Abstract
The human immunodeficiency virus type 1 (HIV-1) fusion peptide serves as a useful model system for understanding viral/target cell fusion, at least to the lipid mixing stage. Previous solid-state NMR studies have shown that the peptide adopts an oligomeric β-strand structure when associated with a lipid and cholesterol mixture close to that of membranes of host cells of the virus. In this study, this structure was further investigated using four different peptide constructs. In aqueous buffer solution, two of the constructs were primarily monomeric whereas the other two constructs had significant populations of oligomers/aggregates. NMR measurements for all membrane-associated peptide constructs were consistent with oligomeric β-strand structure. Thus, constructs that are monomeric in solution can be converted to oligomers as a result of membrane association. In addition, samples prepared by very different methods had very similar NMR spectra, which indicates that the β-strand structure is an equilibrium rather than a kinetically trapped structure. Lipid mixing assays were performed to assess the fusogenicities of the different constructs, and there was not a linear correlation between the solution oligomeric state and fusogenicity. However, the functional assays do suggest that small oligomers may be more fusogenic than either monomers or large aggregates.
INTRODUCTION
Fusion between the membranes of enveloped viruses such as human immunodeficiency virus type 1 (HIV-1) and influenza and the membranes of their target host cells is an essential step in infection (Hernandez et al., 1996; Dimitrov, 2000; Eckert and Kim, 2001; Blumenthal et al., 2003). For the HIV-1 virus, this process is mediated by the integral membrane viral envelope gp41 protein that contains an N-terminal ∼20-residue apolar fusion peptide (FP) domain. This domain is believed to interact with the target cell membrane and to catalyze membrane fusion. The free fusion peptide has also been shown to be a useful model to understand fusion, at least to the lipid mixing stage. For example, the free HIV-1 peptide causes fusion of liposomes and erythrocytes, and numerous mutational studies have shown strong correlations between fusion peptide-induced liposome fusion and viral/host cell fusion (Freed et al., 1990, 1992; Rafalski et al., 1990; Slepushkin et al., 1990; Martin et al., 1993, 1996; Nieva et al., 1994, 1998; Mobley et al., 1995; Schaal et al., 1995; Delahunty et al., 1996; Durell et al., 1997; Kliger et al., 1997; Pereira et al., 1997; Pecheur et al., 1999; Pritsker et al., 1999). Recent studies suggest that envelope protein regions other than the fusion peptide also interact with membranes and play a role in fusion (Epand et al., 1999; Peisajovich et al., 2000a,b, 2003; Suarez et al., 2000; Sackett and Shai, 2002).
A variety of experimental methods have shown that the HIV-1 fusion peptide can assume helical or nonhelical structures in its membrane-associated forms (Rafalski et al., 1990; Gordon et al., 1992, 2002; Martin et al., 1993, 1996; Nieva et al., 1994, 1998; Pereira et al., 1995, 1997; Chang et al., 1997a; Durell et al., 1997; Mobley et al., 1999; Curtain et al., 1999; Yang et al., 2001a, 2002, 2003; Bodner et al., 2004). Models for the helical structure have been developed based on NMR, electron spin resonance (ESR), infrared, and circular dichroism (CD) data, as well as computer simulations (Martin et al., 1993, 1996; Chang et al., 1997a,b, 1999; Chang and Cheng, 1998; Kamath and Wong, 2002; Maddox and Longo, 2002; Wong, 2003).
Fluorescence, ESR, and solid-state NMR data also suggest that nonhelical HIV-1 fusion peptides form oligomeric β-strand structures (Gordon et al., 1992; Kliger et al., 1997; Pritsker et al., 1999; Yang and Weliky, 2003). In particular, solid-state NMR rotational-echo double-resonance (REDOR) experiments were performed on membrane-associated peptide samples in which half of the peptides contained specific 13C carbonyl backbone labels and the other half of the peptides contained specific 15N backbone labels (Yang and Weliky, 2003). Strong evidence for oligomeric β-strand structure was provided by observation of a significant reduction of the 13C signals by the 15N nuclei. Analysis of data from samples that differed in the residue positions of the labels was consistent with approximately equal populations of parallel and antiparallel strand arrangements.
The possible significance of fusion peptide oligomerization is suggested by atomic-resolution structures of the soluble ectodomain of gp41 that show stable gp41 trimers (Chan et al., 1997; Tan et al., 1997; Weissenhorn et al., 1997; Caffrey et al., 1998; Yang et al., 1999). These structures end ∼10 residues C-terminal of the fusion peptide domain, and their three N-termini are close together at the ends of an in-register helical coiled coil. Thus, it appears that at least three fusion peptides are in close proximity when they interact with the target membrane. In the HIV-1 fusion peptide model system, the biologically relevant topology has been mimicked with C-terminal cross-linking, and it has been observed that the cross-linked peptides can induce both a greater final extent and a more rapid rate of fusion than their noncross-linked analogs (Yang et al., 2003). Similar fusogenic enhancements have also been observed for influenza protein domains in which the fusion peptide was thought to assume the biologically relevant oligomeric topology (Kim et al., 1996, 1998; Macosko et al., 1997; Epand et al., 1999, 2001; LeDuc et al., 2000; Leikina et al., 2001). Experiments and modeling studies further suggest that the fusion site contains multiple envelope protein trimers and a corresponding high fusion peptide concentration (Blumenthal et al., 1996; Weissenhorn et al., 1997; Caffrey et al., 1998; Munoz-Barroso et al., 1998; Kuhmann et al., 2000; Bentz, 2000a,b; Mittal and Bentz, 2001; Kliger et al., 2001; Lin et al., 2003). In addition, the functionally disruptive Val-2 to Glu-2 mutation in the gp41 fusion peptide is trans-dominant, i.e., cells expressing 10% mutant protein and 90% wild-type protein exhibit only 40% of the fusion activity of cells with 100% wild-type protein (Freed et al., 1992). One interpretation of these data is that the mutant peptide disrupts the correct assembly of a functionally important fusion peptide oligomer (Kliger et al., 1997; Pritsker et al., 1999).
This article investigates the aqueous oligomerization states of different constructs of model fusion peptides and the relationships between soluble peptide oligomerization, fusion activity, and membrane-associated peptide structure. Soluble peptide oligomerization is considered within the context of previous studies from our and other research groups. Although we use the term oligomerization, in most studies the actual numbers of peptides in a soluble cluster or aggregate have not been well defined. In our group, it was shown that the association state of a 23-residue HIV-1 fusion peptide depends on the buffer type, pH, and salt concentration (Yang et al., 2001a). NMR has also been done on FP in unbuffered aqueous solution at pH 3 and would only be possible if the peptide were monomeric or formed small oligomers (Chang et al., 1997b). By contrast, in buffered neutral pH solutions with 100–150 mM NaCl, FP and related peptides have been shown to form filamentous aggregates (Slepushkin et al., 1992; Pereira et al., 1997). Peptide oligomerization has also been investigated for a construct representing residues 5–55 of gp41 (Chang et al., 1999). In this study, pulsed field gradient NMR and light scattering methods were applied to the peptide in water, and gel filtration chromatography was applied to the peptide in a solution containing 78% H2O/22% acetonitrile/0.1% trifluoracetic acid (TFA). The derived molecular weights were consistent with dimeric or trimeric peptide.
In related studies by the Tamm group on the influenza fusion peptide, oligomerization of the peptide in aqueous buffered solution was probed by CD and ESR measurements (Han and Tamm, 2000a,b). At lower ionic strengths, the CD spectra were not consistent with either helical or β-strand structure, and the ESR spectra of spin-labeled peptides yielded sharp lines, whereas at higher ionic strengths, the CD spectra were consistent with a mixture of helical and β-structures, and the ESR spectra were attenuated. The low and high ionic strength data were interpreted in terms of monomeric and oligomeric/aggregated peptide states, respectively. In addition, binding of the peptide to membranes was measured using 50-nM peptide concentrations and variable concentrations of liposomes composed of 4:1 choline/glycerol lipids. In low ionic strength buffer at pH 5, thermodynamic analysis of the data was consistent with an ∼−1-kcal/mol free energy of peptide self-association in membranes.
Although CD, ESR, light scattering, gel filtration, and NMR can provide information about oligomerization, analytical ultracentrifugation is probably the most definitive probe of peptide oligomerization in aqueous environments and is the method of choice for our studies on the HIV-1 fusion peptide. One specific example of the insight provided by analytical ultracentrifugation is studies on the previously described influenza fusion peptide. We examined two different influenza fusion peptide constructs that both contained C-terminal lysines to improve peptide solubility. In low ionic strength buffer, a construct with three lysines was highly aggregated whereas a construct with six lysines was predominantly monomeric (P. D. Parkanzky, M. Prorok, F. J. Castellino, and D. P. Weliky, unpublished). For reference, the construct in the Tamm studies contained four lysines (Han and Tamm, 2000a,b).
In this article, analytical ultracentrifugation characterization of the soluble oligomerization state of the peptide is also related to functional lipid mixing assays and to solid-state NMR structural studies of membrane-associated fusion peptides. The NMR samples are typically prepared by mixing aqueous solutions of peptide and liposomes, and the conditions are intentionally similar to those used in functional assays so that it is straightforward to correlate structure with function. Previous NMR data have shown that the fusion peptide has an oligomeric β-strand structure when associated with membranes whose lipid headgroup and cholesterol composition is comparable to that of host cells of the virus (Yang et al., 2001a,b, 2002, 2003; Yang and Weliky, 2003; Bodner et al., 2004). However, it was not clear whether oligomer formation was the result of peptide association with the membrane or whether the oligomers preexisted in aqueous solution and then bound to the membrane. Studies of peptide aqueous oligomerization can address this question.
It is also noted that the numbers of peptides in the membrane-associated HIV-1 fusion peptide clusters have not yet been well defined by experiment. There is qualitative evidence from solid-state NMR studies that the clusters are small; in particular, the 13C linewidths are narrower at room temperature than at −50°C, and the signal/noise ratio per 13C at room temperature is ∼1/3 of the value obtained at −50°C (Bodner et al., 2004). Both of these observations suggest that there is significantly greater motion at room temperature than at −50°C, which might be expected for a membrane-associated cluster with a small number of peptides, but which would be less likely for a large solid-like peptide aggregate. In addition, oligomer sizes have been probed in sodium dodecylsulfate (SDS) gels and show masses up to peptide tetramers (Kliger et al., 1997; Pritsker et al., 1999).
In addition to the investigation of HIV-1 fusion peptide oligomerization and its relationship to fusogenicity and membrane-associated structure, this article also considers the separate question of whether the oligomeric β-strand structure observed in solid-state NMR studies represents a thermodynamic equilibrium structure or a kinetically trapped structure. The latter possibility could arise from an activation energy barrier to peptide incorporation into the membrane. If this barrier could be overcome, a different membrane-associated structure might be formed such as the helical monomer observed in detergent at peptide/detergent ∼0.01 (Chang et al., 1997a; Chang and Cheng, 1998). This issue is investigated by comparing the spectra from NMR samples prepared by two very different methods. In one method, peptides bind directly to large unilamellar vesicles (LUVs) in aqueous solution and induce lipid mixing and fusion. For this approach, the initial interaction of peptide and lipid is very similar to that of functional fusion assays. In the second incorporation method, peptide, lipid, and cholesterol are initially cosolubilized in organic solvent, the solvent is then evaporated, and the mixture rehydrated to form membranes. Because the components initially form a clear homogeneous organic solution without lipid bilayers, there may be a reduced barrier to peptide incorporation with the lipid. For other helical peptides such as gramicidin and magainin, it is also known that with peptide/lipid ≤ 0.1, organic dissolution followed by rehydration yields peptides with helical structure (Ketchem et al., 1993; Ramamoorthy et al., 1995; Hirsh et al., 1996; Opella et al., 1999).
MATERIALS AND METHODS
Materials
Rink amide resin was purchased from Advanced Chemtech (Louisville, KY), and 9-fluorenylmethoxycarbonyl (FMOC)-amino acids were obtained from Peptides International (Louisville, KY). Labeled amino acids were purchased from Icon Services (Summit, NJ) and were FMOC-protected using literature methods (Chang et al., 1980; Lapatsanis et al., 1983). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-L-serine) (POPS), phosphatidylinositol (PI), 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-rac-(1-glycerol)) (POPG), sphingomyelin, N-(7-nitro-2,1,3-benzoxadiazol-4-yl)-phosphatidylethanolamine (N-NBD-PE), N-(lissamine Rhodamine B sulfonyl)-phosphatidylethanolamine (N-Rh-PE), and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL). The Micro bicinchoninic acid (BCA) protein assay was obtained from Pierce (Rockford, IL). HEPES and Triton X-100 were obtained from Sigma (St. Louis, MO). All other reagents were analytical grade. The buffer solution used in the study contained 5 mM HEPES (pH 7.0) with 0.01% NaN3.
Peptides
Four different peptide constructs were synthesized. FP (AVGIGALFLGFLGAAGSTMGARS) corresponded to the 23 N-terminal residues of the LAV1a strain of HIV-1 gp41; FPK3 (AVGIGALFLGFLGAAGSTMGARSKKK) contained three additional C-terminal lysines to increase peptide solubility in aqueous solution; FPW (sequence AVGIGALFLGFLGAAGSTMGARSW) contained an additional C-terminal tryptophan whose 280-nm absorbance was useful for the analytical ultracentrifugation studies; and FPK3W (AVGIGALFLGFLGAAGSTMGARSKKKW) contained both the C-terminal lysines and tryptophan. All peptides were synthesized as their C-terminal amides using a peptide synthesizer (ABI 431A, Foster City, CA) equipped for FMOC chemistry. After synthesis, peptides were cleaved from the resin in a 3-h reaction using a mixture of TFA/H2O/phenol/thioanisole/ethanedithiol in a 33:2:2:2:1 vol ratio. Peptides were subsequently purified by reversed-phase high-performance liquid chromatography (HPLC) using a preparative C18 column (Vydac, Hesperia, CA) and a water/acetonitrile gradient containing 0.1% TFA. Mass spectroscopy was used for peptide identification. For the solid-state NMR experiments, peptides were synthesized with a 13C carbonyl label at Phe-8 and a 15N label at Leu-9.
Analytical ultracentrifugation
Sedimentation equilibrium experiments at room temperature were performed on Beckman XL-I and XL-A analytical ultracentrifuges (Palo Alto, CA) using An-60 Ti rotors. The instruments were operated in absorbance mode at either 225, 230, or 280 nm. Samples were loaded into six-channel epon charcoal-filled centerpieces equipped with quartz windows and were equilibrated at rotor speeds between 3000 and 52,000 rpm. Data were fitted with the analysis software supplied by Beckman.
Lipid preparation
A lipid and cholesterol mixture was used that reflected the approximate lipid headgroup and cholesterol composition of membranes of host cells of the HIV-1 virus (Aloia et al., 1993). This mixture (denoted “LM3”) contained POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-L-serine), sphingomyelin, phosphatidylinositol, and cholesterol in a 10:5:2:2:1:10 mol ratio. Preparation of large unilamellar vesicles began with dissolution of lipid and cholesterol powders in chloroform. The chloroform was removed under a stream of nitrogen followed by overnight vacuum pumping. Lipid dispersions were formed by addition of 5 mM pH 7 HEPES buffer followed by homogenization with 10 freeze-thaw cycles. LUVs of 100-nm diameter were subsequently prepared by extrusion (Hope et al., 1985).
Lipid mixing assay of membrane fusion
A resonance energy transfer assay was used to monitor membrane fusion (Struck et al., 1981). Two types of 100-nm diameter LM3 LUVs were prepared. One set contained 2 mol % of the fluorescent lipid N-(7-nitro-2,1,3-benzoxadiazol-4-yl)-phosphatidylethanolamine and 2 mol % of the quenching lipid N-(lissamine Rhodamine B sulfonyl)-phosphatidylethanolamine, whereas the other set only contained unlabeled lipids. Fluorescently labeled and unlabeled vesicles were mixed in a 1:9 ratio. After addition of the fusion peptide, lipid mixing between labeled and unlabeled vesicles caused dilution of the labeled lipids with a resulting increase of fluorescence. Fluorescence was recorded using 4 nm bandwidth on an Instruments S. A. Fluoromax-2 spectrofluorimeter (Edison, NJ) operating at excitation and emission wavelengths of 465 and 530 nm, respectively. A siliconized glass cuvette was used with continuous stirring in a thermostated cuvette holder. Measurements were carried out at 37°C with 2 ml LUVs (150 μM total lipid and 75 μM cholesterol) in buffer solution. The initial residual fluorescence intensity, F0, referenced zero lipid mixing. After quantitation of the peptide concentration with amino acid analysis, the requisite volume of 100 μM peptide in 5 mM pH 7 HEPES was added manually to achieve the desired peptide/lipid mol ratio. After addition and manual mixing of the peptide, the fluorescence (Ft) of the sample was monitored as a function of time and eventually reached a constant value. The maximal fluorescence intensity, Fmax, was then obtained after addition of 10 μl 20% Triton X-100 detergent. Ft and Fmax were corrected for the small fluorophore dilution resulting from the added volumes of peptide and Triton X-100. Percent lipid mixing at time t is denoted as Mt and was calculated using
 |
(1) |
The long-time or final extent of lipid mixing is denoted as Mf and was used to compare the relative fusogenicities of different peptide constructs. For a given set of labeled and unlabeled vesicles, Mf had ±2% reproducibility between different assay trials. Rates of lipid mixing are another measure of fusogenicity but could not be accurately obtained from the experimental data because the time taken for manual addition and mixing of the peptide was significant relative to characteristic fusion times. In future studies, these rates could be determined using data obtained on a stopped-flow fluorimeter.
Preparation method 1 for membrane-associated fusion peptide samples for solid-state NMR studies
Peptide (∼0.4 μmol) was dissolved in ∼35 ml buffer. LUVs of LM3 (40 μmol total lipid and 20 μmol cholesterol) were prepared in ∼2 ml buffer. The vesicle and peptide solutions were mixed and sat overnight at room temperature. The peptide/lipid mixture was then ultracentrifuged at 100,000 × g for 4 h so that the membrane and associated peptide pelleted. FP and FPK3 binding were typically quantitative as determined by bicinchoninic acid assay measurements of negligible peptide concentration in the supernatant. The pellet was transferred by spatula to a 6-mm diameter magic angle spinning (MAS) NMR rotor.
Preparation method 2 for membrane-associated fusion peptide samples for solid-state NMR studies
Peptide, lipid, and cholesterol (0.4 μmol:40 μmol:20 μmol) were codissolved in a solvent mixture made from trifluoroethanol, chloroform, and hexafluoropropanol in a 2:3:2 vol ratio. Solvent was removed by gaseous nitrogen followed by overnight vacuum pumping. The mixture was then hydrated in 2 ml buffer solution and subjected to six freeze-thaw cycles. An additional 34 ml buffer solution was added and the sample was then ultracentrifuged at 100,000 × g for 4 h. The pellet was transferred to the NMR rotor.
Solid-state NMR spectroscopy
In membrane-associated fusion peptide samples, room temperature 13C cross-polarization NMR signals are attenuated, presumably because of slow motion. Because there is three times higher 13C signal/noise at −50°C relative to room temperature, measurements in this study were made at the lower temperature. In previous studies of these types of samples, comparative 13C NMR spectra have been obtained at room temperature and at −50°C, and changes in individual shifts were less than 0.5 ppm over this temperature range (Bodner et al., 2004). Because NMR chemical shifts are diagnostic of peptide structure, it is likely that the peptide structure observed at −50°C is similar to the structure at more physiologically relevant temperature. The temperature independence of the shifts was observed both for helical and β-strand fusion peptide structures.
Experiments were done on a 9.4 T spectrometer (Varian Infinity Plus, Palo Alto, CA) using a triple resonance MAS probe. The NMR detection channel was tuned to 13C at 100.8 MHz, the decoupling channel was tuned to 1H at 400.8 MHz, and the third channel was tuned to 15N at 40.6 MHz. Experiments were carried out using a MAS frequency of 8000 Hz that was stabilized to ±2 Hz. For the NMR experiments, peptides were 13C carbonyl labeled at Phe-8 and 15N amide labeled at Leu-9. NMR spectra were taken using a REDOR filter of the 13C-15N dipolar interaction so that the Phe-8 carbonyl was the only signal observed in the 13C-detected REDOR difference spectrum (Gullion and Schaefer, 1989; Yang et al., 2002). The filtering is important because it allows background-free observation of the distribution of chemical shifts of the labeled Phe-8, which is the spectral feature of interest. Without filtering, this distribution is superimposed on the 13C natural abundance background due to peptide and lipid.
Between 1 and 2 ms of cross-polarization at 50 kHz was followed by a 1-ms REDOR dephasing period and then direct 13C detection. A single 50-kHz 13C refocusing π-pulse was placed at the center of the dephasing time, and 1H TPPM decoupling of 65 kHz was applied during both dephasing and detection (Bennett et al., 1995). 13C chemical shifts were referenced to the methylene resonance of adamantane (38.5 ppm) and the 13C transmitter was set to 155 ppm. 15N chemical shifts were referenced to (NH4)2SO4 (20 ppm) and the 15N transmitter was set to 115 ppm. For the S1 acquisition, the dephasing time contained a 40-kHz 15N π-pulse at the middle and end of each rotor cycle, except for the ends of the fourth and eighth cycles, whereas the S0 acquisition did not contain these pulses. XY-8 phase cycling was used for the 15N pulses (Gullion et al., 1990; Gullion and Schaefer, 1991). During the dephasing period, pulses were not actively synchronized to the rotor phase. To obtain optimal compensation of B0, B1, and spinning frequency drifts, S0 and S1 acquisitions were acquired alternately. The recycle delays were between 1 and 2 s.
RESULTS
Analytical ultracentrifugation
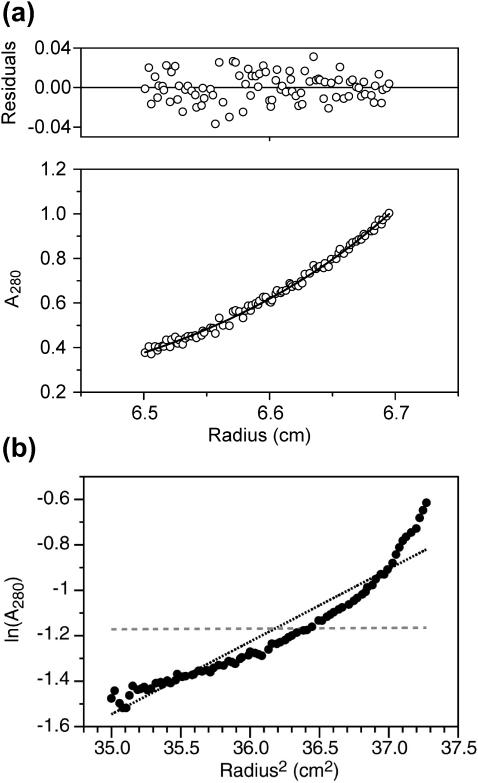
Fig. 1 a displays one example of sedimentation equilibrium data and fitting for an 80-μM solution of FPK3W in 5 mM pH 7 HEPES buffer. The rotor speed was 52,000 rpm, and the peptide concentration was approximately the same as the 100-μM concentration of stock peptide solutions used in the lipid mixing fusion assays. The bottom panel of Fig. 1 a displays 280-nm absorbance (proportional to peptide concentration) as a function of centrifugal radius. The data were fit to a single molar mass (M) using
 |
(2) |
where A and A0 are the experimental absorbances at radius r and reference radius r0, respectively,  is the partial specific volume of FPK3W, ρ is the buffer density, ω is the angular velocity, R is the ideal gas constant, and T is the temperature (Cantor and Shimmel, 1980). This equation assumes no baseline offset and a single value of M, i.e., a single self-association state for all peptides in solution. The value of ν (0.76 ml/g) was calculated from the mass average of the partial specific volumes of the individual amino acids in FPK3W (Laue et al., 1992). The value of ρ was set to 1.0 g/ml. The displayed data were best fit to M = 2600 g/mol, which is close to the monomer mass of 2690 g/mol. As displayed in the top panel of Fig. 1 a, the differences between the experimental and fitted absorbances were small and random as a function of r, which indicates that a single-species model is reasonable. Additionally, a plot of ln(A) versus r2 was linear, as would be expected from Eq. 1. Data were also acquired at 35,000 and 45,000 rpm and could be fit well to Eq. 1 with values of M between 2550 and 2700 g/mol. Thus, these studies demonstrate that FPK3W at ∼100-μM concentration is predominantly monomeric in 5 mM pH 7 HEPES buffer.
is the partial specific volume of FPK3W, ρ is the buffer density, ω is the angular velocity, R is the ideal gas constant, and T is the temperature (Cantor and Shimmel, 1980). This equation assumes no baseline offset and a single value of M, i.e., a single self-association state for all peptides in solution. The value of ν (0.76 ml/g) was calculated from the mass average of the partial specific volumes of the individual amino acids in FPK3W (Laue et al., 1992). The value of ρ was set to 1.0 g/ml. The displayed data were best fit to M = 2600 g/mol, which is close to the monomer mass of 2690 g/mol. As displayed in the top panel of Fig. 1 a, the differences between the experimental and fitted absorbances were small and random as a function of r, which indicates that a single-species model is reasonable. Additionally, a plot of ln(A) versus r2 was linear, as would be expected from Eq. 1. Data were also acquired at 35,000 and 45,000 rpm and could be fit well to Eq. 1 with values of M between 2550 and 2700 g/mol. Thus, these studies demonstrate that FPK3W at ∼100-μM concentration is predominantly monomeric in 5 mM pH 7 HEPES buffer.
FIGURE 1.
(a) Sedimentation equilibrium data and analysis at 20°C of a sample made with 80 μM FPK3W peptide in 5 mM pH 7 HEPES buffer. The bottom panel displays the 280-nm absorbance as a function of centrifugal radius. The data are represented as open circles and were obtained after 20 h spinning at 52,000 rpm. The superimposed curve represents the best fit to the data and was obtained using a molar mass of 2600 g, which is comparable to the 2700-g molar mass of a peptide monomer. The upper panel shows the differences between the experimental and best-fit absorbances. (b) Sedimentation equilibrium data at 5000 rpm of a sample made with 80 μM FPW peptide in 5 mM pH 7 HEPES buffer. The logarithm of 280-nm absorbance is plotted as a function of (radius)2. The data are represented as dark circles and show significant curvature, which indicates that the peptide does not form a single oligomeric species. The dark-dotted and light-dashed lines represent the results that would be expected for 230,000- and 2300-g molar mass species and correspond to FPW 100 mers and monomers, respectively. It appears that the solution contains large FPW oligomers.
Fig. 1 b displays analytical ultracentrifugation data for a solution containing 80-μM FPW in the buffer solution. Peptide pelleting was observed at speeds above 15,000 rpm, so the displayed data were obtained at 5000 rpm. A mean value of M ∼ 230,000 g/mol was estimated from linear fitting and corresponds to ∼100 peptide molecules. However, the experimental data has substantial curvature, which indicates that there is a significant distribution of oligomer/aggregate sizes.
Analytical ultracentrifugation experiments were also attempted on FP and FPK3 with detection of peptide concentration at 225 or 230 nm. Quantitative analysis of the data was difficult because of subtraction of the large buffer absorbance at this wavelength. At high speeds, some pelleting was observed for FP (but not for FPK3), which indicated that there was a population of larger FP oligomers or aggregates.
Some information about the association state of FP can also be gleaned from the results of previous measurements of amide 1H NMR linewidths for neutral aqueous solutions of FP at 100-μM concentration (Yang et al., 2001a). The narrowest linewidths were ∼15 Hz, and through approximate hydrodynamic modeling these linewidths can be correlated to a heptamer peptide mass (Cavanaugh et al., 1996). Solvent exchange also contributes to the observed linewidths, so the true mass may be smaller and might even correspond to the monomer mass. In addition, comparison of solubilities and rates of dissolution of the different peptide constructs yields the ordering FPW < FP < FPK3W, FPK3.
In summary, the analytical ultracentrifugation data show that in the 5-mM HEPES buffer solution 1), FPW is predominantly associated as large oligomers/aggregates; 2), there is a population of FP that forms large oligomers/aggregates; and 3), FPK3W is predominantly monomeric. Because FPK3 contains the same number of charged residues as FPK3W and has comparable solubility properties to FPK3W, we think that FPK3 is also predominantly monomeric in the buffer solution. Because FP has solubility properties that are intermediate between FPW and FPK3W, and because of the observation of solution NMR signals for FP in aqueous solution, we suggest that FP solutions also contain a population of oligomers that are intermediate in size between monomers and large aggregates. FP solutions may also contain a population of monomer peptides.
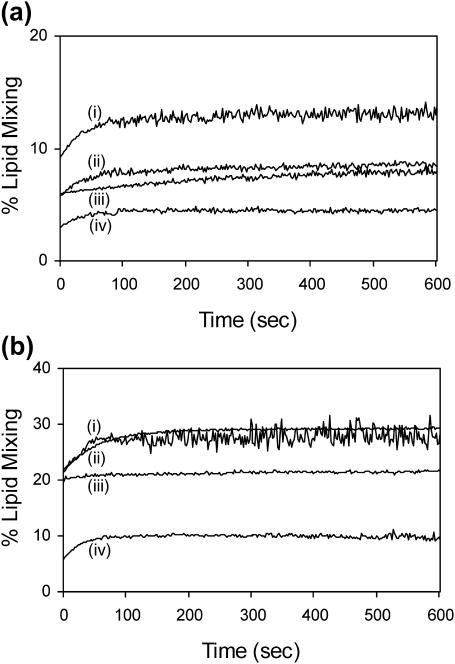
Lipid mixing
Fig. 2 displays plots of Mt versus time for each of the constructs with (a) peptide/lipid = 0.01 and (b) peptide/lipid = 0.02. All peptides induce significant lipid mixing. At peptide/lipid = 0.01, the ordering of Mf values for the different peptides is FPW < FPK3, FPK3W < FP, whereas at peptide/lipid = 0.02, the ordering is FPW < FPK3W < FP, FPK3. Overall, FPW appears to be the least fusogenic whereas FP is the most fusogenic. The data in the analytical ultracentrifugation section suggest that the solution oligomerization states of the peptides are FPK3W and FPK3, monomeric; FP, mixture of small and large oligomers; and FPW, large oligomers/aggregates. Thus, there does not appear to be a linear relationship between solution oligomerization state and fusogenicity, although it may be that small oligomers (FP) promote lipid mixing more efficiently than monomers (FPK3W and FPK3) or large aggregates (FPW).
FIGURE 2.
Lipid mixing induced in 100-nm diameter LM3 vesicles by fusion peptides. The total lipid and cholesterol concentrations were 150 and 75 μM, respectively, and the same batches of vesicles were used for all assays. In a and b, the curves correspond to the following peptides: (i) FP; (ii) FPK3; (iii) FPK3W; and (iv) FPW. In a, the peptide concentration was 1.5 μM, and in b the peptide concentration was 3.0 μM. Data acquisition began after manual addition and mixing of the peptide in the vesicle solution. Because the time for these steps is significant relative to characteristic lipid mixing times, the apparent lipid mixing is nonzero at 0 s.
Solid-state NMR spectra
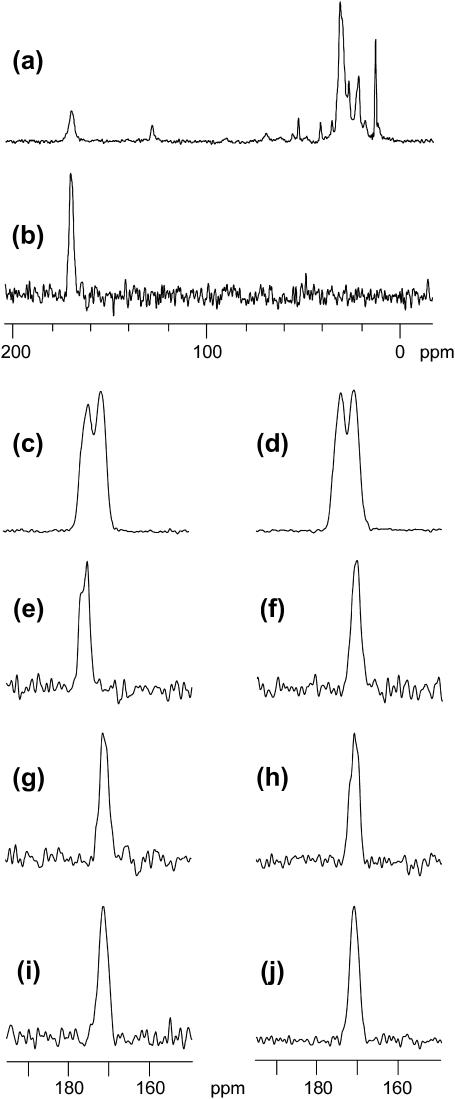
Fig. 3 displays 13C NMR spectra of HIV-1 fusion peptides. Panels a and b, respectively, represent unfiltered (S0) and REDOR-filtered (S0 − S1) 13C NMR spectra of a membrane-associated peptide sample prepared by method 1 and containing FPK3 (∼0.4 μmol) and LM3 (40 μmol total lipid and 20 μmol cholesterol). The REDOR-filtered spectrum is dominated by the Phe-8 carbonyl signal with near-quantitative suppression of natural abundance 13C signals. Panels c–j display the carbonyl regions of REDOR-filtered spectra. The Phe-8 chemical shift distribution in each spectrum provides information about the local Phe-8 conformation in the sample, as will be described subsequently. Spectra c–d are for lyophilized peptides, spectrum e is for peptide associated with detergent, and spectra f–j are for peptides associated with LM3 membranes.
FIGURE 3.
13C solid-state NMR spectra of fusion peptide samples. Spectrum a is the unfiltered S0 spectrum of an FPK3/LM3 sample containing ∼0.4 μmol peptide, 40 μmol total lipid, and 20 μmol cholesterol, and spectrum b is the REDOR-filtered difference spectrum of this sample. Because the peptide was 13C carbonyl labeled at Phe-8 and 15N labeled at Leu-9, the difference spectrum is dominated by the Phe-8 carbonyl signal. The vertical scale in spectrum b is expanded by a factor of ∼6.5 relative to the scale in spectrum a. In c–j, REDOR-filtered difference spectra are displayed for samples containing different fusion peptide constructs: (c) FP freeze-dried from a ∼500-μM aqueous solution; (d) FPK3 freeze-dried from a ∼100-μM aqueous solution; (e) FPK3 (2 mM) in dodecylphosphocholine detergent solution (200 mM); (f) FP, (g and j) FPK3, (h) FPW, and (i) FPK3W associated with LM3 (∼0.4 μmol peptide, 40 μmol total lipid, and 20 μmol cholesterol). The samples used for spectra f–i were made by method 1, with mixing of aqueous peptide and vesicle solutions. The sample used for spectrum j was made by method 2, with organic cosolubilization of peptide, lipid, and cholesterol, followed by evaporation of the organic solvent and hydration with aqueous buffer. The spectra in c and d display two peaks and indicate conformational heterogeneity for the lyophilized peptide. The spectrum in e is peaked in the 175–176-ppm range, and the spectra in f–j are peaked in the 171–172-ppm range. The chemical shifts are consistent with helical structure near Phe-8 for the detergent sample and β-strand structure near Phe-8 for the LM3 samples. Data were acquired using cross-polarization, 8-kHz MAS frequency, and temperatures of −50°C for spectra a–d and f–j and −80°C for spectrum e. Each spectrum was processed with 50-Hz Gaussian line broadening and polynomial baseline correction. There were 12,288 transients summed in spectrum a, and the total (S0 + S1) numbers of transients in the REDOR-filtered difference spectra were (b) 24,576; (c) 20,000; (d) 31,488; (e) 132,524; (f) 22,336; (g) 24,576; (h) 159,808; (i) 186,560; and (j) 166,336.
It would be very interesting to have solid-state NMR spectra of the peptides in frozen solutions at the ∼100-μM concentrations that were used in the analytical ultracentrifugation studies and that were used in the stock peptide solutions of the fusion assays. These frozen solution spectra could be compared with the solid-state NMR spectra of detergent- and membrane-associated peptides and could provide some information about conformational distributions in solution. However, for our 400-MHz NMR spectrometer, the detection limit is ∼0.2 μmol 13C labeled peptide, and in the 160-μL rotor volume a 100-μM peptide sample would only contain ∼0.02 μmol peptide. Although the frozen solution experiments are unfeasible, we made some effort to probe the solution state by obtaining spectra of lyophilized aqueous solutions of FP or FPK3. The peptide concentrations before lyophilization were ∼500 and 100 μM, respectively, and the lyophilized peptide NMR spectra are displayed in Fig. 3, c and d. Both spectra show distinct peaks at 174–175 and 171–172 ppm, which indicate that there are at least two Phe-8 conformations in the lyophilized state. Possible assignments for these conformations will be given after the following discussion of the spectra of the detergent- and membrane-associated peptides.
To obtain information about the fusion peptide monomer in a lipid-like environment, we made a sample containing ∼0.3 μmol FPK3 and ∼30 μmol dodecylphosphocholine (DPC) in buffer. In this sample, each micelle contained approximately one peptide and ∼100 detergent molecules. Fig. 3 e displays the NMR spectrum of this sample (obtained at −80°C) and contains a signal peaked at 175–176 ppm. Panels f–j display spectra of different fusion peptide constructs associated with LM3, and each contained ∼0.4 μmol peptide, 40 μmol lipid, and 20 μmol cholesterol. The samples in f–i were made by method 1 and contained FP, FPK3, FPW, and FPK3W, respectively, and the sample in j was made by method 2 and contained FPK3. All of the spectra in f–j have similar appearances with symmetric lineshapes, peak chemical shifts of 171–172 ppm, and full-width at half-maximum (FWHM) linewidths of ∼2.5 ppm.
Previous studies have provided information about the local secondary structure near Phe-8 in the HIV-1 fusion peptide in different environments. We use these studies in conjunction with known correlations between chemical shifts and conformation to understand the spectra and local Phe-8 conformations in the NMR samples. For FP associated with detergent at peptide/detergent ∼ 0.01, a solution NMR study showed predominant helical structure from Ile-4 to Gly-16 whereas, for FP associated with LM3, 2D solid-state NMR measurements were consistent with predominant β-strand structure from Val-2 to Ala-15 (Chang et al., 1997a; Yang et al., 2001a; Gabrys et al., 2003). Considering these results, it is most reasonable to assign the 175–176-ppm Phe-8 signal in Fig. 3 e to helical structure and the 171–172-ppm signal in f to β-strand structure. These assignments are generally consistent with the 175- and 169-ppm carbonyl shifts of solid α-helical and solid β-sheet polyphenylalanine, respectively, and with the 175.1- ± 1.4-ppm (helical) and 172.2- ± 1.6-ppm (β-strand) shift distributions observed for Phe carbonyls in soluble proteins (Kricheldorf and Muller, 1983; Saito, 1986; Zhang et al., 2003). These distributions are derived from a chemical shift database of soluble proteins of known structures, and each distribution contains >50 shifts. The referencing for both the solid-state shifts and for the solution shift distributions can be traced to neat tetramethylsilane at 0 ppm (Wishart et al., 1995; Morcombe and Zilm, 2003).
The previously described 175–176-ppm assignment to helical Phe-8 and 171–172-ppm assignment to β-strand Phe-8 are used to interpret the other spectra displayed in Fig. 3. In particular, the similar shifts observed for FP, FPK3, FPW, and FPK3W associated with LM3 membranes (f–j) suggest that Phe-8 has β-strand structure in all of these samples. In addition, the β-strand structure is dominant for either aqueous (g) or organic (j) incorporation of FPK3 peptide with lipid.
For LM3-associated FP and FPK3, interpeptide REDOR measurements have shown that the peptide forms β-strand oligomers, and we expect the Phe-8 β-strand structure observed for LM3-associated FPW and FPK3W also correlates with oligomer formation (Yang and Weliky, 2003). In the FPK3/detergent sample, there was about one peptide per micelle, so it is likely that the peptide is monomeric. For a monomeric peptide embedded in an apolar environment, hydrogen bonding with water is improbable, and helical structure will likely be favored because it maximizes intrapeptide hydrogen bonding. In the membrane samples, there are 102–103 peptides per unfused vesicle, and there is the possibility of interpeptide hydrogen bonding and formation of oligomeric β-strand structure either before or after fusion has occurred. For detergent micelles, it has also been shown that there is an increase in β-strand structure with increased peptide/detergent ratio (Gordon et al., 1992). At FP/detergent = 0.005, the CD spectrum was deconvolved as 55% α-helical and 45% random coil, whereas at FP/detergent = 0.1 (several peptides per micelle) the spectrum was deconvolved as 40% α-helical, 35% β-strand, and 25% random coil. The increased β-strand content may be due to formation of β-strand oligomers within the micelle.
The presence of two components in the lyophilized FP and FPK3 peptide spectra (panels c–d) is consistent with a mixture of conformations. The component centered at 171–172 ppm likely corresponds to β-strand structure, whereas the component centered at 174–175 ppm likely corresponds to helical or other non-β-strand structure. This latter component is not observed in the spectra of any of the membrane-associated peptides and suggests that there is a significant conformational change near Phe-8 for a large fraction of the FP and FPK3 peptides when they bind to LM3 membranes.
There are at least two explanations for the origin of the 171–172-ppm β-strand component in the lyophilized peptides. It may be that the β-strand component is not found in aqueous solution and results instead from β-strand formation during lyophilization. An alternative explanation is that the β-strand conformation exists in aqueous solution and is in rapid interconversion with other conformations. The possibility of rapid interconversion is supported by a fairly narrow (∼1 ppm) distribution of amide 1H shifts for these peptides in solution (Cavanaugh et al., 1996; Yang, 2003). The interconversion would then be stopped by freezing and lyophilization. In this model, each peptide in the lyophilized state has a conformation close to the one that it had just before freezing, and the distribution of conformations among all of the lyophilized peptides is approximately the same as the conformational distribution in solution. It is known that there is a good correlation between the solution and lyophilized conformational distribution for a 17-residue predominantly α-helical peptide (Long and Tycko, 1998; Balbach et al., 2000; Yang and Weliky, 2003).
As described in the previous paragraph, there are two interpretations of the 171–172 ppm β-strand component of the lyophilized peptides: 1), formation of β-sheet structure during lyophilization; and 2), a β-strand conformation that interconverts rapidly with other conformations in solution and that is trapped by lyophilization. If the first interpretation is correct, then the 174–175 component would likely correspond to peptides that had not converted to β-strand during lyophilization. It is likely that this latter population of peptides was conformationally disordered in solution, as evidenced by solution CD spectra that did not correlate well with model spectra of either α-helical, β-strand, or random coil conformations (Chang et al., 1997b; Yang, 2003). However, analysis of the lyophilized FPK3 spectrum yielded a 3-ppm linewidth (full-width at half-maximum) for the 174–175-ppm component, which is narrower than the 5–6-ppm linewidths of labeled carbonyl resonances in ∼10 mM frozen solutions of conformationally disordered antibody epitope peptides, as well as the 5–6-ppm linewidths of solid peptides lyophilized from these solutions (Weliky et al., 1999; Yang et al., 2001a; D. P. Weliky, unpublished data). Given the 5–6-ppm linewidth expected for disordered structure, it may be more reasonable to assign the total 170–176-ppm signals in Fig. 3, c and d, to the aqueous peptide conformation rather than just the 174–175-ppm component.
In summary, 1), comparison of spectra c and d and f–i in Fig. 3 suggests that a significant fraction of the peptides convert from non-β-strand conformation in solution to β-strand conformation after association with the membrane; and 2), the β-strand structural component in the lyophilized peptide may reflect some β-strand conformation in aqueous solution or may be an artifact of lyophilization.
DISCUSSION
The importance of fusion peptide oligomers in viral/target cell fusion has been implied by the large reduction in fusion observed for viruses containing predominantly wild-type fusion peptide and small amounts of mutant fusion peptide (Freed et al., 1992). A similar relationship has been observed in the fusion peptide model system, and formation of a correctly assembled HIV-1 fusion peptide oligomer has also been postulated as an important step in membrane fusion catalysis (Kliger et al., 1997; Pritsker et al., 1999). Detection of oligomers in the membrane-associated forms of FP, FPK3, and other peptide constructs has been made by solid-state NMR and other techniques, but it has not been clear whether these oligomers preexisted in aqueous solution or whether they formed upon membrane association. In this study, we demonstrate that a peptide construct (FPK3W) that is predominantly monomeric in aqueous solution adopts β-strand structure upon membrane association. Previous solid-state NMR studies have shown that the β-strand structure is oligomeric, so it appears that oligomerization can occur upon membrane association.
The β-strand structure was also observed for samples prepared with initial cosolubilization of FPK3, lipid, and cholesterol in organic solvent. Observation of the same structure with both organic and aqueous incorporation of peptide with lipid suggests that the β-strand is an equilibrium rather than a kinetically trapped structure for peptide associated with LM3 membranes. In particular, for the organic approach, a homogeneous solution of lipid and peptide was initially made without lipid bilayer structure, which may make it easier to achieve final equilibrium peptide incorporation. In future work, equilibrium incorporation could also be attempted by solubilization of peptide, lipid, and cholesterol in surfactant solution, followed by removal of the surfactant (Marassi et al., 1997).
Figs. 1 and 3 indicate that the membrane-associated oligomeric β-strand structure can also result from peptide that is initially oligomeric in aqueous solution. A remaining question is whether differences in aqueous association state correlate with more subtle structural differences in the membrane-associated peptide, e.g., relative populations of parallel and antiparallel strand arrangements and turn structures. Although this study does not address this issue, it is noted that a previous solid-state NMR study provided evidence for membrane-induced dissociation of putative soluble FP oligomers (Yang and Weliky, 2003). Such dissociation has also been inferred from CD and infrared studies of the influenza fusion peptide (Han and Tamm, 2000b). Thus, the membrane-associated oligomeric structure of the HIV-1 fusion peptide may be somewhat independent of the soluble peptide association state.
As discussed in the Introduction, the HIV-1 fusion peptide as well as other fusion peptides can adopt α-helical or β-strand conformation in membranes. At peptide/lipid ∼ 0.01, one of the strong determinants of the conformational distribution is the lipid headgroup and cholesterol composition of the membrane. For LM3 membranes, solid-state NMR data for both the HIV-1 and influenza fusion peptide are consistent with predominant β-strand structure, whereas in membranes composed of POPC/POPG (4:1) lipids without cholesterol, α-helical structure is predominant (Yang et al., 2001a; Bodner et al., 2004; P. D. Parkanzky, M. Prorok, F. J. Castellino, and D. P. Weliky, unpublished). The latter observation is supported by infrared, circular dichroism, and electron spin resonance data from the Tamm group (Han and Tamm, 2000a; Han et al., 2001). In an effort to address the question of the conformation of the fusion-active state, we have carried out studies of lipid mixing induced by HIV-1 or influenza fusion peptides between vesicles composed of LM3 or between vesicles composed of POPC/POPG. Comparable lipid mixing is observed for both vesicle compositions, which suggests that both the α-helical and the β-strand conformations are fusion active at least with respect to lipid mixing (P. D. Parkanzky, M. Prorok, F. J. Castellino, and D. P. Weliky, unpublished).
It is also possible that there is a transient fusogenic structure that is unstructured or flexible rather than a well-defined α-helical or β-strand conformation. The Ulrich group recently argued in favor of this hypothesis based on comparison of lipid mixing rates obtained for 18-residue internal fusion peptides of the sea urchin bindin fertilization protein (Afonin et al., 2003). In this study, similar rates were observed for epimers containing either a L- or a D-4-fluorophenylglycine residue, and this result was contrary to the strong steric effect on rate expected for a well-defined structure. In our group, the question of a transient structure has also been examined using experiments that rely on the increased fusogenicity of the influenza fusion peptide at pH 5.0 relative to pH 7.4 (P. D. Parkanzky, M. Prorok, F. J. Castellino, and D. P. Weliky, unpublished). The peptide was bound to either LM3 or POPC/POPG vesicles at pH 7.4, and then in each sample lipid mixing was monitored after lowering the pH to 5.0. For both the LM3 and POPC/POPG samples, there was a significant increase in lipid mixing after lowering the pH. Solid-state NMR chemical shift measurements were consistent with a predominant β-strand conformation in LM3 at pH 5.0 and pH 7.4 and with predominant α-helical conformation in POPC/POPG at pH 5.0 and pH 7.4. Taken together, the lipid mixing and solid-state NMR data argue in favor of lipid mixing induction by both the α-helical and β-strand influenza fusion peptide conformations and argue against a single transient fusogenic conformation. Clearly, further experiments are required to clarify the nature of the fusogenic structure.
In the next few paragraphs, we consider a possible model that may explain the results of Fig. 2. As described in the previous paragraphs, the structure(s) of the fusogenic form of the peptide is not yet clearly understood, and our model is not based on a particular structure. In Fig. 2, it is clear that lipid mixing can be induced by both FPK3W and FPK3 (which are predominantly monomeric) and by FP and FPW (which have significant populations of oligomers/aggregates). FP appears to be the most fusogenic construct, and FPW is the least fusogenic construct. Both FP and FPK3 bind quantitatively to LM3 in the NMR sample preparation, so it is not likely that the different fusogenicities are correlated with variations in binding affinities of the different constructs. It may be possible to understand the fusogenicity data using a Nc model based on the hypothesis that lipid mixing requires membrane-associated clusters whose number of peptides, N, exceeds a critical number of peptides, Nc (Nieva et al., 1998). The value of Nc could be based on the number of peptides needed for a particular oligomeric structure (which might be α-helical or β-strand ) and/or the free energy released when Nc peptides bind to the membrane in close proximity (Han and Tamm, 2000a). In the simplest model, membrane disruption may be correlated with the size of the cluster, and fusion may require a degree of disruption that is only achieved for N ≥ Nc clusters. As the structures of the α-helical and β-strand forms of fusion peptides become better elucidated, it may become possible to test for an underlying structural basis of the Nc model.
Within this Nc model, the membrane-associated cluster sizes for peptide/lipid = 0.01 are postulated: NFP ∼ Nc; NFPK3, NFPK3W < Nc; and NFPW ≫ Nc. These sizes qualitatively correlate with the putative soluble peptide oligomerization states. In the model, FP would then have the largest number of fusogenic clusters and the highest fusogenicity (Mf). This prediction is consistent with the experimental fusogenicities at peptide/lipid = 0.01.
Other aspects of the experimental data displayed in Fig. 2 can be explained with this model. For example, comparison of data at peptide/lipid = 0.01 and 0.02 shows that the fusogenicities of FP and FPW are approximately proportional to peptide/lipid. This might be expected when the cluster N ≥ Nc at both peptide/lipid ratios and the number of fusogenic clusters simply increases with peptide/lipid. By contrast, the fusogenicities of FPK3 and FPK3W have nonlinear (greater than proportional) dependences on peptide/lipid. This can be understood in terms of a change in average cluster size from N < Nc at peptide/lipid = 0.01 to N ∼ Nc at peptide/lipid = 0.02.
Our model is also supported by a previous lipid mixing study done on cross-linked peptides and their noncross-linked analogs (Yang et al., 2003). At peptide strand/lipid ≤ 0.01, the cross-linked peptides had greater Mf than their noncross-linked analogs, but at higher peptide strand/lipid the Mf of all constructs were much more comparable. These observations can be understood by the increase in local peptide concentration due to cross-linking and the resulting larger number of clusters with N > Nc. In accord with experiment, the fusogenic enhancement of cross-linking should be more important at lower strand/lipid (where relatively few of the clusters have N > Nc) than at higher strand/lipid (where most of the clusters have N > Nc). The Nc model is also supported by lipid mixing studies of FP that had been incubated for different lengths of time in 100 mM NaCl solution (Pereira et al., 1997). In these studies, there was an inverse relationship between Mf and incubation time that can be understood in terms of an increase of N beyond Nc as a function of incubation time. This latter prediction correlates with experimental observations that higher ionic strength leads to increased peptide aggregation in solution (Han and Tamm, 2000b; Yang et al., 2001a).
In this study, incorporation of C-terminal lysines and tryptophan was critical for synthesis and for analytical ultracentrifugation verification of a monomeric peptide state. The overall synthetic and analytical ultracentrifugation approach may also be generally useful for clarification of oligomerization questions concerning different fusion peptide constructs such as those that include large fusion protein domains. For example, it has been common in structural and functional studies of HIV-1 fusion peptides to first dissolve peptide in dimethylsulfoxide (DMSO) and then add an aliquot of the dimethylsulfoxide solution to a liposome solution (Martin et al., 1996; Pereira et al., 1997; Sackett and Shai, 2002; Haque and Lentz, 2002). Because the peptide concentration is much higher than the liposome concentration, the rate of peptide/peptide collisions is much greater than the rate of peptide/liposome collisions, and the oligomeric state of the peptide is not known before binding to the membrane (Steinfeld et al., 1999). Using the approach of this article, it may be possible to find conditions for which peptide constructs that vary in length and/or sequence have well-defined oligomeric states in the buffer solution. This may clarify the contribution of oligomerization to structure/function relationships in these peptides.
CONCLUSIONS
Structural and functional comparisons were made between four different HIV-1 fusion peptide constructs, two of which were predominantly monomeric in aqueous solution and two of which had significant populations of oligomers/aggregates in aqueous solution. When associated with membranes comparable to those of host cells of the virus, all constructs formed β-strand structure at Phe-8, and experiments suggest that this is an equilibrium rather than a kinetically trapped structure. The membrane-associated β-strand structure is oligomeric, and this oligomerization can result from association of soluble peptide monomers with membranes. There is not a linear correlation between the solution oligomerization state and the fusogenicity of the peptide construct, but the functional data suggest that small peptide oligomers may be more fusogenic than either peptide monomers or large peptide aggregates.
Acknowledgments
We acknowledge Dr. Mark Lemmon for assistance with acquisition and analysis of the analytical ultracentrifugation data. We acknowledge Dr. Francesca Marassi for suggesting the organic solvent mixture used for preparation method 2. We acknowledge Drs. Honggao Yan and David DeWitt for use of their fluorimeter and ultracentrifuge, respectively. We acknowledge the Michigan State University Mass Spectrometry and Macromolecular Structure facilities for assistance with mass spectrometric analysis and amino acid analysis, respectively. We acknowledge Paul Parkanzky for useful discussions and Brian Dodds, Andrew Goetz, and Michele Bodner for assistance with experiments. We thank Charles Gabrys for a critical reading of this article.
This work was supported in part by a Camille and Henry Dreyfus Foundation New Faculty award, a Michigan State University Intramural Research Grants Program award, National Institutes of Health award R01-AI47153, and National Science Foundation award 9977650 to D.P.W. A Michigan State University Center for Biological Modeling fellowship provided partial salary support for J.Y.
References
- Afonin, S., R. W. Glaser, M. Berditchevskaia, P. Wadhwani, K. H. Guhrs, U. Mollmann, A. Perner, and A. S. Ulrich. 2003. 4-Fluorophenylglycine as a label for 19F NMR structure analysis of membrane-associated peptides. ChemBioChem. 4:1151–1163. [DOI] [PubMed] [Google Scholar]
- Aloia, R. C., H. Tian, and F. C. Jensen. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA. 90:5181–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbach, J. J., J. Yang, D. P. Weliky, P. J. Steinbach, V. Tugarinov, J. Anglister, and R. Tycko. 2000. Probing hydrogen bonds in the antibody-bound HIV-1 gp120 V3 loop by solid state NMR REDOR measurements. J. Biomol. NMR. 16:313–327. [DOI] [PubMed] [Google Scholar]
- Bennett, A. E., C. M. Rienstra, M. Auger, K. V. Lakshmi, and R. G. Griffin. 1995. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 103:6951–6958. [Google Scholar]
- Bentz, J. 2000a. Membrane fusion mediated by coiled coils: a hypothesis. Biophys. J. 78:886–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz, J. 2000b. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys. J. 78:227–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, R., M. J. Clague, S. R. Durell, and R. M. Epand. 2003. Membrane fusion. Chem. Rev. 103:53–69. [DOI] [PubMed] [Google Scholar]
- Blumenthal, R., D. P. Sarkar, S. Durell, D. E. Howard, and S. J. Morris. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodner, M. L., C. M. Gabrys, P. D. Parkanzky, J. Yang, C. A. Duskin, and D. P. Weliky. 2004. Temperature dependence and resonance assignment of 13C NMR spectra of selectively and uniformly labeled fusion peptides associated with membranes. Magn. Reson. Chem. 42:187–194. [DOI] [PubMed] [Google Scholar]
- Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor, C. R., and P. R. Shimmel. 1980. Biophysical Chemistry Part II: Techniques for the Study of Biological Structure and Function. W. H. Freeman, New York.
- Cavanaugh, J., W. J. Fairbrother, A. G. Palmer, and N. J. Skelton. 1996. Protein NMR Spectroscopy Principles and Practice. Academic Press, San Diego, CA.
- Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 89:263–273. [DOI] [PubMed] [Google Scholar]
- Chang, D. K., and S. F. Cheng. 1998. Determination of the equilibrium micelle-inserting position of the fusion peptide of gp41 of human immunodeficiency virus type 1 at amino acid resolution by exchange broadening of amide proton resonances. J. Biomol. NMR. 12:549–552. [DOI] [PubMed] [Google Scholar]
- Chang, D. K., S. F. Cheng, and W. J. Chien. 1997a. The amino-terminal fusion domain peptide of human immunodeficiency virus type 1 gp41 inserts into the sodium dodecyl sulfate micelle primarily as a helix with a conserved glycine at the micelle-water interface. J. Virol. 71:6593–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D. K., S. F. Cheng, and V. D. Trivedi. 1999. Biophysical characterization of the structure of the amino-terminal region of gp41 of HIV-1. Implications on viral fusion mechanism. J. Biol. Chem. 274:5299–5309. [DOI] [PubMed] [Google Scholar]
- Chang, D. K., W. J. Chien, and S. F. Cheng. 1997b. The FLG motif in the N-terminal region of glucoprotein 41 of human immunodeficiency virus type 1 adopts a type-I beta turn in aqueous solution and serves as the initiation site for helix formation. Eur. J. Biochem. 247:896–905. [DOI] [PubMed] [Google Scholar]
- Chang, C. D., M. Waki, M. Ahmad, J. Meienhofer, E. O. Lundell, and J. D. Haug. 1980. Preparation and properties of N-alpha-9-fluorenylmethyloxycarbonylamino acids bearing tert-butyl side-chain protection. Int. J. Pept. Prot. Res. 15:59–66. [DOI] [PubMed] [Google Scholar]
- Curtain, C., F. Separovic, K. Nielsen, D. Craik, Y. Zhong, and A. Kirkpatrick. 1999. The interactions of the N-terminal fusogenic peptide of HIV-1 gp41 with neutral phospholipids. Eur. Biophys. J. 28:427–436. [DOI] [PubMed] [Google Scholar]
- Delahunty, M. D., I. Rhee, E. O. Freed, and J. S. Bonifacino. 1996. Mutational analysis of the fusion peptide of the human immunodeficiency virus type 1: identification of critical glycine residues. Virology. 218:94–102. [DOI] [PubMed] [Google Scholar]
- Dimitrov, D. S. 2000. Cell biology of virus entry. Cell. 101:697–702. [DOI] [PubMed] [Google Scholar]
- Durell, S. R., I. Martin, J. M. Ruysschaert, Y. Shai, and R. Blumenthal. 1997. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion (review). Mol. Membr. Biol. 14:97–112. [DOI] [PubMed] [Google Scholar]
- Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810. [DOI] [PubMed] [Google Scholar]
- Epand, R. F., J. C. Macosko, C. J. Russell, Y. K. Shin, and R. M. Epand. 1999. The ectodomain of HA2 of influenza virus promotes rapid pH dependent membrane fusion. J. Mol. Biol. 286:489–503. [DOI] [PubMed] [Google Scholar]
- Epand, R. F., C. M. Yip, L. V. Chernomordik, D. L. LeDuc, Y. K. Shin, and R. M. Epand. 2001. Self-assembly of influenza hemagglutinin: studies of ectodomain aggregation by in situ atomic force microscopy. Biochim. Biophys. Acta. 1513:167–175. [DOI] [PubMed] [Google Scholar]
- Freed, E. O., E. L. Delwart, G. L. Buchschacher, Jr., and A. T. Panganiban. 1992. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc. Natl. Acad. Sci. USA. 89:70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed, E. O., D. J. Myers, and R. Risser. 1990. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc. Natl. Acad. Sci. USA. 87:4650–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrys, C. M., J. Yang, and D. P. Weliky. 2003. Analysis of local conformation of membrane-bound and polycrystalline peptides by two-dimensional slow-spinning rotor-synchronized MAS exchange spectroscopy. J. Biomol. NMR. 26:49–68. [DOI] [PubMed] [Google Scholar]
- Gordon, L. M., C. C. Curtain, Y. C. Zhong, A. Kirkpatrick, P. W. Mobley, and A. J. Waring. 1992. The amino-terminal peptide of HIV-1 glycoprotein 41 interacts with human erythrocyte membranes: peptide conformation, orientation and aggregation. Biochim. Biophys. Acta. 1139:257–274. [DOI] [PubMed] [Google Scholar]
- Gordon, L. M., P. W. Mobley, R. Pilpa, M. A. Sherman, and A. J. Waring. 2002. Conformational mapping of the N-terminal peptide of HIV-1 gp41 in membrane environments using 13C-enhanced Fourier transform infrared spectroscopy. Biochim. Biophys. Acta. 1559:96–120. [DOI] [PubMed] [Google Scholar]
- Gullion, T., D. B. Baker, and M. S. Conradi. 1990. New, compensated Carr-Purcell sequences. J. Magn. Reson. 89:479–484. [Google Scholar]
- Gullion, T., and J. Schaefer. 1989. Rotational-echo double-resonance NMR. J. Magn. Reson. 81:196–200. [DOI] [PubMed] [Google Scholar]
- Gullion, T., and J. Schaefer. 1991. Elimination of resonance offset effects in rotational-echo, double-resonance NMR. J. Magn. Reson. 92:439–442. [Google Scholar]
- Han, X., J. H. Bushweller, D. S. Cafiso, and L. K. Tamm. 2001. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 8:715–720. [DOI] [PubMed] [Google Scholar]
- Han, X., and L. K. Tamm. 2000a. A host-guest system to study structure-function relationships of membrane fusion peptides. Proc. Natl. Acad. Sci. USA. 97:13097–13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., and L. K. Tamm. 2000b. pH-dependent self-association of influenza hemagglutinin fusion peptides in lipid bilayers. J. Mol. Biol. 304:953–965. [DOI] [PubMed] [Google Scholar]
- Haque, M. E., and B. R. Lentz. 2002. Influence of gp4l fusion peptide on the kinetics of poly(ethylene glycol)-mediated model membrane fusion. Biochemistry. 41:10866–10876. [DOI] [PubMed] [Google Scholar]
- Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627–661. [DOI] [PubMed] [Google Scholar]
- Hirsh, D. J., J. Hammer, W. L. Maloy, J. Blazyk, and J. Schaefer. 1996. Secondary structure and location of a magainin analogue in synthetic phospholipid bilayers. Biochemistry. 35:12733–12741. [DOI] [PubMed] [Google Scholar]
- Hope, M. J., M. B. Bally, G. Webb, and P. R. Cullis. 1985. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane-potential. Biochim. Biophys. Acta. 812:55–65. [DOI] [PubMed] [Google Scholar]
- Kamath, S., and T. C. Wong. 2002. Membrane structure of the human immunodeficiency virus gp41 fusion domain by molecular dynamics simulation. Biophys. J. 83:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchem, R. R., W. Hu, and T. A. Cross. 1993. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 261:1457–1460. [DOI] [PubMed] [Google Scholar]
- Kim, C. H., J. C. Macosko, and Y. K. Shin. 1998. The mechanism for low-pH-induced clustering of phospholipid vesicles carrying the HA2 ectodomain of influenza hemagglutinin. Biochemistry. 37:137–144. [DOI] [PubMed] [Google Scholar]
- Kim, C. H., J. C. Macosko, Y. G. Yu, and Y. K. Shin. 1996. On the dynamics and confirmation of the HA2 domain of the influenza virus hemagglutinin. Biochemistry. 35:5359–5365. [DOI] [PubMed] [Google Scholar]
- Kliger, Y., A. Aharoni, D. Rapaport, P. Jones, R. Blumenthal, and Y. Shai. 1997. Fusion peptides derived from the HIV type 1 glycoprotein 41 associate within phospholipid membranes and inhibit cell-cell fusion. Structure-function study. J. Biol. Chem. 272:13496–13505. [DOI] [PubMed] [Google Scholar]
- Kliger, Y., S. A. Gallo, S. G. Peisajovich, I. Munoz-Barroso, S. Avkin, R. Blumenthal, and Y. Shai. 2001. Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J. Biol. Chem. 276:1391–1397. [DOI] [PubMed] [Google Scholar]
- Kricheldorf, H. R., and D. Muller. 1983. Secondary structure of peptides.3. 13C NMR cross polarization/magic angle spinning spectroscopic characterization of solid polypeptides. Macromolecules. 16:615–623. [Google Scholar]
- Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapatsanis, L., G. Milias, K. Froussios, and M. Kolovos. 1983. Synthesis of N-2,2,2-(trichloroethoxycarbonyl)-L-amino acids and N-(9-fluorenylmethoxycarbonyl)-L-amino acids involving succinimidoxy anion as a leaving group in amino-acid protection. Synthesis. 8:671–673. [Google Scholar]
- Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins. In Analytical Ultracentrifugation in Biochemistry and Polymer Science. S. E. Harding, A. J. Rowe, and J. C. Horton, editors. Royal Society of Chemistry, Cambridge, UK. 90–125.
- LeDuc, D. L., Y. K. Shin, R. F. Epand, and R. M. Epand. 2000. Factors determining vesicular lipid mixing induced by shortened constructs of influenza hemagglutinin. Biochemistry. 39:2733–2739. [DOI] [PubMed] [Google Scholar]
- Leikina, E., D. L. LeDuc, J. C. Macosko, R. Epand, Y. K. Shin, and L. V. Chernomordik. 2001. The 1–127 HA2 construct of influenza virus hemagglutinin induces cell-cell hemifusion. Biochemistry. 40:8378–8386. [DOI] [PubMed] [Google Scholar]
- Lin, X. X., C. A. Derdeyn, R. Blumenthal, J. West, and E. Hunter. 2003. Progressive truncations C terminal to the membrane-spanning domain of simian immunodeficiency virus Env reduce fusogenicity and increase concentration dependence of Env for fusion. J. Virol. 77:7067–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, H. W., and R. Tycko. 1998. Biopolymer conformational distributions from solid-state NMR: alpha-helix and 310-helix contents of a helical peptide. J. Am. Chem. Soc. 120:7039–7048. [Google Scholar]
- Macosko, J. C., C. H. Kim, and Y. K. Shin. 1997. The membrane topology of the fusion peptide region of influenza hemagglutinin determined by spin-labeling EPR. J. Mol. Biol. 267:1139–1148. [DOI] [PubMed] [Google Scholar]
- Maddox, M. W., and M. L. Longo. 2002. Conformational partitioning of the fusion peptide of HIV-1 gp41 and its structural analogs in bilayer membranes. Biophys. J. 83:3088–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi, F. M., A. Ramamoorthy, and S. J. Opella. 1997. Complete resolution of the solid-state NMR spectrum of a uniformly 15N-labeled membrane protein in phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 94:8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, I., F. Defrise-Quertain, E. Decroly, M. Vandenbranden, R. Brasseur, and J. M. Ruysschaert. 1993. Orientation and structure of the NH2-terminal HIV-1 gp41 peptide in fused and aggregated liposomes. Biochim. Biophys. Acta. 1145:124–133. [DOI] [PubMed] [Google Scholar]
- Martin, I., H. Schaal, A. Scheid, and J. M. Ruysschaert. 1996. Lipid membrane fusion induced by the human immunodeficiency virus type 1 gp41 N-terminal extremity is determined by its orientation in the lipid bilayer. J. Virol. 70:298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, A., and J. Bentz. 2001. Comprehensive kinetic analysis of influenza hemagglutinin-mediated membrane fusion: role of sialate binding. Biophys. J. 81:1521–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley, P. W., H. F. Lee, C. C. Curtain, A. Kirkpatrick, A. J. Waring, and L. M. Gordon. 1995. The amino-terminal peptide of HIV-1 glycoprotein 41 fuses human erythrocytes. Biochim. Biophys. Acta. 1271:304–314. [DOI] [PubMed] [Google Scholar]
- Mobley, P. W., A. J. Waring, M. A. Sherman, and L. M. Gordon. 1999. Membrane interactions of the synthetic N-terminal peptide of HIV-1 gp41 and its structural analogs. Biochim. Biophys. Acta. 1418:1–18. [DOI] [PubMed] [Google Scholar]
- Morcombe, C. R., and K. W. Zilm. 2003. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 162:479–486. [DOI] [PubMed] [Google Scholar]
- Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva, J. L., S. Nir, A. Muga, F. M. Goni, and J. Wilschut. 1994. Interaction of the HIV-1 fusion peptide with phospholipid vesicles: different structural requirements for fusion and leakage. Biochemistry. 33:3201–3209. [DOI] [PubMed] [Google Scholar]
- Nieva, J. L., S. Nir, and J. Wilschut. 1998. Destabilization and fusion of zwitterionic large unilamellar lipid vesicles induced by a beta-type structure of the HIV-1 fusion peptide. J. Liposome Res. 8:165–182. [Google Scholar]
- Opella, S. J., F. M. Marassi, J. J. Gesell, A. P. Valente, Y. Kim, M. Oblatt-Montal, and M. Montal. 1999. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat. Struct. Biol. 6:374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecheur, E., J. Sainte-Marie, A. Bienvenue, and D. Hoekstra. 1999. Peptides and membrane fusion: towards an understanding of the molecular mechanism of protein-induced fusion. J. Membr. Biol. 167:1–17. [DOI] [PubMed] [Google Scholar]
- Peisajovich, S. G., L. Blank, R. F. Epand, R. M. Epand, and Y. Shai. 2003. On the interaction between gp41 and membranes: the immunodominant loop stabilizes gp41 helical hairpin conformation. J. Mol. Biol. 326:1489–1501. [DOI] [PubMed] [Google Scholar]
- Peisajovich, S. G., R. F. Epand, M. Pritsker, Y. Shai, and R. M. Epand. 2000a. The polar region consecutive to the HIV fusion peptide participates in membrane fusion. Biochemistry. 39:1826–1833. [DOI] [PubMed] [Google Scholar]
- Peisajovich, S. G., O. Samuel, and Y. Shai. 2000b. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol. 296:1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, F. B., F. M. Goni, A. Muga, and J. L. Nieva. 1997. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: dose and sequence effects. Biophys. J. 73:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, F. B., F. M. Goni, and J. L. Nieva. 1995. Liposome destabilization induced by the HIV-1 fusion peptide effect of a single amino acid substitution. FEBS Lett. 362:243–246. [DOI] [PubMed] [Google Scholar]
- Pritsker, M., J. Rucker, T. L. Hoffman, R. W. Doms, and Y. Shai. 1999. Effect of nonpolar substitutions of the conserved Phe11 in the fusion peptide of HIV-1 gp41 on its function, structure, and organization in membranes. Biochemistry. 38:11359–11371. [DOI] [PubMed] [Google Scholar]
- Rafalski, M., J. D. Lear, and W. F. DeGrado. 1990. Phospholipid interactions of synthetic peptides representing the N-terminus of HIV gp41. Biochemistry. 29:7917–7922. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy, A., F. M. Marassi, M. Zasloff, and S. J. Opella. 1995. 3-Dimensional solid-state NMR spectroscopy of a peptide oriented in membrane bilayers. J. Biomol. NMR. 6:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackett, K., and Y. Shai. 2002. The HIV-1 gp41 N-terminal heptad repeat plays an essential role in membrane fusion. Biochemistry. 41:4678–4685. [DOI] [PubMed] [Google Scholar]
- Saito, H. 1986. Conformation-dependent 13C chemical-shifts - a new means of conformational characterization as obtained by high-resolution solid-state 13C NMR. Magn. Reson. Chem. 24:835–852. [Google Scholar]
- Schaal, H., M. Klein, P. Gehrmann, O. Adams, and A. Scheid. 1995. Requirement of N-terminal amino acid residues of gp41 for human immunodeficiency virus type 1-mediated cell fusion. J. Virol. 69:3308–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepushkin, V. A., S. M. Andreev, M. V. Sidorova, G. B. Melikyan, V. B. Grigoriev, V. M. Chumakov, A. E. Grinfeldt, R. A. Manukyan, and E. V. Karamov. 1992. Investigation of human immunodeficiency virus fusion peptides. Analysis of interrelations between their structure and function. AIDS Res. Hum. Retroviruses. 8:9–18. [DOI] [PubMed] [Google Scholar]
- Slepushkin, V. A., G. B. Melikyan, M. S. Sidorova, V. M. Chumakov, S. M. Andreev, R. A. Manulyan, and E. V. Karamov. 1990. Interaction of human immunodeficiency virus (HIV-1) fusion peptides with artificial lipid membranes. Biochem. Biophys. Res. Commun. 172:952–957. [DOI] [PubMed] [Google Scholar]
- Steinfeld, J. I., J. S. Francisco, and W. L. Hase. 1999. Chemical Kinetics and Dynamics. Prentice Hall, Upper Saddle River, NJ.
- Struck, D. K., D. Hoekstra, and R. E. Pagano. 1981. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 20:4093–4099. [DOI] [PubMed] [Google Scholar]
- Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA. 94:12303–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 387:426–430. [DOI] [PubMed] [Google Scholar]
- Weliky, D. P., A. E. Bennett, A. Zvi, J. Anglister, P. J. Steinbach, and R. Tycko. 1999. Solid-state NMR evidence for an antibody-dependent conformation of the V3 loop of HIV-1 gp120. Nat. Struct. Biol. 6:141–145. [DOI] [PubMed] [Google Scholar]
- Wishart, D. S., C. G. Bigam, J. Yao, F. Abildgaard, H. J. Dyson, E. Oldfield, J. L. Markley, and B. D. Sykes. 1995. 1H, 13C and 15N chemical-shift referencing in biomolecular NMR. J. Biomol. NMR. 6:135–140. [DOI] [PubMed] [Google Scholar]
- Wong, T. C. 2003. Membrane structure of the human immunodeficiency virus gp41 fusion peptide by molecular dynamics simulation II. The glycine mutants. Biochim. Biophys. Acta. 1609:45–54. [DOI] [PubMed] [Google Scholar]
- Yang, J. 2003. Solid-state nuclear magnetic resonance structural studies of the HIV-1 fusion peptide in the membrane environment. PhD thesis. Michigan State University, East Lansing, MI.
- Yang, J., C. M. Gabrys, and D. P. Weliky. 2001a. Solid-state nuclear magnetic resonance evidence for an extended beta strand conformation of the membrane-bound HIV-1 fusion peptide. Biochemistry. 40:8126–8137. [DOI] [PubMed] [Google Scholar]
- Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126:131–144. [DOI] [PubMed] [Google Scholar]
- Yang, J., P. D. Parkanzky, M. L. Bodner, C. G. Duskin, and D. P. Weliky. 2002. Application of REDOR subtraction for filtered MAS observation of labeled backbone carbons of membrane-bound fusion peptides. J. Magn. Reson. 159:101–110. [DOI] [PubMed] [Google Scholar]
- Yang, J., P. D. Parkanzky, B. A. Khunte, C. G. Canlas, R. Yang, C. M. Gabrys, and D. P. Weliky. 2001b. Solid state NMR measurements of conformation and conformational distributions in the membrane-bound HIV-1 fusion peptide. J. Mol. Graph. Model. 19:129–135. [DOI] [PubMed] [Google Scholar]
- Yang, J., and D. P. Weliky. 2003. Solid state nuclear magnetic resonance evidence for parallel and antiparallel strand arrangements in the membrane-associated HIV-1 fusion peptide. Biochemistry. 42:11879–11890. [DOI] [PubMed] [Google Scholar]
- Yang, R., J. Yang, and D. P. Weliky. 2003. Synthesis, enhanced fusogenicity, and solid state NMR measurements of cross-linked HIV-1 fusion peptides. Biochemistry. 42:3527–3535. [DOI] [PubMed] [Google Scholar]
- Zhang, H. Y., S. Neal, and D. S. Wishart. 2003. RefDB: a database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 25:173–195. [DOI] [PubMed] [Google Scholar]