscience, Piscataway, NJ) with an average length of 724 bp was used. For preparation, the DNA was diluted in 10 mM Tris buffer (Sigma, Steinheim, Germany) at pH 8.3 containing 150 mM NaCl and 1 mM EDTA (Sigma) to a concentration of 1 mg ml−1.
The peptides were prepared by solid phase synthesis on aliphatic safety-catch resin (Advanced ChemTech, Louisville, KY) using Fmoc-protected amino acids. Ac-(Leu-Ala-Arg-Leu)3-NH-linker and Ac-(Aib-Leu-Arg)4-NH-linker, respectively, were obtained upon activation of the safety-catch resin with iodoacetonitrile followed by reaction of Ac-(Leu-Ala-Arg-Leu)3-resin and Ac-(Aib-Leu-Arg)4-resin with the linker 1,8-diamino-3,6-dioxaoctane. The synthesized peptide derivatives were purified by high-pressure liquid chromatography on a reverse phase column (218 TP 1022 Efficiency, protein & peptide C18, 250 × 22 mm, Vydac, Columbia, MD), using acetonitrile/water/TFA gradients. The final products were identified by elemental analysis and matrix-assisted laser desorption ionization mass spectroscopy using a Voyager DE MALDI (PerSeptive Biosystems, Framingham, MA) apparatus.
The DNA-binding agents daunomycin (Sigma), ethidium bromide (Merck, Darmstadt, Germany), distamycin A (Sigma), YO (Molecular Probes, Eugene, OR), YOYO (Molecular Probes), Ac-(Leu-Ala-Arg-Leu)3-NH-linker and Ac-(Aib-Leu-Arg)4-NH-linker were added to 10 μl of the DNA solution in a concentration of 150 μM, corresponding to a 1:10 ratio of agent molecules per base pair. Constant molar ratios were applied because reliable binding constants are not available yet for the binding of the peptides to DNA. The solution was incubated for 24 h at 4°C. For immobilization, the solution was incubated for 24 h on a freshly evaporated gold surface (30 nm on glass slides) at ambient temperature. Before use, the samples were rinsed with buffer solution to remove excess DNA-ligand complexes from the surface.
Force spectroscopy
Force spectroscopy measurements were performed on a commercial AFM (Multimode, Veeco Instruments, Santa Barbara, CA). The acquisition of the cantilever deflection force signal and the vertical movement of the piezo electric elements was controlled by a 16 bit AD/DA card (PCI-6052E, National Instruments, Austin, TX) and a high-voltage amplifier (600H, NanoTechTools, Echandens, Switzerland) via a home-built software based on Labview (National Instruments). The deflection signal was low pass filtered (<10 kHz) and averaged by a factor of 5.
The spring constants of all AFM cantilevers (Si3Ni4-Microlever, Thermomicroscopes, Sunnyvale, CA) were calibrated by the thermal fluctuation method (Hutter and Bechhoefer, 1993) with an absolute uncertainty of 15%. All given measurements were performed with different cantilevers with spring constants ranging from 12 pN nm−1 to 14 pN nm−1.
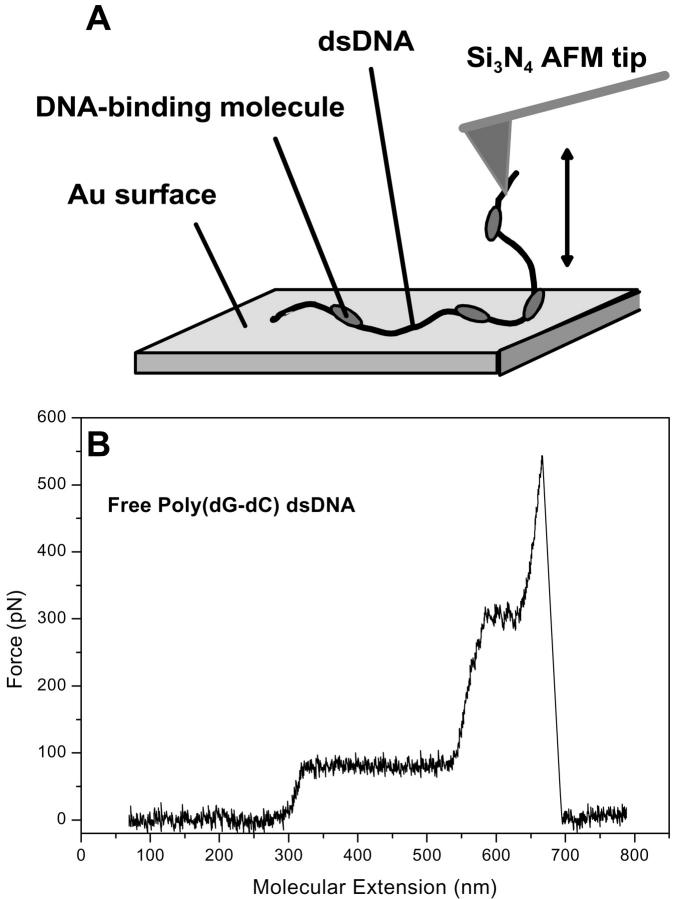
The dsDNA strands were mechanically contacted with the tip from the gold surface by applying a contact force of 1–2 nN (Rief et al., 1999; Clausen-Schaumann et al., 2000) and extended with a piezo velocity of 1000 nm s−1. All measurements were performed under identical Tris buffer solution (see above) at 20°C. The experimental setup is sketched schematically in Fig. 1 A.
FIGURE 1.
(A) Experimental setup; (B) Force-extension trace for free poly(dG-dC) dsDNA.
RESULTS AND DISCUSSION
Free dsDNA
The quantitative results of the force spectroscopy measurements on the reference molecule free poly(dG-dC) dsDNA basically confirm the findings previously reported by Rief et al. (1999; Clausen-Schaumann et al., 2000). Fig. 1 B shows the respective force-extension curve. Clearly discernible is the plateau at 75 pN due to overstretching of the double helix to >170% its B-DNA contour length, which corresponds to the reported 65 pN (Rief et al., 1999; Clausen-Schaumann et al., 2000) within the error of measurement dominated by the uncertainty in the cantilever spring constant calibration. The value for the fractional elongation of the fully extended S-DNA over the B-DNA contour length, 70%, has also been reported by other groups performing DNA stretching by means of optical or magnetic tweezers (Smith et al., 1992, 1996; Cluzel et al., 1996).
Force-induced melting of the double helix begins at an extension of 550 nm up to a force of 300 pN. The melting transition is followed by single-strand stretching. At a force of 540 pN and an extension of 660 nm, the single strand is detached from the tip and the cantilever relaxes. In our experiments, the contour length strongly depends on where the DNA molecule was picked up by the tip and thus varied from molecule to molecule. However, the forces at which the force-induced structural transitions occurred were independent of the specific value of the contour length.
Minor groove binding
Binding of the peptide distamycin A in the minor groove obviously does not have a large impact on the conformation of the DNA, as can be inferred from the qualitative agreement between the force-extension curves of free DNA and the complex (Fig. 2 A). The complex of DNA with the minor groove binding agent still exhibits the internal transitions due to overstretching and melting of the double helix characteristic for free DNA. The B-S transition plateau remains distinct from the melting transition. In contrast to the results for free DNA, we observed a considerable lowering of the B-S plateau to 50 pN. To avoid the uncertainty in the cantilever calibration, we performed comparative experiments on free DNA and the DNA-distamycin complex using the same AFM tip. This direct comparison showed that the decrease in force was highly reproducible. As reported by Krautbauer et al. (2002a,b), binding of the minor groove binder netropsin to λ-DNA led to an increase of the plateau force, a finding which we could reproduce also for distamycin in our group by means of optical tweezers measurements (Sischka et al., unpublished results). Thus, the decrease in the B-S transition force found for the complex of distamycin A to poly(dG-dC) dsDNA must be due to the sequence differences between λ-DNA and poly(dG-dC) dsDNA, distamycin showing a preferred binding to A-T-rich regions in a mixed sequence.
FIGURE 2.
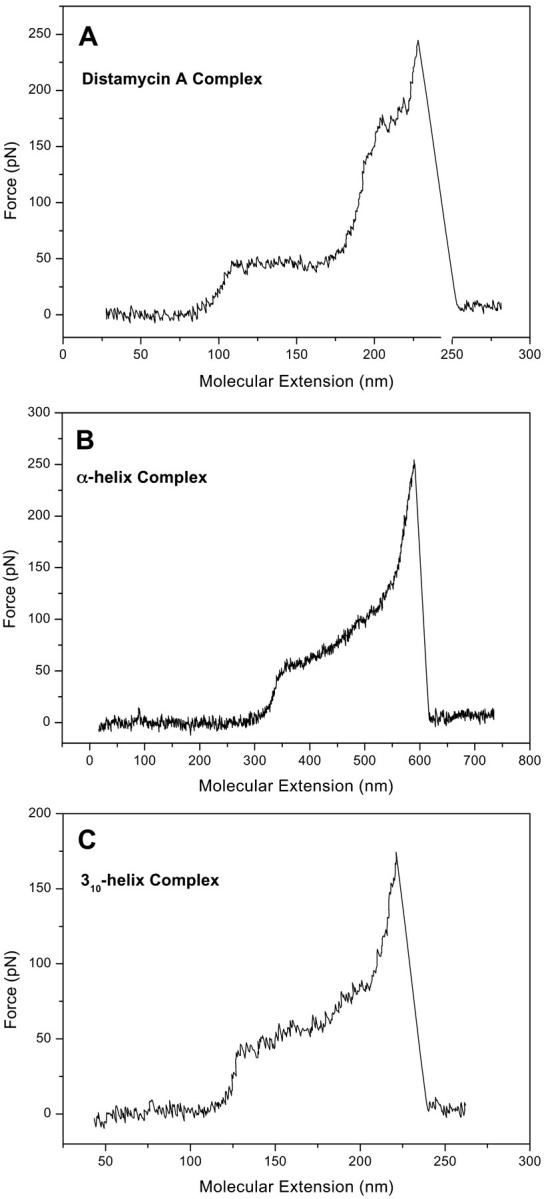
Force-extension traces for (A) the DNA-distamycin A complex; (B) the DNA complex with the α-helical peptide Ac-(Leu-Ala-Arg-Leu)3-NH-linker; (C) the complex of DNA with the 310-helical peptide Ac-(Aib-Leu-Arg)4-NH-linker.
Major groove binding
In the force-extension trace for the complex of poly(dG-dC) dsDNA with the α-helical peptide (Fig. 2 B), no B-S transition separate from the melting transition is observed. Thus, the B-S transition does not appear as a plateau. Nevertheless, the point of maximum B-DNA elongation can still be discriminated. This finding suggests that the peptide, showing a force-extension characteristic that differs from both the one for the minor groove binder distamycin and the curves of the intercalants (Fig. 3), adopts a binding mechanism different from the one of the minor groove binding peptide distamycin A. Regarding the chemistry of the system, it was supposed that a possible binding of the α-helix to DNA should be based upon unspecific electrostatic interactions between the guanidino groups of the peptide and the negatively charged DNA backbone and should occur in the major groove of the double helix (Niidome et al., 1996). Force spectroscopy measurements now indicate a mechanism that differs from both intercalation and minor groove binding, supporting the interpretation that the peptide binds to the major groove. The internal transition onset of the complex starts at a pulling force of ∼60 pN.
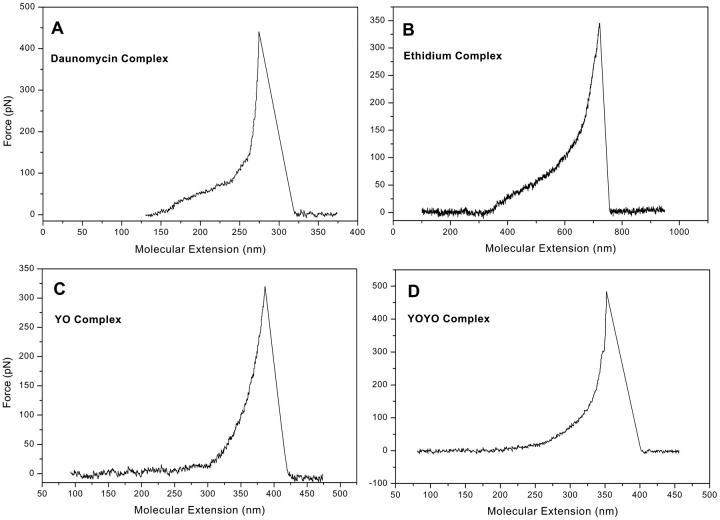
FIGURE 3.
Force-extension traces for the complexes of poly(dG-dC) dsDNA with the intercalants (A) daunomycin; (B) ethidium bromide; (C) YO; (D) YOYO.
Binding of the 310-helical peptide to poly(dG-dC) dsDNA has almost the same effects as the binding of the α-helix: The force-extension curve in Fig. 2 C is very similar to the one in Fig. 2 B, with the exception that the onset of the structural transition starts at a pulling force of ∼50 pN.
Intercalation
Daunomycin as an intercalant inserts into DNA via a stacking interaction of its aromatic ring system with the base pairs. Intercalated DNA should display a considerable resistance against pulling with an external force. The force-extension curve in Fig. 3 A confirms this assumption: The force-extension trace for the complex of poly(dG-dC) dsDNA with the intercalant shows that, upon intercalation, the overstretching plateau vanishes. An intramolecular transition can only be distinguished from the double helix melting as a region of minor slope compared to the actual melting and only gives rise to a slight flattening of the curve up to forces of 75 pN.
Fig. 3 B shows the corresponding force-extension curve for the complex consisting of poly(dG-dC) dsDNA and the intercalant ethidium bromide. As for daunomycin, the elastic properties of DNA are significantly changed upon intercalation, so there is no distinct B-S transition observable. The slope of the force-extension curve is steeper than for the daunomycin complex. Up to 100 pN, there is still a flatter region discernible. These findings correlate well with previously reported results (Anselmetti et al., 2000; Krautbauer et al., 2002a,b).
Intercalation and major groove binding of DNA by the intercalants YO and YOYO leads to a uniform force-distance curve with an increasing gradient. The force-extension curve for the YO-DNA complex can be seen in Fig. 3 C. There is no indication for an intramolecular change in conformation. Binding of excess YO results in a force-extension trace with a steadily rising slope. Finally, as can be inferred from the force-extension curve (Fig. 3 D), the bis-intercalative binding mode of YOYO does not cause significant deviations in the overstretching behavior of the DNA complex in comparison to the corresponding YO-DNA complex.
CONCLUSIONS
We have shown that AFM-based force spectroscopy measurements on single molecules provide a powerful tool to distinguish between different binding modes of small sequence-unspecific ligands to dsDNA such as intercalation, minor and major groove binding. Distinct characteristics could be detected in the force-extension traces of free poly(dG-dC) dsDNA and the DNA complexes with the minor groove binder distamycin A, supposed major groove binding α-helical and 310-helical peptides, the intercalants daunomycin and ethidium bromide, the intercalant and major groove binder YO and the bis-intercalant and major groove binder YOYO. It could be demonstrated that the intramolecular B-S transition visible in the force-extension curves of free DNA also occurs in complexes of DNA with groove binders, whereas it vanishes as a distinct plateau in the corresponding DNA-intercalant curves. B-S transition of poly(dG-dC) dsDNA complexes with groove binding molecules like distamycin A and the synthetic helical peptides studied here showed a tendency to occur at forces slightly lowered by 10 pN compared to free poly(dG-dC) dsDNA. Intercalation of dsDNA by the agents daunomycin and ethidium bromide has the effect that an intramolecular transition besides the melting can only be discerned as a slight flattening of the force-extension curve slope. The intercalants act to withstand the partial unwinding of the DNA that accompanies the B-S transition. This effect was even more pronounced for the intercalant YO and the bis-intercalant YOYO.
Advanced theoretical models explaining our results should lead to a quantitative analysis of the binding behavior. An improved understanding of binding process and mechanisms will provide interesting perspectives for fundamental research in the fields of biochemistry, pharmacy, and genetics. In this context, AFM force spectroscopy will gain importance as a molecular biosensor that allows for the identification of distinct binding motives in the interaction of biological systems at the single molecule level.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 613).
References
- Anselmetti, D., J. Fritz, B. Smith, and X. Fernandez-Busquets. 2000. Single molecule DNA biophysics with atomic force microscopy. Single Molecules. 1:17–23. [Google Scholar]
- Aubel-Sadron, G., and D. Londos-Gagliardi. 1984. Daunorubicin and doxorubicin, anthracycline antibiotics, a physicochemical and biological review. Biochimie. 66:333–352. [DOI] [PubMed] [Google Scholar]
- Baguley, B. C. 1982. Nonintercalative DNA-binding antitumour compounds. Mol. Cell. Biochem. 43:167–181. [DOI] [PubMed] [Google Scholar]
- Bailly, C., and J. B. Chaires. 1998. Sequence-specific DNA minor groove binders. Design and synthesis of netropsin and distamycin analogues. Bioconjug. Chem. 9:513–538. [DOI] [PubMed] [Google Scholar]
- Clausen-Schaumann, H., M. Rief, C. Tolksdorf, and H. E. Gaub. 2000. Mechanical stability of single DNA molecules. Biophys. J. 78:1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel, P., A. Lebrun, C. Heller, R. Lavery, J.-L. Viovy, D. Chatenay, and F. Caron. 1996. DNA: An extensible molecule. Science. 271:792–794. [DOI] [PubMed] [Google Scholar]
- Dammer, U., M. Hegner, D. Anselmetti, P. Wagner, M. Dreier, W. Huber, and H.-J. Güntherodt. 1996. Specific antigen/antibody interactions measured by force microscopy. Biophys. J. 70:2437–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer, U., O. Popescu, P. Wagner, D. Anselmetti, H.-J. Güntherodt, and G. N. Misevic. 1995. Binding strength between cell adhesion proteoglycans measured by atomic force microscopy. Science. 267:1173–1175. [DOI] [PubMed] [Google Scholar]
- Florin, E.-L., V. T. Moy, and H. E. Gaub. 1994. Adhesion forces between individual ligand-receptor pairs. Science. 264:415–417. [DOI] [PubMed] [Google Scholar]
- Florin, E.-L., M. Rief, H. Lehmann, M. Ludwig, C. Dornmair, V. T. Moy, and H. E. Gaub. 1995. Sensing specific molecular interactions with the atomic force microscope. Biosens. Bioelectron. 10:895–901. [Google Scholar]
- Fritz, J., A. G. Katopodis, F. Kolbinger, and D. Anselmetti. 1998. Force-mediated kinetics of single P-selectin/ligand complexes observed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 95:12283–12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer, A. N., and H. S. Rye. 1992. Stable dye-DNA intercalation complexes as reagents for high-sensitivity fluorescence detection. Nature. 359:859–861. [DOI] [PubMed] [Google Scholar]
- Hinterdorfer, P., W. Baumgartner, H. Gruber, K. Schilcher, and H. Schindler. 1996. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 93:3477–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortobagyi, G. N. 1997. Anthracyclines in the treatment of cancer. Drugs. 54:1–7. [DOI] [PubMed] [Google Scholar]
- Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 7:1868–1873. [Google Scholar]
- Krautbauer, R., S. Fischerländer, S. Allen, and H. E. Gaub. 2002a. Mechanical fingerprints of DNA drug complexes. Single Molecules. 3:97–103. [Google Scholar]
- Krautbauer, R., L. H. Pope, T. E. Schrader, S. Allen, and H. E. Gaub. 2002b. Discriminating small molecule DNA binding modes by single molecule force spectroscopy. FEBS Lett. 510:154–158. [DOI] [PubMed] [Google Scholar]
- Larsson, A., C. Carlsson, M. Jonsson, and B. Albinsson. 1994. Characterization of the binding of the fluorescent dyes YO and YOYO to DNA by polarized light spectroscopy. J. Am. Chem. Soc. 116:8459–8465. [Google Scholar]
- Lee, G. U., L. A. Chrisey, and R. J. Colton. 1994a. Direct measurement of the forces between complementary strands of DNA. Science. 266:771–773. [DOI] [PubMed] [Google Scholar]
- Lee, G. U., D. A. Kidwell, and R. J. Colton. 1994b. Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir. 10:354–357. [Google Scholar]
- Morgan, A. R., D. H. Evans, G. U. Lee, and D. E. Pulleyblank. 1979. Review: Ethidium fluorescence assays, part 2: Enzymatic studies and DNA-protein interactions. Nucleic Acids Res. 10:571–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy, V. T., E.-L. Florin, and H. E. Gaub. 1994. Intermolecular forces and energies in between ligands and receptors. Science. 266:257–259. [DOI] [PubMed] [Google Scholar]
- Niidome, T., N. Ohmori, A. Ichinose, A. Wada, H. Mihara, T. Hirayama, and H. Aoyagi. 1996. Binding of cationic alpha-helical peptides to plasmid DNA and their gene-transfer abilities into cells. J. Biol. Chem. 272:15307–15312. [DOI] [PubMed] [Google Scholar]
- Rief, M., H. Clausen-Schaumann, and H. E. Gaub. 1999. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 6:346–349. [DOI] [PubMed] [Google Scholar]
- Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997a. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- Rief, M., F. Oesterhelt, B. Heymann, and H. E. Gaub. 1997b. Single molecule force spectroscopy on polsaccharides by atomic force microscopy. Science. 275:1295–1297. [DOI] [PubMed] [Google Scholar]
- Ros, R., F. Schwesinger, D. Anselmetti, M. Kubon, R. Schäfer, A. Plückthun, and L. Tiefenauer. 1998. Antigen binding forces of individually addressed single-chain Fv antibody molecules. Proc. Natl. Acad. Sci. USA. 95:7402–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzina, J., and V. A. Bloomfield. 2001a. Force-induced melting of the DNA double helix. 1. Thermodynamic analysis. Biophys. J. 80:882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzina, J., and V. A. Bloomfield. 2001b. Force-induced melting of the DNA double helix. 2. Effect of solution conditions. Biophys. J. 80:894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. B., Y. Cui, and C. Bustamante. 1996. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 271:795–799. [DOI] [PubMed] [Google Scholar]
- Smith, S. B., L. Finzi, and C. Bustamante. 1992. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 258:1122–1126. [DOI] [PubMed] [Google Scholar]
- Strick, T. R., J.-F. Allemand, D. Bensimon, and V. Croquette. 1998. Behaviour of supercoiled DNA. Biophys. J. 74:2016–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz, T., K. Oroszlan, R. Schäfer, and H.-J. Güntherodt. 1999. Dynamic force spectroscopy of single DNA molecules. Proc. Natl. Acad. Sci. USA. 96:11277–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, P. R., and W. A. Denny. 1996. The mutagenic properties of DNA minor groove binding ligands. Mutat. Res. 355:141–169. [DOI] [PubMed] [Google Scholar]
- Wenner, J. R., M. C. Williams, J. Rouzina, and V. A. Bloomfield. 2002. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 82:3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]