Abstract
A time trajectory of an observable that fluctuates between two values (say, on and off), stemming from some unknown multisubstate kinetic scheme, is the output of many single-molecule experiments. Here we show that when all successive waiting times along the trajectory are uncorrelated the on and the off waiting time probability density functions contain all the information. By relating the lack of correlation in the trajectory to the topology of kinetic schemes, we can immediately specify those kinetic schemes that are equally consistent with experiment, and cannot be differentiated by any sophisticated analyses of the trajectory. Correlated trajectories, however, contain additional information about the underlying kinetic scheme, and we consider the strategy that one should use to extract it.
INTRODUCTION
Since the first patch-clamp measurements (Neher and Sakmann, 1976), great advances have been made in our ability to look at complex systems on the single-molecule level (Moerner and Orrit, 1999; Weiss, 1999; Nie et al., 1994; Shera et al., 1990; Mets and Rigler, 1994; Ha et al., 1999; Schuler et al., 2002; Yang et al., 2003; Rhoades et al., 2003; Wennmalm et al., 1997; Bokinsky et al., 2003; Lu et al., 1998; Edman et al., 1999; Edman and Rigler, 2000; Velonia et al., 2005; Flomenbom et al., 2005; Kasianowicz et al., 1996). In an important class of such experiments, the output is a time series (trajectory) of on-off events (Fig. 1, A and B). For example, in patch-clamp measurements (Neher and Sakmann, 1976), one records the ion current through a membrane pore under an applied electric field for a long time. The fluctuations between two values of the current are attributed to conformational changes that result in opening and closing the membrane pore. From the two-state current trajectory, one wishes to learn about the dynamics of conformational changes of the membrane pore. In single enzyme activity measurements (Lu et al., 1998; Edman et al., 1999; Edman and Rigler, 2000; Velonia et al., 2005; Flomenbom et al., 2005), one monitors photon counts as a function of time, and collects the counts into bins giving rise to the trajectory. A two-state trajectory is obtained when either the enzyme itself switches between a fluorescent state and a nonfluorescent state (Lu et al., 1998), or a nonfluorescent substrate is transformed into a fluorescent product (Edman et al., 1999; Edman and Rigler, 2000; Velonia et al., 2005; Flomenbom et al., 2005). By studying this system, one wishes to deduce the mechanism of the enzymatic activity.
FIGURE 1.
On-off trajectories as a function of time. These trajectories were obtained by simulating the kinetic schemes shown in Fig. 2 K (A) and Fig. 2 L (B). The transition rate values are given in Fig. 3.
In practice, noise-induced fluctuations in the signal occur around the on and the off values. The ability to restore reliably the noiseless trajectory from the experimental output (i.e., to deconvolute the noise) depends roughly on the difference between these values relative to the sum of the amplitudes of the noise in each of the states. Here, we assume that we are given a noiseless two-state trajectory.
Such a two-state trajectory contains information about the underlying mechanism, which we describe by a kinetic scheme in which each substate belongs either to the on state or to the off state. The kinetic scheme may have a large number of substates (Fig. 2, A–F), and a net flow at steady state along some of the connections (Fig. 2, G–I) (i.e., a nonequilibrium steady state), when an external source of energy is present (Hill, 1985). The goal is to learn as much as possible about the underlying kinetic scheme.
FIGURE 2.

A set of kinetic schemes containing black-circled off substates and red-squared on substates, which can be used to produce on-off trajectories. (A–F) Reducible schemes with only reversible connections. As discussed in the text, these schemes are reducible (i.e., generate two-state trajectories that can be made equivalent to trajectories generated by scheme J), because the on and the off states are connected in each of these cases through a single substate. Schemes B and C lead to identical trajectories, in statistical sense, when both the on and the off waiting time PDFs of the two schemes are made the same. The same is true for schemes D–F. (G–I) Reducible schemes with irreversible connections. (J) The two-state semi-Markovian scheme is described completely by the waiting time PDFs 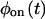 and
and 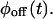 (K) The simplest irreducible model is a four-substate model. (L) An example of a reducible four-substate model.
(K) The simplest irreducible model is a four-substate model. (L) An example of a reducible four-substate model.
FROM TRAJECTORIES TO KINETIC SCHEMES
The basic functions that are easily obtained from single-molecule two-state time series are the waiting time (or, lifetime) probability density functions (PDFs) of the on state, 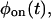 and of the off state,
and of the off state, 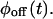 These functions, which cannot be found from bulk experiments, can be calculated for any kinetic scheme (Cao, 2000). Clearly, any proposed kinetic scheme must reproduce
These functions, which cannot be found from bulk experiments, can be calculated for any kinetic scheme (Cao, 2000). Clearly, any proposed kinetic scheme must reproduce 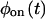 and
and 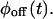 However, when
However, when 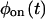 and
and 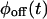 are multiexponentials, several models will fulfill this requirement, and their number increases with the complexity of the waiting time PDFs (the trajectories on Fig. 1, A and B, have the same waiting time PDFs, but were produced from different kinetic schemes). Can one discriminate between kinetic schemes that lead to the same
are multiexponentials, several models will fulfill this requirement, and their number increases with the complexity of the waiting time PDFs (the trajectories on Fig. 1, A and B, have the same waiting time PDFs, but were produced from different kinetic schemes). Can one discriminate between kinetic schemes that lead to the same 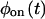 and
and 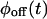 by looking at the trajectory in more detail?
by looking at the trajectory in more detail?
A trajectory is completely described by 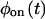 and
and 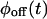 only when waiting times along the trajectory are uncorrelated. Therefore, kinetic schemes that lead to uncorrelated trajectories with the same
only when waiting times along the trajectory are uncorrelated. Therefore, kinetic schemes that lead to uncorrelated trajectories with the same 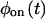 and
and 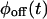 cannot be distinguished by the trajectory analysis. This means that the trajectory from such a kinetic scheme does not contain information about the connectivity of substates within each of the two states, which, as shown below, is a consequence of a specific connectivity between substates of different states. We say that such schemes are “reducible” to a two-state semi-Markovian (TSSM) scheme (Fig. 2 J). A TSSM process is one where the on [off] waiting times are drawn randomly and independently out of a nonexponential
cannot be distinguished by the trajectory analysis. This means that the trajectory from such a kinetic scheme does not contain information about the connectivity of substates within each of the two states, which, as shown below, is a consequence of a specific connectivity between substates of different states. We say that such schemes are “reducible” to a two-state semi-Markovian (TSSM) scheme (Fig. 2 J). A TSSM process is one where the on [off] waiting times are drawn randomly and independently out of a nonexponential 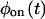 [
[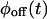 ]. In the literature, the term non-Markovian is often used for any process with nonexponential waiting time PDFs. However, here we reserve this term to describe a trajectory of correlated waiting times.
]. In the literature, the term non-Markovian is often used for any process with nonexponential waiting time PDFs. However, here we reserve this term to describe a trajectory of correlated waiting times.
The most straightforward test for correlation in the trajectory is based on the two successive waiting times PDFs, 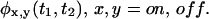 A trajectory shows no correlations when
A trajectory shows no correlations when 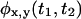 can be written, for every x and y, as a product of the individual waiting time PDFs,
can be written, for every x and y, as a product of the individual waiting time PDFs, 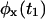 and
and 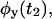
 |
(1) |
When all two successive waiting times PDFs are factorized, higher order successive waiting times PDFs, e.g., 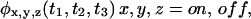 will also be factorized. Since higher order successive waiting times PDFs determine all the statistical properties of the trajectory and these factorize when Eq. 1 is fulfilled, it follows that for uncorrelated trajectories
will also be factorized. Since higher order successive waiting times PDFs determine all the statistical properties of the trajectory and these factorize when Eq. 1 is fulfilled, it follows that for uncorrelated trajectories 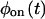 and
and 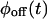 contain all the information in the time series.
contain all the information in the time series.
Kinetic schemes are reducible (i.e., Eq. 1 is fulfilled) regardless of the system parameters if and only if after every transition from the on state to the off state, the off substates are populated with the same initial probabilities, and vice versa. This occurs only for a very specific connectivity between the on and the off substates, and we now give a full characterization of the reducible schemes. When only reversible connections between substates are present, a scheme is reducible when the on and the off regions are connected through one substate (Fig. 2, A–F), called a gateway substate. In general, there are two types of gateway substates. A type 1 gateway substate is one where all the transitions from the other state enter it (the on substate 1 in Fig. 2 G). A type 2 gateway substate is one where all the transitions to the other state originate from it (the on substate 2 in Fig. 2 G). Thus, for a reducible scheme with only reversible connections, the gateway substate is of both types simultaneously. For a kinetic scheme with a nonequilibrium steady state, there are three combinations of gateway substates that lead to a reducible scheme: a), two gateway substates of different types in the same state (Fig. 2 G), and b) and c), two gateway substates of the same type, either type 1 (Fig. 2 H) or type 2 (Fig. 2 I), in different states. Note that the above requirements are the minimal ones and a reducible scheme can possess more than two gateway substates. Because our argument relies only on the connectivity of the scheme, the reducible schemes can be characterized by any substate waiting time PDFs and not just the Markovian (exponential) one. Finally, it should be pointed out that other less general schemes can fulfill Eq. 1, thus are reducible, because of symmetry for special choices of the transition rates.
As an example of reducible kinetic schemes, consider the two schemes shown in Fig. 2, B and C, each containing n off substates and one on substate. Both schemes are reducible because there is only one substate in the on state. Even though they reflect very different mechanisms, it is possible to find transition rates that make 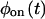 and
and 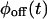 of the two schemes the same (e.g., by equating coefficients of the powers of the Laplace variable s of
of the two schemes the same (e.g., by equating coefficients of the powers of the Laplace variable s of 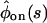 and
and 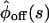 from the two schemes (
from the two schemes (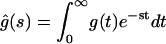 ), and solving the resulting set of equations relating the transition rates of the two models). The trajectories generated from the two schemes will then be identical (in a statistical sense). Contrary to our results, in the context of enzyme kinetics, it has been previously suggested that it is possible to distinguish between schemes, Fig. 2, B and C, using more sophisticated analyses of the trajectory (Edman and Rigler, 2000). The simplest equivalent reducible schemes are shown in Fig. 2, D–F. Recently, Witkoskie and Cao (2004) pointed out that counter to intuition two of those schemes (Fig. 2, E and F) can be made indistinguishable using similarity transformation arguments.
), and solving the resulting set of equations relating the transition rates of the two models). The trajectories generated from the two schemes will then be identical (in a statistical sense). Contrary to our results, in the context of enzyme kinetics, it has been previously suggested that it is possible to distinguish between schemes, Fig. 2, B and C, using more sophisticated analyses of the trajectory (Edman and Rigler, 2000). The simplest equivalent reducible schemes are shown in Fig. 2, D–F. Recently, Witkoskie and Cao (2004) pointed out that counter to intuition two of those schemes (Fig. 2, E and F) can be made indistinguishable using similarity transformation arguments.
For irreducible kinetic schemes 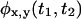 is not factorized for at least one combination of
is not factorized for at least one combination of 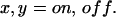 In these cases, functions other than the waiting time PDFs contain additional information. Such functions are: i),
In these cases, functions other than the waiting time PDFs contain additional information. Such functions are: i),  itself (Lu et al., 1998; Cao, 2000; McManus et al., 1985; Colquhoun et al., 1996), as used in the pioneering work of Xie and collaborators (1998), and calculated for any kinetic scheme by Cao (2000); ii), the x-y propagator for stationary processes, which is the probability density to be in state y at time t given that the process was in state x at time 0 (Lu et al., 1998; Edman et al., 1999; Edman and Rigler, 2000; Flomenbom et al., 2005; Schenter et al., 1999; Boguñá et al., 2000), and determines the normalized state-correlation function, which is the bulk relaxation function; iii), higher order state propagators (Edman and Rigler, 2000; Schenter et al., 1999; Wang and Wolynes, 1995), or the corresponding higher order state-correlation functions; iv), higher order successive waiting times PDFs, e.g.
itself (Lu et al., 1998; Cao, 2000; McManus et al., 1985; Colquhoun et al., 1996), as used in the pioneering work of Xie and collaborators (1998), and calculated for any kinetic scheme by Cao (2000); ii), the x-y propagator for stationary processes, which is the probability density to be in state y at time t given that the process was in state x at time 0 (Lu et al., 1998; Edman et al., 1999; Edman and Rigler, 2000; Flomenbom et al., 2005; Schenter et al., 1999; Boguñá et al., 2000), and determines the normalized state-correlation function, which is the bulk relaxation function; iii), higher order state propagators (Edman and Rigler, 2000; Schenter et al., 1999; Wang and Wolynes, 1995), or the corresponding higher order state-correlation functions; iv), higher order successive waiting times PDFs, e.g. 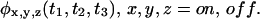 Note that the functions in i, iii, and iv can be obtained only from single-molecule experiments.
Note that the functions in i, iii, and iv can be obtained only from single-molecule experiments.
Which of these functions is the most useful in differentiating among irreducible schemes is still an open question. In practice, a function that involves many arguments will be noisy due to the limited number of events in the time series. We have found that the PDF of the sum of (or, binned) successive waiting times, e.g., 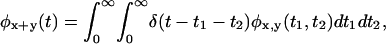 can not only be more accurately obtained from finite trajectories, but is more discriminatory than the equal successive waiting times PDF (Supplementary Material), e.g.
can not only be more accurately obtained from finite trajectories, but is more discriminatory than the equal successive waiting times PDF (Supplementary Material), e.g.  (Cao, 2000).
(Cao, 2000). 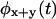 can be easily constructed from the trajectory by building the histogram of the random variable
can be easily constructed from the trajectory by building the histogram of the random variable 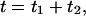 obtained from all adjacent waiting times in the time series. One can also calculate, in addition to the functions themselves, the difference between them and the product of the individual waiting time PDFs, e.g.,
obtained from all adjacent waiting times in the time series. One can also calculate, in addition to the functions themselves, the difference between them and the product of the individual waiting time PDFs, e.g., 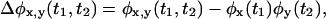 and
and  where
where 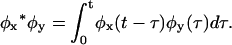 These differences vanish for reducible schemes.
These differences vanish for reducible schemes.
DISCUSSION AND CONCLUDING REMARKS
Given a two-state trajectory, after constructing 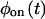 and
and 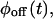 one should immediately determine whether the underlying kinetic scheme is reducible using Eq. 1. Due to the finite length of the trajectory, the moments of
one should immediately determine whether the underlying kinetic scheme is reducible using Eq. 1. Due to the finite length of the trajectory, the moments of 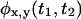 can be more accurately calculated than the PDF, and should be compared to the corresponding products of the moments of
can be more accurately calculated than the PDF, and should be compared to the corresponding products of the moments of  and
and 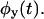 Another test compares the bulk relaxation function (the state-correlation function) obtained directly from the trajectory, with the corresponding theoretical result for a TSSM process (Flomenbom et al., 2005). The expression for the bulk relaxation function for a stationary TSSM is known, in Laplace space, for arbitrary waiting time PDFs (Cox, 1962; also see Eq. 3.15 in Boguñá et al., 2000), so one can plug in the Laplace transforms of the experimental
Another test compares the bulk relaxation function (the state-correlation function) obtained directly from the trajectory, with the corresponding theoretical result for a TSSM process (Flomenbom et al., 2005). The expression for the bulk relaxation function for a stationary TSSM is known, in Laplace space, for arbitrary waiting time PDFs (Cox, 1962; also see Eq. 3.15 in Boguñá et al., 2000), so one can plug in the Laplace transforms of the experimental 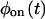 and
and 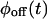 into this expression, and invert the result, either analytically or numerically, back into the time domain. If the experimental bulk relaxation function and the theoretical one for a TSSM process with the experimental
into this expression, and invert the result, either analytically or numerically, back into the time domain. If the experimental bulk relaxation function and the theoretical one for a TSSM process with the experimental 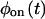 and
and 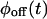 coincide, the scheme is reducible, and no further analysis is required. Another simple and informative analysis method involves the trajectory of the waiting times as a function of the occurrence index. Correlations between waiting times can be detected more easily from this trajectory than the on-off trajectory (compare Figs. 1 and 3), and used to learn about the scheme transition rate values (see the caption of Fig. 3).
coincide, the scheme is reducible, and no further analysis is required. Another simple and informative analysis method involves the trajectory of the waiting times as a function of the occurrence index. Correlations between waiting times can be detected more easily from this trajectory than the on-off trajectory (compare Figs. 1 and 3), and used to learn about the scheme transition rate values (see the caption of Fig. 3).
FIGURE 3.
Off waiting times trajectories as a function of the occurrence index corresponding to the on-off trajectories in Fig. 1, generated from the irreducible (Fig. 2 K), and reducible (Fig. 2 L) four-substate schemes. 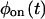 and
and  for the two schemes are the same, by setting (
for the two schemes are the same, by setting (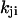 is the transition rate from substate i to j),
is the transition rate from substate i to j), 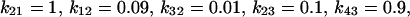 and
and 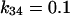 for the irreducible one, and
for the irreducible one, and 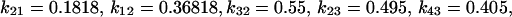 and
and 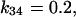 for the reducible one. These values are found by comparing
for the reducible one. These values are found by comparing 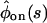 and
and 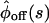 of the two kinetic schemes. In the ordered waiting times trajectory generated from the irreducible scheme similar waiting times tend to follow each other (A), whereas from the reducible one, the waiting times are randomly distributed (B). By applying a threshold on this trajectory, which separates the fast from the slow events, one can estimate the transition rates
of the two kinetic schemes. In the ordered waiting times trajectory generated from the irreducible scheme similar waiting times tend to follow each other (A), whereas from the reducible one, the waiting times are randomly distributed (B). By applying a threshold on this trajectory, which separates the fast from the slow events, one can estimate the transition rates 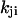 by calculating the average of the fast and slow off waiting times, given by
by calculating the average of the fast and slow off waiting times, given by 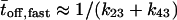 and
and 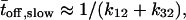 and the average number of successive fast and slow off waiting times, given by,
and the average number of successive fast and slow off waiting times, given by, 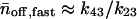 and
and 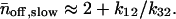
Finally, we note that some of the fundamental concepts presented in this work have already been used in the analyses of the catalytic activity of individual lipase molecules (Flomenbom et al., 2005). In this case, the (immobilized) enzyme converted a nonfluorescent substrate molecule into a fluorescent product molecule. The two-state trajectories were constructed from the photon count trajectories. The off state was associated with the conversion substrate→product, whereas the on state was associated with the diffusion of the product molecule away from the enzyme and its vicinity. The off waiting time PDF was best fitted to a stretched exponential. This functional behavior was interpreted as stemming from a spectrum of active enzymatic conformations. The bulk relaxation function test was then applied, and the kinetic scheme was shown to be irreducible. Additionally, clusters of fast events were detected in the ordered off waiting times trajectory (similar to Fig. 3 A), indicating that single lipase molecules display correlations in their activity. These findings were combined to build a kinetic scheme that involves reaction and conformational changes simultaneously, and to extract some of the conformational and reaction rate values.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
We thank A. M. Berezhkovskii and I. Gopich for stimulating discussions.
References
- Boguñá, M., A. M. Berezhkovskii, and G. H. Weiss. 2000. Residence time densities for non-Markovian systems. Physica A. 282:475–485. [Google Scholar]
- Bokinsky, G., D. Rueda, V. K. Misra, A. Gordus, M. M. Rhodes, H. P. Babcock, N. G. Walter, and X. Zhuang. 2003. Single-molecule transition state of RNA folding. Proc. Natl. Acad. Sci. USA. 100:9302–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. 2000. Event-averaged measurements of single-molecule kinetics. Chem. Phys. Lett. 327:38–44. [Google Scholar]
- Colquhoun, D., A. G. Hawkes, and K. Srodzinski. 1996. Joint distributions of apparent open times and shut times of single ion channels and the maximum likelihood fitting of mechanisms. Philos. Trans. R. Soc. Lond. A. 354:2555–2590. [Google Scholar]
- Cox, D. R. 1962. Renewal Theory. Methuen, London, UK.
- Edman, L., Z. Földes-Papp, S. Wennmalm, and R. Rigler. 1999. The fluctuating enzyme: a single molecule approach. Chem. Phys. 247:11–22. [Google Scholar]
- Edman, L., and R. Rigler. 2000. Memory landscapes of single-enzyme molecules. Proc. Natl. Acad. Sci. USA. 97:8266–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flomenbom, O., K. Velonia, D. Loos, S. Masuo, M. Cotlet, Y. Engelborghs, J. Hofkens, A. E. Rowan, R. J. M. Nolte, M. Van der Auweraer, F. C. de Schryver, and J. Klafter. 2005. Stretched exponential decay and correlations in the catalytic activity of fluctuating single lipase molecules. Proc. Natl. Acad. Sci. USA. 102:2368–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, T., A. Y. Ting, J. Liang, W. B. Caldwell, A. A. Deniz, D. S. Chemla, P. G. Schultz, and S. Weiss. 1999. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. USA. 96:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, T. L. 1985. Cooperativity Theory in Biochemistry. Springer-Verlag, New York, NY.
- Kasianowicz, J. J., E. Brandin, D. Branton, and D. W. Deamer. 1996. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA. 93:13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., L. Xun, and X. S. Xie. 1998. Single-molecule enzymatic dynamics. Science. 282:1877–1882. [DOI] [PubMed] [Google Scholar]
- McManus, O. B., A. L. Blaz, and K. L. Magleby. 1985. Inverse relationship of the durations of the open and shut intervals for Cl and K channels. Nature. 317:625–628. [DOI] [PubMed] [Google Scholar]
- Mets, U., and R. Rigler. 1994. Submillisecond detection of single rhodamine molecules in water. J. Fluorescence. 4:259–264. [DOI] [PubMed] [Google Scholar]
- Moerner, W. E., and M. Orrit. 1999. Illuminating single molecules in condensed matter. Science. 283:1670–1676. [DOI] [PubMed] [Google Scholar]
- Neher, E., and B. Sakmann. 1976. Single-channel currents recorded from membrane of denervated frog muscle fibers. Nature. 260:799–802. [DOI] [PubMed] [Google Scholar]
- Nie, S., D. T. Chiu, and R. N. Zare. 1994. Probing individual molecules with confocal fluorescence microscopy. Science. 266:1018–1021. [DOI] [PubMed] [Google Scholar]
- Rhoades, E., E. Gussakovsky, and G. Haran. 2003. Watching proteins fold one molecule at a time. Proc. Natl. Acad. Sci. USA. 100:3197–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenter, G. K., H. P. Lu, and X. S. Xie. 1999. Statistical analyses and theoretical models of single-molecule enzymatic dynamics. J. Phys. Chem. A. 103:10477–10488. [Google Scholar]
- Schuler, B., E. A. Lipman, and W. A. Eaton. 2002. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 419:743–747. [DOI] [PubMed] [Google Scholar]
- Shera, E. B., N. K. Seitzinger, L. M. Davis, R. A. Keller, and S. A. Soper. 1990. Detection of single fluorescent molecules. Chem. Phys. Lett. 174:553–557. [Google Scholar]
- Velonia, K., O. Flomenbom, D. Loos, S. Masuo, M. Cotlet, Y. Engelborghs, J. Hofkens, A. E. Rowan, J. Klafter, R. J. M. Nolte, and F. C. de Schryver. 2005. Single enzyme kinetics of CALB catalyzed hydrolysis. Angew. Chem. Int. Ed. Engl. 44:560–564. [DOI] [PubMed] [Google Scholar]
- Wang, J., and P. Wolynes. 1995. Intermittency of single molecule reaction dynamics in fluctuating environment. Phys. Rev. Lett. 74:4317–4320. [DOI] [PubMed] [Google Scholar]
- Weiss, S. 1999. Fluorescence spectroscopy of single biomolecules. Science. 283:1676–1683. [DOI] [PubMed] [Google Scholar]
- Wennmalm, S., L. Edman, and R. Rigler. 1997. Conformational fluctuations in single DNA molecules. Proc. Natl. Acad. Sci. USA. 94:10641–10646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkoskie, J. B., and J. Cao. 2004. Single molecule kinetics. I. Theoretical analysis of indicators. J. Chem. Phys. 121:6361–6372. [DOI] [PubMed] [Google Scholar]
- Yang, H., G. Luo, P. Karnchanaphanurach, T.-M. Louie, I. Rech, S. Cova, L. Xun, and X. S. Xie. 2003. Protein conformational dynamics probed by single-molecule electron transfer. Science. 302:262–266. [DOI] [PubMed] [Google Scholar]




