Abstract
Genomic DNA copy number alterations are key genetic events in the development and progression of human cancers. Here we report a genome-wide microarray comparative genomic hybridization (array CGH) analysis of DNA copy number variation in a series of primary human breast tumors. We have profiled DNA copy number alteration across 6,691 mapped human genes, in 44 predominantly advanced, primary breast tumors and 10 breast cancer cell lines. While the overall patterns of DNA amplification and deletion corroborate previous cytogenetic studies, the high-resolution (gene-by-gene) mapping of amplicon boundaries and the quantitative analysis of amplicon shape provide significant improvement in the localization of candidate oncogenes. Parallel microarray measurements of mRNA levels reveal the remarkable degree to which variation in gene copy number contributes to variation in gene expression in tumor cells. Specifically, we find that 62% of highly amplified genes show moderately or highly elevated expression, that DNA copy number influences gene expression across a wide range of DNA copy number alterations (deletion, low-, mid- and high-level amplification), that on average, a 2-fold change in DNA copy number is associated with a corresponding 1.5-fold change in mRNA levels, and that overall, at least 12% of all the variation in gene expression among the breast tumors is directly attributable to underlying variation in gene copy number. These findings provide evidence that widespread DNA copy number alteration can lead directly to global deregulation of gene expression, which may contribute to the development or progression of cancer.
Conventional cytogenetic techniques, including comparative genomic hybridization (CGH) (1), have led to the identification of a number of recurrent regions of DNA copy number alteration in breast cancer cell lines and tumors (2–4). While some of these regions contain known or candidate oncogenes [e.g., FGFR1 (8p11), MYC (8q24), CCND1 (11q13), ERBB2 (17q12), and ZNF217 (20q13)] and tumor suppressor genes [RB1 (13q14) and TP53 (17p13)], the relevant gene(s) within other regions (e.g., gain of 1q, 8q22, and 17q22–24, and loss of 8p) remain to be identified. A high-resolution genome-wide map, delineating the boundaries of DNA copy number alterations in tumors, should facilitate the localization and identification of oncogenes and tumor suppressor genes in breast cancer. In this study, we have created such a map, using array-based CGH (5–7) to profile DNA copy number alteration in a series of breast cancer cell lines and primary tumors.
An unresolved question is the extent to which the widespread DNA copy number changes that we and others have identified in breast tumors alter expression of genes within involved regions. Because we had measured mRNA levels in parallel in the same samples (8), using the same DNA microarrays, we had an opportunity to explore on a genomic scale the relationship between DNA copy number changes and gene expression. From this analysis, we have identified a significant impact of widespread DNA copy number alteration on the transcriptional programs of breast tumors.
Materials and Methods
Tumors and Cell Lines.
Primary breast tumors were predominantly large (>3 cm), intermediate-grade, infiltrating ductal carcinomas, with more than 50% being lymph node positive. The fraction of tumor cells within specimens averaged at least 50%. Details of individual tumors have been published (8, 9), and are summarized in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Breast cancer cell lines were obtained from the American Type Culture Collection. Genomic DNA was isolated either using Qiagen genomic DNA columns, or by phenol/chloroform extraction followed by ethanol precipitation.
DNA Labeling and Microarray Hybridizations.
Genomic DNA labeling and hybridizations were performed essentially as described in Pollack et al. (7), with slight modifications. Two micrograms of DNA was labeled in a total volume of 50 microliters and the volumes of all reagents were adjusted accordingly. “Test” DNA (from tumors and cell lines) was fluorescently labeled (Cy5) and hybridized to a human cDNA microarray containing 6,691 different mapped human genes (i.e., UniGene clusters). The “reference” (labeled with Cy3) for each hybridization was normal female leukocyte DNA from a single donor. The fabrication of cDNA microarrays and the labeling and hybridization of mRNA samples have been described (8).
Data Analysis and Map Positions.
Hybridized arrays were scanned on a GenePix scanner (Axon Instruments, Foster City, CA), and fluorescence ratios (test/reference) calculated using scanalyze software (available at http://rana.lbl.gov). Fluorescence ratios were normalized for each array by setting the average log fluorescence ratio for all array elements equal to 0. Measurements with fluorescence intensities more than 20% above background were considered reliable. DNA copy number profiles that deviated significantly from background ratios measured in normal genomic DNA control hybridizations were interpreted as evidence of real DNA copy number alteration (see Estimating Significance of Altered Fluorescence Ratios in the supporting information). When indicated, DNA copy number profiles are displayed as a moving average (symmetric 5-nearest neighbors). Map positions for arrayed human cDNAs were assigned by identifying the starting position of the best and longest match of any DNA sequence represented in the corresponding UniGene cluster (10) against the “Golden Path” genome assembly (http://genome.ucsc.edu/; Oct 7, 2000 Freeze). For UniGene clusters represented by multiple arrayed elements, mean fluorescence ratios (for all elements representing the same UniGene cluster) are reported. For mRNA measurements, fluorescence ratios are “mean-centered” (i.e., reported relative to the mean ratio across the 44 tumor samples). The data set described here can be accessed in its entirety in the supporting information.
Results
We performed CGH on 44 predominantly locally advanced, primary breast tumors and 10 breast cancer cell lines, using cDNA microarrays containing 6,691 different mapped human genes (Fig. 1a; also see Materials and Methods for details of microarray hybridizations). To take full advantage of the improved spatial resolution of array CGH, we ordered (fluorescence ratios for) the 6,691 cDNAs according to the “Golden Path” (http://genome.ucsc.edu/) genome assembly of the draft human genome sequences (11). In so doing, arrayed cDNAs not only themselves represent genes of potential interest (e.g., candidate oncogenes within amplicons), but also provide precise genetic landmarks for chromosomal regions of amplification and deletion. Parallel analysis of DNA from cell lines containing different numbers of X chromosomes (Fig. 1b), as we did before (7), demonstrated the sensitivity of our method to detect single-copy loss (45, XO), and 1.5- (47,XXX), 2- (48,XXXX), or 2.5-fold (49,XXXXX) gains (also see Fig. 5, which is published as supporting information on the PNAS web site). Fluorescence ratios were linearly proportional to copy number ratios, which were slightly underestimated, in agreement with previous observations (7). Numerous DNA copy number alterations were evident in both the breast cancer cell lines and primary tumors (Fig. 1a), detected in the tumors despite the presence of euploid non-tumor cell types; the magnitudes of the observed changes were generally lower in the tumor samples. DNA copy-number alterations were found in every cancer cell line and tumor, and on every human chromosome in at least one sample. Recurrent regions of DNA copy number gain and loss were readily identifiable. For example, gains within 1q, 8q, 17q, and 20q were observed in a high proportion of breast cancer cell lines/tumors (90%/69%, 100%/47%, 100%/60%, and 90%/44%, respectively), as were losses within 1p, 3p, 8p, and 13q (80%/24%, 80%/22%, 80%/22%, and 70%/18%, respectively), consistent with published cytogenetic studies (refs. 2–4; a complete listing of gains/losses is provided in Tables 2 and 3, which are published as supporting information on the PNAS web site). The total number of genomic alterations (gains and losses) was found to be significantly higher in breast tumors that were high grade (P = 0.008), consistent with published CGH data (3), estrogen receptor negative (P = 0.04), and harboring TP53 mutations (P = 0.0006) (see Table 4, which is published as supporting information on the PNAS web site).
Figure 1.
Genome-wide measurement of DNA copy number alteration by array CGH. (a) DNA copy number profiles are illustrated for cell lines containing different numbers of X chromosomes, for breast cancer cell lines, and for breast tumors. Each row represents a different cell line or tumor, and each column represents one of 6,691 different mapped human genes present on the microarray, ordered by genome map position from 1pter through Xqter. Moving average (symmetric 5-nearest neighbors) fluorescence ratios (test/reference) are depicted using a log2-based pseudocolor scale (indicated), such that red luminescence reflects fold-amplification, green luminescence reflects fold-deletion, and black indicates no change (gray indicates poorly measured data). (b) Enlarged view of DNA copy number profiles across the X chromosome, shown for cell lines containing different numbers of X chromosomes.
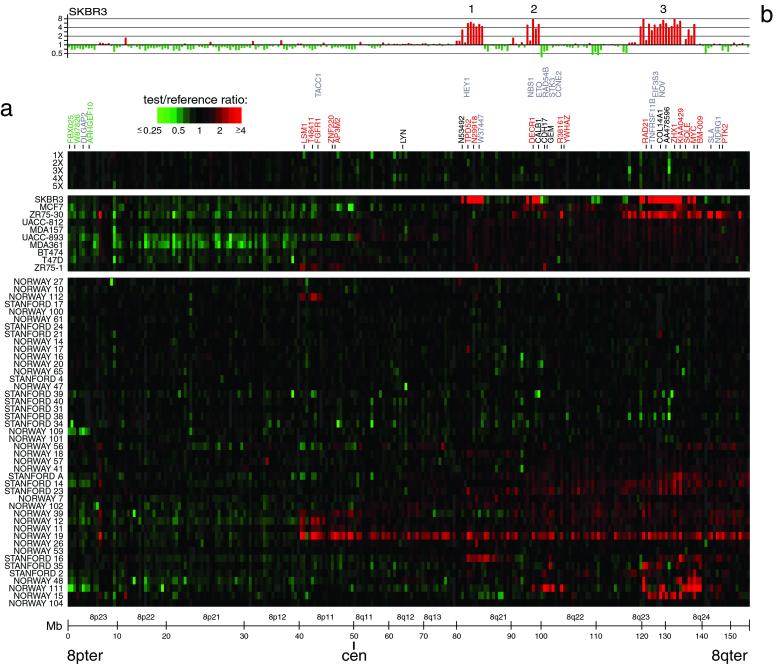
The improved spatial resolution of our array CGH analysis is illustrated for chromosome 8, which displayed extensive DNA copy number alteration in our series. A detailed view of the variation in the copy number of 241 genes mapping to chromosome 8 revealed multiple regions of recurrent amplification; each of these potentially harbors a different known or previously uncharacterized oncogene (Fig. 2a). The complexity of amplicon structure is most easily appreciated in the breast cancer cell line SKBR3. Although a conventional CGH analysis of 8q in SKBR3 identified only two distinct regions of amplification (12), we observed three distinct regions of high-level amplification (labeled 1–3 in Fig. 2b). For each of these regions we can define the boundaries of the interval recurrently amplified in the tumors we examined; in each case, known or plausible candidate oncogenes can be identified (a description of these regions, as well as the recurrently amplified regions on chromosomes 17 and 20, can be found in Figs. 6 and 7, which are published as supporting information on the PNAS web site).
Figure 2.
DNA copy number alteration across chromosome 8 by array CGH. (a) DNA copy number profiles are illustrated for cell lines containing different numbers of X chromosomes, for breast cancer cell lines, and for breast tumors. Breast cancer cell lines and tumors are separately ordered by hierarchical clustering to highlight recurrent copy number changes. The 241 genes present on the microarrays and mapping to chromosome 8 are ordered by position along the chromosome. Fluorescence ratios (test/reference) are depicted by a log2 pseudocolor scale (indicated). Selected genes are indicated with color-coded text (red, increased; green, decreased; black, no change; gray, not well measured) to reflect correspondingly altered mRNA levels (observed in the majority of the subset of samples displaying the DNA copy number change). The map positions for genes of interest that are not represented on the microarray are indicated in the row above those genes represented on the array. (b) Graphical display of DNA copy number profile for breast cancer cell line SKBR3. Fluorescence ratios (tumor/normal) are plotted on a log2 scale for chromosome 8 genes, ordered along the chromosome.
For a subset of breast cancer cell lines and tumors (4 and 37, respectively), and a subset of arrayed genes (6,095), mRNA levels were quantitatively measured in parallel by using cDNA microarrays (8). The parallel assessment of mRNA levels is useful in the interpretation of DNA copy number changes. For example, the highly amplified genes that are also highly expressed are the strongest candidate oncogenes within an amplicon. Perhaps more significantly, our parallel analysis of DNA copy number changes and mRNA levels provides us the opportunity to assess the global impact of widespread DNA copy number alteration on gene expression in tumor cells.
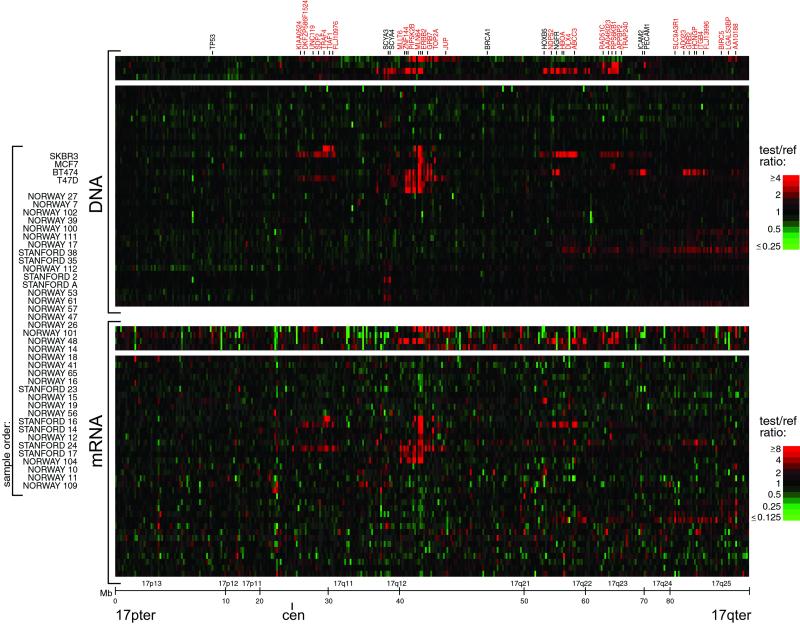
A strong influence of DNA copy number on gene expression is evident in an examination of the pseudocolor representations of DNA copy number and mRNA levels for genes on chromosome 17 (Fig. 3). The overall patterns of gene amplification and elevated gene expression are quite concordant; i.e., a significant fraction of highly amplified genes appear to be correspondingly highly expressed. The concordance between high-level amplification and increased gene expression is not restricted to chromosome 17. Genome-wide, of 117 high-level DNA amplifications (fluorescence ratios >4, and representing 91 different genes), 62% (representing 54 different genes; see Table 5, which is published as supporting information on the PNAS web site) are found associated with at least moderately elevated mRNA levels (mean-centered fluorescence ratios >2), and 42% (representing 36 different genes) are found associated with comparably highly elevated mRNA levels (mean-centered fluorescence ratios >4).
Figure 3.
Concordance between DNA copy number and gene expression across chromosome 17. DNA copy number alteration (Upper) and mRNA levels (Lower) are illustrated for breast cancer cell lines and tumors. Breast cancer cell lines and tumors are separately ordered by hierarchical clustering (Upper), and the identical sample order is maintained (Lower). The 354 genes present on the microarrays and mapping to chromosome 17, and for which both DNA copy number and mRNA levels were determined, are ordered by position along the chromosome; selected genes are indicated in color-coded text (see Fig. 2 legend). Fluorescence ratios (test/reference) are depicted by separate log2 pseudocolor scales (indicated).
To determine the extent to which DNA deletion and lower-level amplification (in addition to high-level amplification) are also associated with corresponding alterations in mRNA levels, we performed three separate analyses on the complete data set (4 cell lines and 37 tumors, across 6,095 genes). First, we determined the average mRNA levels for each of five classes of genes, representing DNA deletion, no change, and low-, medium-, and high-level amplification (Fig. 4a). For both the breast cancer cell lines and tumors, average mRNA levels tracked with DNA copy number across all five classes, in a statistically significant fashion (P values for pair-wise Student's t tests comparing adjacent classes: cell lines, 4 × 10−49, 1 × 10−49, 5 × 10−5, 1 × 10−2; tumors, 1 × 10−43, 1 × 10−214, 5 × 10−41, 1 × 10−4). A linear regression of the average log(DNA copy number), for each class, against average log(mRNA level) demonstrated that on average, a 2-fold change in DNA copy number was accompanied by 1.4- and 1.5-fold changes in mRNA level for the breast cancer cell lines and tumors, respectively (Fig. 4a, regression line not shown). Second, we characterized the distribution of the 6,095 correlations between DNA copy number and mRNA level, each across the 37 tumor samples (Fig. 4b). The distribution of correlations forms a normal-shaped curve, but with the peak markedly shifted in the positive direction from zero. This shift is statistically significant, as evidenced in a plot of observed vs. expected correlations (Fig. 4c), and reflects a pervasive global influence of DNA copy number alterations on gene expression. Notably, the highest correlations between DNA copy number and mRNA level (the right tail of the distribution in Fig. 4b) comprise both amplified and deleted genes (data not shown). Third, we used a linear regression model to estimate the fraction of all variation measured in mRNA levels among the 37 tumors that could be attributed to underlying variation in DNA copy number. From this analysis, we estimate that, overall, about 7% of all of the observed variation in mRNA levels can be explained directly by variation in copy number of the altered genes (Fig. 4d). We can reduce the effects of experimental measurement error on this estimate by using only that fraction of the data most reliably measured (fluorescence intensity/background >3); using that data, our estimate of the percent variation in mRNA levels directly attributed to variation in gene copy number increases to 12% (Fig. 4d). This still undoubtedly represents a significant underestimate, as the observed variation in global gene expression is affected not only by true variation in the expression programs of the tumor cells themselves, but also by the variable presence of non-tumor cell types within clinical samples.
Figure 4.
Genome-wide influence of DNA copy number alterations on mRNA levels. (a) For breast cancer cell lines (gray) and tumor samples (black), both mean-centered mRNA fluorescence ratio (log2 scale) quartiles (box plots indicate 25th, 50th, and 75th percentile) and averages (diamonds; Y-value error bars indicate standard errors of the mean) are plotted for each of five classes of genes, representing DNA deletion (tumor/normal ratio < 0.8), no change (0.8–1.2), low- (1.2–2), medium- (2–4), and high-level (>4) amplification. P values for pair-wise Student's t tests, comparing averages between adjacent classes (moving left to right), are 4 × 10−49, 1 × 10−49, 5 × 10−5, 1 × 10−2 (cell lines), and 1 × 10−43, 1 × 10−214, 5 × 10−41, 1 × 10−4 (tumors). (b) Distribution of correlations between DNA copy number and mRNA levels, for 6,095 different human genes across 37 breast tumor samples. (c) Plot of observed versus expected correlation coefficients. The expected values were obtained by randomization of the sample labels in the DNA copy number data set. The line of unity is indicated. (d) Percent variance in gene expression (among tumors) directly explained by variation in gene copy number. Percent variance explained (black line) and fraction of data retained (gray line) are plotted for different fluorescence intensity/background (a rough surrogate for signal/noise) cutoff values. Fraction of data retained is relative to the 1.2 intensity/background cutoff. Details of the linear regression model used to estimate the fraction of variation in gene expression attributable to underlying DNA copy number alteration can be found in the supporting information (see Estimating the Fraction of Variation in Gene Expression Attributable to Underlying DNA Copy Number Alteration).
Discussion
This genome-wide, array CGH analysis of DNA copy number alteration in a series of human breast tumors demonstrates the usefulness of defining amplicon boundaries at high resolution (gene-by-gene), and quantitatively measuring amplicon shape, to assist in locating and identifying candidate oncogenes. By analyzing mRNA levels in parallel, we have also discovered that changes in DNA copy number have a large, pervasive, direct effect on global gene expression patterns in both breast cancer cell lines and tumors. Although the DNA microarrays used in our analysis may display a bias toward characterized and/or highly expressed genes, because we are examining such a large fraction of the genome (approximately 20% of all human genes), and because, as detailed above, we are likely underestimating the contribution of DNA copy number changes to altered gene expression, we believe our findings are likely to be generalizable (but would nevertheless still be remarkable if only applicable to this set of ∼6,100 genes).
In budding yeast, aneuploidy has been shown to result in chromosome-wide gene expression biases (13). Two recent studies have begun to examine the global relationship between DNA copy number and gene expression in cancer cells. In agreement with our findings, Phillips et al. (14) have shown that with the acquisition of tumorigenicity in an immortalized prostate epithelial cell line, new chromosomal gains and losses resulted in a statistically significant respective increase and decrease in the average expression level of involved genes. In contrast, Platzer et al. (15) recently reported that in metastatic colon tumors only ∼4% of genes within amplified regions were found more highly (>2-fold) expressed, when compared with normal colonic epithelium. This report differs substantially from our finding that 62% of highly amplified genes in breast cancer exhibit at least 2-fold increased expression. These contrasting findings may reflect methodological differences between the studies. For example, the study of Platzer et al. (15) may have systematically under-measured gene expression changes. In this regard it is remarkable that only 14 transcripts of many thousand residing within unamplified chromosomal regions were found to exhibit at least 4-fold altered expression in metastatic colon cancer. Additionally, their reliance on lower-resolution chromosomal CGH may have resulted in poorly delimiting the boundaries of high-complexity amplicons, effectively overcalling regions with amplification. Alternatively, the contrasting findings for amplified genes may represent real biological differences between breast and metastatic colon tumors; resolution of this issue will require further studies.
Our finding that widespread DNA copy number alteration has a large, pervasive and direct effect on global gene expression patterns in breast cancer has several important implications. First, this finding supports a high degree of copy number-dependent gene expression in tumors. Second, it suggests that most genes are not subject to specific autoregulation or dosage compensation. Third, this finding cautions that elevated expression of an amplified gene cannot alone be considered strong independent evidence of a candidate oncogene's role in tumorigenesis. In our study, fully 62% of highly amplified genes demonstrated moderately or highly elevated expression. This highlights the importance of high-resolution mapping of amplicon boundaries and shape [to identify the “driving” gene(s) within amplicons (16)], on a large number of samples, in addition to functional studies. Fourth, this finding suggests that analyzing the genomic distribution of expressed genes, even within existing microarray gene expression data sets, may permit the inference of DNA copy number aberration, particularly aneuploidy (where gene expression can be averaged across large chromosomal regions; see Fig. 3 and supporting information). Fifth, this finding implies that a substantial portion of the phenotypic uniqueness (and by extension, the heterogeneity in clinical behavior) among patients' tumors may be traceable to underlying variation in DNA copy number. Sixth, this finding supports a possible role for widespread DNA copy number alteration in tumorigenesis (17, 18), beyond the amplification of specific oncogenes and deletion of specific tumor suppressor genes. Widespread DNA copy number alteration, and the concomitant widespread imbalance in gene expression, might disrupt critical stochiometric relationships in cell metabolism and physiology (e.g., proteosome, mitotic spindle), possibly promoting further chromosomal instability and directly contributing to tumor development or progression. Finally, our findings suggest the possibility of cancer therapies that exploit specific or global imbalances in gene expression in cancer.
Supplementary Material
Acknowledgments
We thank the many members of the P.O.B. and D.B. labs for helpful discussions. J.R.P. was a Howard Hughes Medical Institute Physician Postdoctoral Fellow during a portion of this work. P.O.B. is a Howard Hughes Medical Institute Associate Investigator. This work was supported by grants from the National Institutes of Health, the Howard Hughes Medical Institute, the Norwegian Cancer Society, and the Norwegian Research Council.
Abbreviation
- CGH
comparative genomic hybridization
References
- 1.Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 2.Kallioniemi A, Kallioniemi O P, Piper J, Tanner M, Stokke T, Chen L, Smith H S, Pinkel D, Gray J W, Waldman F M. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O P. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 4.Forozan F, Mahlamaki E H, Monni O, Chen Y, Veldman R, Jiang Y, Gooden G C, Ethier S P, Kallioniemi A, Kallioniemi O P. Cancer Res. 2000;60:4519–4525. [PubMed] [Google Scholar]
- 5.Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 6.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W L, Chen C, Zhai Y, et al. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 7.Pollack J R, Perou C M, Alizadeh A A, Eisen M B, Pergamenschikov A, Williams C F, Jeffrey S S, Botstein D, Brown P O. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 8.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 9.Sorlie T, Perou C M, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen M B, van de Rijn M, Jeffrey S S, et al. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuler G D. J Mol Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- 11.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Fejzo M S, Godfrey T, Chen C, Waldman F, Gray J W. Genes Chromosomes Cancer. 1998;22:105–113. doi: 10.1002/(sici)1098-2264(199806)22:2<105::aid-gcc4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 13.Hughes T R, Roberts C J, Dai H, Jones A R, Meyer M R, Slade D, Burchard J, Dow S, Ward T R, Kidd M J, et al. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 14.Phillips J L, Hayward S W, Wang Y, Vasselli J, Pavlovich C, Padilla-Nash H, Pezullo J R, Ghadimi B M, Grossfeld G D, Rivera A, et al. Cancer Res. 2001;61:8143–8149. [PubMed] [Google Scholar]
- 15.Platzer P, Upender M B, Wilson K, Willis J, Lutterbaugh J, Nosrati A, Willson J K, Mack D, Ried T, Markowitz S. Cancer Res. 2002;62:1134–1138. [PubMed] [Google Scholar]
- 16.Albertson D G, Ylstra B, Segraves R, Collins C, Dairkee S H, Kowbel D, Kuo W L, Gray J W, Pinkel D. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 17.Li R, Yerganian G, Duesberg P, Kraemer A, Willer A, Rausch C, Hehlmann R. Proc Natl Acad Sci USA. 1997;94:14506–14511. doi: 10.1073/pnas.94.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasnick D, Duesberg P H. Biochem J. 1999;340:621–630. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.