Abstract
The 3.31-Mb genome sequence of the intracellular pathogen and potential bioterrorism agent, Brucella suis, was determined. Comparison of B. suis with Brucella melitensis has defined a finite set of differences that could be responsible for the differences in virulence and host preference between these organisms, and indicates that phage have played a significant role in their divergence. Analysis of the B. suis genome reveals transport and metabolic capabilities akin to soil/plant-associated bacteria. Extensive gene synteny between B. suis chromosome 1 and the genome of the plant symbiont Mesorhizobium loti emphasizes the similarity between this animal pathogen and plant pathogens and symbionts. A limited repertoire of genes homologous to known bacterial virulence factors were identified.
Species in the genus Brucella (Brucella spp.) are the etiological agents of brucellosis, a zoonotic disease endemic in many areas of the world, characterized by chronic infections in animals leading to abortion and infertility, and a systemic, febrile illness in humans (1). Human infection frequently occurs via direct contact with tissues and fluids from infected animals, but can also be contracted by consumption of contaminated foods or by inhalation (2). Brucella suis was the first pathogenic organism weaponized by the U.S. military during the 1950s (3). It constitutes a potential bioterrorism threat that could be targeted against military personnel, civilians, or food supplies (4, 5). Early diagnosis of brucellosis is problematic, no acceptable vaccines are currently available for human immunization, and the current treatment regimen is prolonged antibiotic therapy (6).
Brucella spp. are facultative intracellular pathogens that enter the host through mucosal surfaces and are able to survive inside macrophages. The primary strategy for survival in macrophages appears to be inhibition of phagosome–lysosome fusion (7–9). Localization and proliferation within autophagosome-like compartments associated with the rough endoplasmic reticulum has also been demonstrated in placental trophoblasts and other nonprofessional phagocytes (10, 11). The complete genome sequence of B. suis provides insight into the lifestyle, pathogenesis, and evolution of this pathogen.
Methods
ORF Prediction and Gene Identification.
ORFs likely to encode proteins were predicted by glimmer (12, 13). This program, based on interpolated Markov models, was trained with ORFs larger than 600 bp from the genomic sequence, as well as with the B. suis genes available in GenBank. All predicted proteins larger than 30 aa were searched against a nonredundant protein database as described (14). Frameshifts and point mutations were detected and corrected where appropriate. Remaining frameshifts and point mutations are considered to be authentic and were annotated as “authentic frameshift” or “authentic point mutation.” Protein membrane-spanning domains were identified by toppred (15, 16). The 5′ regions of each ORF were inspected to define initiation codons using homologies, position of ribosomal binding sites, and transcriptional terminators. Two sets of hidden Markov models were used to determine ORF membership in families and superfamilies: PFAM V5.5 (17) and TIGRFAMS 1.0 H (18). PFAM V5.5 hidden Markov models were also used with a constraint of minimum two hits to find repeated domains within proteins and mask them. Domain-based paralogous families were then built by performing all-versus-all searches on the remaining protein sequences by using a modified version of a previously described method (19).
Comparative Genomics.
The B. suis and Brucella melitensis genomes were compared at the nucleotide level by suffix tree analysis using mummer to identify exact matches of at least 20 base pairs (13), and their ORF sets were compared using FASTA3 (20). The protein sets of B. suis, Sinorhizobium meliloti, Mesorhizobium loti, and Agrobacterium tumefaciens were also compared using FASTA3. Shared genes were defined using a FASTA3 P value cutoff of 10−15.
Trinucleotide Composition.
Regions of atypical nucleotide composition were identified by the χ2 analysis: the distribution of all 64 trinucleotides (3-mers) was computed for the complete genome in all six reading frames, followed by the 3-mer distribution in 2,000-bp windows. Windows overlapped by 1,000-bp. For each window, the χ2 statistic on the difference between its 3-mer content and that of the whole genome was computed.
Results and Discussion
General Features of the Genome.
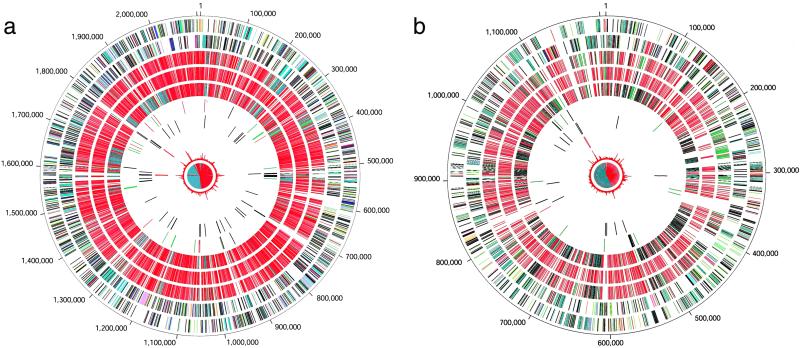
The genome of B. suis strain 1330, a swine isolate and standard reference strain for B. suis biovar 1 (21), was sequenced by the whole genome sequencing method (22). The B. suis 1330 genome consists of two circular chromosomes of 2,107,792 bp (Chr I) and 1,207,381 bp (Chr II) (see Fig. 1, Table 1, and Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). A total of 2,185 and 1,203 ORFs were identified on Chr I and II, respectively.
Figure 1.
Circular representation of the two chromosomes of B. suis strain 1330. The outer circle shows predicted coding regions on the plus strand color-coded by role categories: salmon, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; green, energy metabolism; purple, fatty acid and phospholipid metabolism; pink, protein fate/synthesis; orange, purines, pyrimidines, nucleosides, nucleotides; blue, regulatory functions; gray, transcription; teal, transport and binding proteins; black, hypothetical and conserved hypothetical proteins. Second circle, predicted coding regions on the minus strand color-coded by role categories. Third circle, top hits to S. meliloti, according to replicon: red, main chromosome; green, plasmid pSymA; gray, plasmid pSymB. Fourth circle, top hits to M. loti, according to replicon: red, main chromosome; green, plasmid pMLa; gray, plasmid pMLb. Fifth circle, top hits to A. tumefaciens, according to replicon: red, circular chromosome; gray, linear chromosome; green, plasmid pAtC58; blue, plasmid pTiC58. Sixth circle, locations of B. suis-specific regions greater than 100 base pairs, green; B. melitensis-specific regions greater than 100 base pairs, red; and transposase genes, black. Seventh circle, tRNAs in brown. Eighth circle, rRNA operons in red. Ninth circle, atypical nucleotide composition curve in red. Tenth circle, GC-skew curve in red/black.
Table 1.
General features of the B. suis genome
| Chr I | Chr II | |
|---|---|---|
| Size, bp | 2,107,792 | 1,207,381 |
| G+C content, % | 57.2 | 57.3 |
| Protein-coding genes | ||
| No. similar to known proteins | 1,178 | 730 |
| No. similar to proteins of unknown function | 224 | 122 |
| No. of conserved hypotheticals | 418 | 163 |
| No. of hypothetical proteins* | 365 | 188 |
| Total | 2,185 | 1,203 |
| Average ORF size (base pairs) | 842 | 897 |
| Coding, % | 87.3 | 89.4 |
| Stable RNAs | ||
| rRNA operons | 2 | 1 |
| tRNA | 41 | 14 |
Hypothetical proteins were defined as those with no database match, with the exception of database matches to B. melitensis hypothetical proteins.
Strains of the four B. suis biovars have been shown to be variable in chromosome number and size, having either one 3.3-Mb chromosome (biovar 3) or 2 chromosomes of smaller size (biovars 1, 2, and 4), possibly because of recombination events involving the three rRNA operons (23). It has been proposed that the different Brucella strains evolved from an ancestor with a single circular chromosome equivalent to the structure seen in biovar 3 (23). Genome analysis indicates that the two chromosomes probably have distinct evolutionary origins.
Chr I resembles a classic bacterial circular chromosome with the likely origin of replication adjacent to a gene cluster consisting of dnaA, dnaN, and recF (24). In contrast, Chr II possesses a cluster of plasmid-like replication genes including a replication initiation protein RepC (BRA0001) and partitioning proteins RepA and RepB (BRA1202 and BRA1203), similar to plasmid replication genes from Agrobacterium Ti plasmids, and plasmids from other organisms including Rhizobium spp. The partitioning proteins ParA and ParB on Chr I also cluster phylogenetically with chromosomal ParAs and ParBs from alpha proteobacteria.
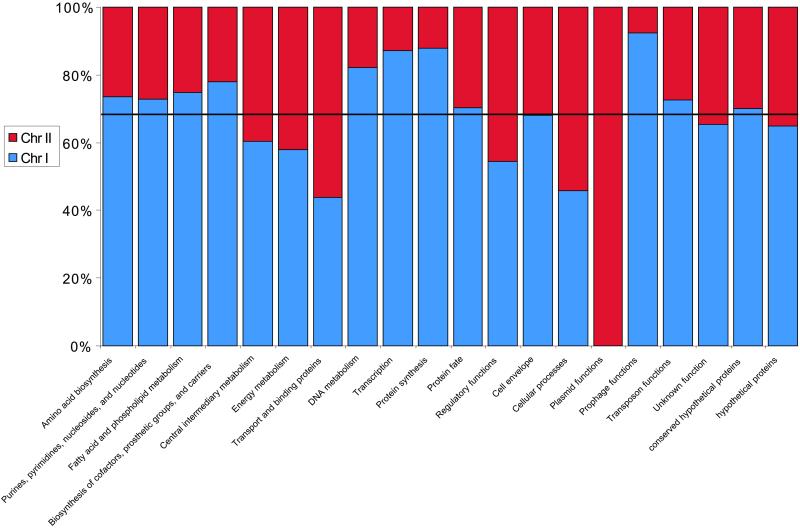
There is a pronounced asymmetry of genes in different functional categories between the two chromosomes (Fig. 2). Chr I encodes the majority of the core metabolic machinery for processes such as transcription, translation, and protein synthesis. For instance, 51 of 53 ribosomal proteins and 41 of 55 tRNAs are located on Chr I. Chr I also possesses a high percentage of phage-related proteins, because of the presence of inserted phage remnants. In contrast, Chr II is overrepresented in genes involved in processes such as membrane transport, central intermediary and energy metabolism, and regulation (Fig. 2); these appear to largely represent auxiliary pathways for utilization of specific substrates. Cellular processes and plasmid functions are also concentrated on Chr II, primarily because of the presence of three clusters of flagellar biosynthesis and secretion genes and the clusters of conjugation-associated and plasmid-like replication genes on Chr II, respectively. Chr II is not predicted to be dispensable, because it includes a number of essential genes such as the solitary tRNA-Cys and three tRNA synthetases.
Figure 2.
Percentage of B. suis ORFs encoded on Chr I (blue) and II (red) in biological role categories. The horizontal line represents the percentage of ORFs encoded on each of the two chromosomes.
The differences in gene content and plasmid-like replication are consistent with Chr II being derived from a megaplasmid that was captured by an ancestral Brucella, as has been suggested for Vibrio cholerae (25). The acquisition of such a megaplasmid was presumably a very ancient event because the G+C percentage of the two chromosomes are very similar and essentially identical chromosomal structures are found in B. melitensis and B. abortus.
Brucella Comparative Genomics.
Comparison of the B. suis genome with that of the published B. melitensis genome (26) revealed extensive similarity and gene synteny. Comparison of the two genomes by mummer suffix tree analysis (13) revealed a total of 7,307 single nucleotide polymorphisms (SNPs) within a shared backbone of 3,237,820 bp (see Table 3, which is published as supporting information on the PNAS web site). The frequency of interspecies SNP occurrence between B. suis and B. melitensis is lower than the intraspecies frequency for some other sequenced organisms for which multiple strains have been sequenced (see Table 3).
The majority (>90%) of B. suis and B. melitensis genes share 98–100% identity at the nucleotide level. The more variable genes (<95% identity) consist primarily of hypothetical genes, as well as a UreE urease component, and probable surface-exposed genes such as outer membrane proteins, membrane transporters, a putative invasin, and ShdA-like adhesins. These more variable genes may significantly contribute to the differences in pathogenicity or host preference between these two organisms. The high degree of similarity between the B. suis and B. melitensis genomes at both the gene and nucleotide level is consistent with the proposition that the Brucella species should be grouped as biovars of a single species (27, 28).
Thirty-three regions greater than 100 bp unique to either B. suis (22 regions) or B. melitensis (11 regions) were identified (Table 2) and are referred to hereafter as islands as per the usage in Perna et al. (29). Based on the whole genome nucleotide alignments, only 42 B. suis and 32 B. melitensis genes were identified that are completely absent in the other genome. The annotated protein data sets of these two organisms also contain additional differences due to frame-shifted or truncated genes and different predictions of hypothetical genes. The largest B. melitensis- and B. suis-specific islands appear to be due to phage-mediated integration events. The largest B. suis-specific island consists of 18 genes, flanked at one end by two phage genes including an integrase homologue (BRA0362) and containing conjugal transfer genes homologous to those of IncP plasmids such as RP4 and a probable plasmid replication gene. Another B. suis-specific island is of phage origin; BR0584–BR0593 are related to lambda-like phage proteins or are conserved hypothetical proteins, of these, BR0588–BR0593 are absent from B. melitensis, possibly because of a deletion event. The largest B. melitensis-specific island consists of 29 complete genes, flanked at one end by a phage integrase homologue (BMEI1702), and containing 27 hypothetical or conserved hypothetical genes, and a putative peptidoglycan hydrolase (Table 2). All of these three regions have atypical trinucleotide content, the two B. suis-specific islands are adjacent to tRNAs, highlighting the importance of these elements as recombinational hotspots. Two other probable integrated phage remnants in B. suis (BR1083–BR1078 and BR0260–BR0256), including phage integrase genes, are also located adjacent to tRNAs and are present in B. melitensis. Approximately 50% of the hypothetical proteins in these unique phage-associated regions are predicted to be surface-exposed and could contribute to differences in host preference and disease manifestation between these organisms (30, 31).
Table 2.
Regions unique to B. suis or B. melitensis
| Coordinates
|
Size, bp | ORFs in region*
|
Putative function(s) | ||
|---|---|---|---|---|---|
| B. suis | B. melitensis | Partial ORFs | Intact ORFs | ||
| Unique B. suis regions | |||||
| Chromosome 1 | |||||
| 1830260–1830371 | 163542 | 111 | BR1895 | Cell division protein, FtsK | |
| 1782549–1786501 | 207313 | 3952 | BR1855 | BR1852–BR1854 | Transposase, hypothetical, conserved hypothetical (2) |
| 1618082–1618926 | 370350 | 844 | BR1671–BR1673 | Transposase (2), HlyD family secretion protein | |
| 1568658–1568890 | 419471 | 232 | BR1622 | OMP31-like outer membrane protein | |
| 1158710–1158859 | 829930 | 149 | |||
| 1050997–1051159 | 937906 | 162 | |||
| 1030464–1031244 | 957888 | 780 | BR1059, BR1060 | HlyD family protein, Multidrug-resistance transporter | |
| 924517–927170 | 1061223 | 2653 | BR0951 | BR0952–BR0955 | Amino acid ABC transporter-binding protein, hypothetical, ABC transporter-permease (2), putative GST |
| 581357–584895 | 1401689 | 3538 | BR0588–BR0593 | Hypothetical, conserved hypothetical (3), major capsid protein, protease | |
| 408365–408478 | 1574445 | 113 | BR0404 | Glycyl-tRNA synthetase, beta subunit | |
| 397748–398173 | 1584637 | 425 | BR0391 | BR0389, BR0390 | Hypothetical |
| 366793–366928 | 1615531 | 135 | BR0355 | Hydroxypyruvate reductase | |
| 271721–271883 | 1710693 | 162 | |||
| 233098–233272 | 1769806 | 174 | BR0221 | Transcriptional regulator | |
| Chromosome 2 | |||||
| 157505–158033 | 1112570 | 528 | BRA0173 | Outer membrane protein | |
| 343385–361675 | 927290 | 18,290 | BRA0362–BRA0379 | Phage integrase, DNA-binding protein, RepA, trbL, trbJ, traC, traJ, traI, hypothetical (8), conserved hypothetical, DnaJ | |
| 521895–522864 | 767132 | 969 | BRA0541, BRA0542 | Hypothetical, NAD-dependent epimerase family | |
| 610838–618576 | 679160 | 7738 | BRA0630, BRA0636 | BRA0631–BRA0635 | Oxidoreductase, amino acid ABC transporter-binding protein (2), transcriptional regulator, b-ketoadipyl CoA thiolase, conserved hypothetical (2) |
| 731653–732234 | 566167 | 581 | BRA0749, BRA0750 | ABC transporter-permease | |
| 889144–889843 | 409215 | 699 | BRA0907 | Conserved hypothetical | |
| 1062798–1062987 | 236185 | 189 | BRA1080 | Dipeptide ABC transporter-permease | |
| 1082827–1083058 | 216360 | 231 | BRA1096 | Transcriptional regulator | |
| Unique B. melitensis regions | |||||
| Chromosome 1 | |||||
| 1766571 | 222996–223107 | 107 | |||
| 1161964 | 826061–826825 | 764 | BMEI0801 | Propionyl-CoA carboxylase beta chain | |
| 1055172 | 933462–933738 | 276 | BMEI0899 | Phage-related DNA binding protein | |
| 1050978 | 937920–938199 | 238 | |||
| 776056 | 1209608–1210453 | 845 | BMEI1163, BMEI1164 | Transposase | |
| 383374 | 1598986–1599091 | 105 | BMEI1555 | BMEI1554 | Transporter, MFS superfamily |
| 219685 | 1783211–1783405 | 194 | BMEI1742 | ABC transporter ATP-binding protein | |
| 79687 | 1923357–1923696 | 339 | BMEI1873 | Cell surface protein | |
| 271941 | 1710394–1710672 | 240 | BMEI1659 | Hypothetical | |
| 258912 | 1723285–1744168 | 20,883 | BMEI1703 | BMEI1674–BMEI1702 | Hypothetical (24), phage integrase, transposase, conserved hypothetical (4) |
| Chromosome 2 | |||||
| 809098 | 688410–688518 | 108 | BMEII0466 | Tetratricopeptide repeat family | |
Intact ORFs are full-length ORFs located in the respective unique region while partial ORFs are those that are partly contained within the unique region. No ORFs are listed for regions that are intergenic.
Other B. suis- and B. melitensis-specific regions are smaller in size and are not located in regions of atypical nucleotide content. Some of these probably represent deletion rather than insertion events. For instance, BR0955–BR0952 encode an amino acid ABC transporter in B. suis, but two of the three subunits of this transporter are missing in B. melitensis. Other differences related to metabolism include a B. suis-specific island containing BRA0630–BRA0635, which includes two ABC amino acid periplasmic binding proteins, and an amino acid dehydrogenase, suggesting B. suis may be able to use an amino acid or related derivative that B. melitensis cannot. These two B. suis-specific islands may explain the capability of B. suis, unlike B. melitensis, to oxidize ornithine, citrulline, arginine, and lysine, in part the basis for differential biotyping of the Brucella spp. (31).
Comparative Genomics with Other Alpha-Proteobacteria.
B. suis is a member of the alpha-proteobacteria, related to other human/animal pathogens such as Bartonella spp. and more distantly to plant pathogens such as A. tumefaciens and to plant symbionts such as S. meliloti (32). Analysis of the complete set of predicted B. suis proteins revealed that it is very similar to that of species in the Rhizobium/Agrobacterium group. A total of 1,902 B. suis ORFs were conserved in all three of M. loti, S. meliloti, and A. tumefaciens, and 2,408 B. suis ORFs were conserved in at least one of these three genomes (see Table 4, which is published as supporting information on the PNAS web site).
Like B. suis, M. loti, S. meliloti, and A. tumefaciens have multiple replicons (33). The majority of B. suis proteins from Chr I have significant hits (P < 10−15) to proteins encoded on the main circular chromosomes of these three organisms (see Table 4). In contrast, proteins encoded on B. suis Chr II have significant numbers of hits to proteins encoded on the linear chromosome of A. tumefaciens (339 proteins) and to the pSymA (88 proteins) and pSymB (162 proteins) megaplasmids of S. meliloti. Fig. 1 shows that these best hits to these other replicons cluster in specific regions.
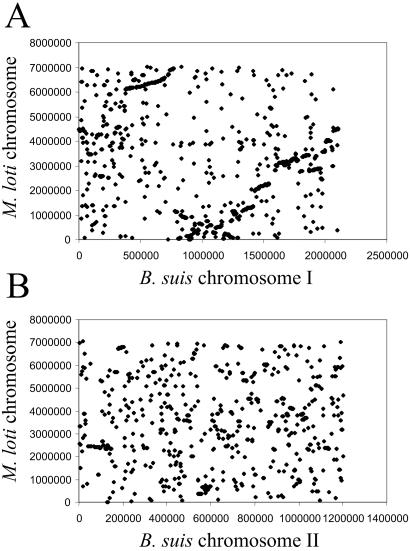
A. tumefaciens and S. meliloti have been reported to share extensive conservation of gene order (gene synteny) with each other, but only short regions of limited synteny with M. loti (34). B. suis Chr I shares extensive regions of gene synteny with the M. loti chromosome (Fig. 3) despite the latter being almost three times larger, but only short regions of limited synteny with any replicon from S. meliloti or A. tumefaciens. A plot of the synteny with M. loti shows an X-alignment pattern with symmetry around the terminus of replication (35), probably indicative of small-scale inversion events. The absence of synteny near the origin of replication is likely indicative of more extensive gene rearrangement events in this region. B. suis Chr II shares limited regions of gene synteny with M. loti and the linear plasmid of A. tumefaciens and the pSym megaplasmids of S. meliloti. These analyses reveal that the chromosomes and megaplasmids of these organisms share a very complex evolutionary history including gene exchange, rearrangements, and replicon fusion.
Figure 3.
Whole genome proteome alignment (35) between proteins encoded on (A) B. suis chromosome I (horizontal axis) and the M. loti chromosome (vertical axis) and (B) B. suis chromosome II (horizontal axis) and the M. loti chromosome (vertical axis). Plots show the chromosome locations of pairs of predicted proteins that have significant similarity (P < 10−15).
Metabolism and Transport.
Based on genome analysis, B. suis is predicted to use at least thirteen sugars, and 18 aa as carbon/nitrogen/energy sources, with more than two-thirds of these pathways encoded on the small chromosome. The genes necessary for glycolysis (except phosphofructokinase) and a complete pentose phosphate pathway, which would enable usage of glucose by circumventing the missing step in glycolysis, are split between the two chromosomes. The Entner–Doudoroff pathway is encoded on the large chromosome. Genes are present that would enable the reduction of nitrous oxide, nitric oxide, nitrate, and nitrite.
B. suis is predicted to have an unexpected capacity to use plant-derived compounds. There is an intact beta-ketoadipate pathway (BRA1155–BRA1162), and homoprotocatechuate pathway (BRA0636–BRA0647), encoded on Chr II. These two pathways are widely distributed among diverse soil microorganisms and play a central role in the processing and degradation of plant-derived aromatic compounds, including the breakdown products of lignins. Additionally, the B. suis genome encodes four monooxygenases (BR0746, BR0960, BR1292, BR1899), an extradiol ring cleaving dioxygenase (BRA1001), two nitroreductases (BR1069, BRA1054), and 39 oxidoreductases of unknown specificity that are probably involved in the reduction of aromatic or aliphatic compounds. The derivatives of these reactions are probably funneled into the beta-ketoadipate and homoprotocatechuate pathways. It is known that soil contaminated with Brucella can remain infectious for 7–10 weeks (36, 37). These pathways may contribute to the survival of B. suis outside of its host.
B. suis possesses broader transport capabilities than other sequenced intracellular human pathogens (38). Over two-thirds of the B. suis transporters are ABC transporters (91 systems, 284 genes), primarily for amino acids (22 transporters), sugars (16 transporters), and peptides (11 transporters), as was also observed in A. tumefaciens, S. meliloti, and M. loti (39). The concentration of amino acid and peptide transporters could be related to the trafficking of B. suis to a compartment associated with the ER, where peptides and amino acids might be plentiful. The systems for peptide and sugar uptake are preferentially distributed on the small chromosome (see Fig. 5, which is published as supporting information on the PNAS web site), as was also seen for their catabolic genes.
Transposable Elements.
Twenty-nine insertion sequences (IS) were identified in the B. suis genome, including two clusters of IS elements (Fig. 1). The first cluster (11 elements) is associated with a gene cluster involved in lipopolysaccharide biosynthesis on Chr I (BR0512–BR0536), the second, smaller cluster of IS elements (4 elements) surrounds a putative cell-wall protein on Chr II (BRA0553). Both lipopolysaccharide and the cell-wall surface protein are probably surface-exposed and therefore potentially under selective pressure from the host immune response. The presence of the IS elements may indicate the occurrence of multiple gene replacement or recombinational events.
Chr II includes a 50-kb region (BRA1072–BRA1116) that may represent a putative composite transposon. This region is flanked by two essentially identical IS3-like elements in inverted orientation, each adjacent to tRNA genes, and has an atypical trinucleotide composition, suggesting a possible foreign origin (Fig. 1). It does not contain any obvious predicted virulence genes, but does have a large number of peptide ABC uptake genes (16 of 43 genes).
Pathogenicity.
Adhesion, invasion, immune evasion, and intracellular trafficking are all poorly understood in B. suis, but genome analysis has implicated a variety of genes in these processes (see Table 5, which is published as supporting information on the PNAS web site). Three proteins (BRA1148, BRA0173, and BR2013) containing a domain homologous to autotransporters, such as the Salmonella typhimurium fibronectin binding protein ShdA (40), were identified that may serve as putative adhesins mediating attachment before phagocytosis (41). Fibronectin has been reported inside endocytic vacuoles containing Brucella, suggesting that binding to fibronectin may facilitate uptake (42). All three of these genes are encoded in regions with atypical nucleotide composition, suggesting possible acquisition by lateral gene transfer. Six additional putative adhesins related to the Bartonella Pap31/HbpA proteins (43) and to Brucella Omp25 protein were identified (see Table 5). Homologues (BR1836 and BR0340) of the Bartonella bacilliformis invasion-associated proteins IalA and IalB (44, 45) are present.
There are two clusters of genes (BR0510–BR0540, BR0981–BR0982) involved in lipopolysaccharide biosynthesis, which has been implicated in virulence (46). Both of these are located in regions of atypical nucleotide composition. The first of these regions is also associated with multiple IS elements (see above). Two additional gene clusters (BRA0542–BRA0547 and BRA0135–BRA0137) are also involved in polysaccharide biosynthesis, possibly of an unknown polysaccharide.
As described for B. melitensis (26), B. suis possesses a type IV secretion system that has been implicated in intracellular trafficking (47) and three clusters of genes encoding flagellar biosynthesis. No putative effectors such as secreted phospholipases or other toxins were detected on the B. suis genome, which is consistent with its limited cytopathogenicity. Two putative hemolysins (BR0437 and BR0068) were identified.
B. suis encodes two distinct gene clusters on the large chromosome encoding urease UreA-G subunits. Phylogenetic analysis (see Fig. 6, which is published as supporting information on the PNAS web site) indicates that the seven subunits of one of the ureases (BR0267–BR0273) consistently branch with subunits from alpha-proteobacteria, such as M. loti and A. tumefaciens, whereas subunits of the second urease (BR1356–BR1361) consistently branch with subunits from gamma-proteobacteria, such as Yersinia pestis, suggesting that it may have been laterally acquired by an ancestral Brucella. Ureases of several bacteria have been implicated in their virulence in different animal models of infection (48). For example, the M. tuberculosis urease has been implicated in inhibition of phagolysosomal fusion and modulation of pH of the phagosome, facilitating intracellular survival (49). Only one other completely sequenced genome, E. coli O157:H7, possesses two ureases, but these are identical and apparently the result of a recent duplication event. One or both of the Brucella ureases may play a role in the ability of these organisms to either survive or prevent phagolysosomal acidification. However, it should be noted that a nickel transporter mutant with reduced urease activity was not attenuated during intracellular growth (50).
Conclusions.
Comparison of the closely related B. suis and B. melitensis genomes has defined a finite set of differences that could be responsible for the differences in virulence and host preference between these organisms. Based on the genomic and functional similarities between B. suis and organisms from the Rhizobium/Agrobacterium group, it seems likely that the Brucella evolved from a soil/plant-associated ancestral bacteria and speculatively, it may be metabolically active outside of a mammalian host. Furthermore, there may be fundamental similarities in parasitic/symbiotic strategies between animal pathogens such as B. suis, plant symbionts such as S. meliloti, and plant pathogens such as A. tumefaciens. A variety of predicted virulence factors were identified whose experimental characterization may enhance our understanding of Brucella pathogenesis.
Supplementary Material
Acknowledgments
We thank M. Heaney, S. Lo, M. Holmes, B. Lee, R. Karamchedu, and V. Sapiro for database and IT support at The Institute for Genomic Research (TIGR), and the TIGR faculty and sequencing core for expert advice and assistance. This work was supported by the Defense Advanced Research Projects Agency and National Institutes of Health, National Institute of Allergy and Infectious Disease Grant 1U01AI49036-01.
Abbreviations
- Chr
chromosome
- IS
insertion sequence
Footnotes
References
- 1.Young E J. Clin Infect Dis. 1995;21:283–289. doi: 10.1093/clinids/21.2.283. [DOI] [PubMed] [Google Scholar]
- 2.Franz D R. Ann NY Acad Sci. 1999;894:100–104. doi: 10.1111/j.1749-6632.1999.tb08051.x. [DOI] [PubMed] [Google Scholar]
- 3.Regis E. The Biology of Doom: The History of America's Secret Germ Warfare Project. New York: Henry Holt and Associates; 1999. [Google Scholar]
- 4.Kortepeter M G, Parker G W. Emerg Infect Dis. 1999;5:523–527. doi: 10.3201/eid0504.990411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horn F P, Breeze R G. Ann NY Acad Sci. 1999;894:9–17. doi: 10.1111/j.1749-6632.1999.tb08037.x. [DOI] [PubMed] [Google Scholar]
- 6.Nicoletti P. Adv Biotechnol Processes. 1990;13:147–168. [PubMed] [Google Scholar]
- 7.Arenas G N, Staskevich A S, Aballay A, Mayorga L S. Infect Immun. 2000;68:4255–4263. doi: 10.1128/iai.68.7.4255-4263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldwin C L, Winter A J. Immunol Ser. 1994;60:363–380. [PubMed] [Google Scholar]
- 9.Naroeni A, Jouy N, Ouahrani-Bettache S, Liautard J P, Porte F. Infect Immun. 2001;69:486–493. doi: 10.1128/IAI.69.1.486-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson T D, Cheville N F, Meador V P. Vet Pathol. 1986;23:227–239. doi: 10.1177/030098588602300302. [DOI] [PubMed] [Google Scholar]
- 11.Pizarro-Cerda J, Meresse S, Parton R G, van der Goot G, Sola-Landa A, Lopez-Goni I, Moreno E, Gorvel J P. Infect Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzberg S L, Delcher A L, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcher A L, Harmon D, Kasif S, White O, Salzberg S L. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haft D H, Loftus B J, Richardson D L, Yang F, Eisen J A, Paulsen I T, White O. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nierman W C, Feldblyum T V, Laub M T, Paulsen I T, Nelson K E, Eisen J, Heidelberg J F, Alley M R, Ohta N, Maddock J R, et al. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson W R. Methods Mol Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 21.Morgan W J, Corbel M J. Dev Biol Stand. 1976;31:27–37. [PubMed] [Google Scholar]
- 22.Tettelin H, Nelson K E, Paulsen I T, Eisen J A, Read T D, Peterson S, Heidelberg J, DeBoy R T, Haft D H, Dodson R J, et al. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 23.Jumas-Bilak E, Michaux-Charachon S, Bourg G, O'Callaghan D, Ramuz M. Mol Microbiol. 1998;27:99–106. doi: 10.1046/j.1365-2958.1998.00661.x. [DOI] [PubMed] [Google Scholar]
- 24.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 25.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature (London) 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DelVecchio V G, Kapatral V, Redkar R J, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, Lykidis A, et al. Proc Natl Acad Sci USA. 2002;99:443–448. doi: 10.1073/pnas.221575398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandara B, Merino A L, Rogel M A, Martinez-Romero E. J Clin Microbiol. 2001;39:235–240. doi: 10.1128/JCM.39.1.235-240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verger J-M, Grimont F, Grimont P A, Grayon M. Int J Syst Bacteriol. 1985;35:292–295. [Google Scholar]
- 29.Perna N T, Plunkett G, III, Burland V, Mau B, Glasner J D, Rose D J, Mayhew G F, Evans P S, Gregor J, Kirkpatrick H A, et al. Nature (London) 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 30.Smith L D, Ficht T A. Crit Rev Microbiol. 1990;17:209–230. doi: 10.3109/10408419009105726. [DOI] [PubMed] [Google Scholar]
- 31.Timoney J F. Hagan and Bruner's Microbiology and Infectious Diseases of Domestic Animals. Ithaca, NY: Comstock Publishing Associates; 1988. [Google Scholar]
- 32.Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. J Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, et al. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 34.Galibert F, Finan T M, Long S R, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett M J, Becker A, Boistard P, et al. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 35. Eisen, J. A., Heidelberg, J. F., White, O. & Salzberg, S. L. (2000) Genome Biol.1, research0011.1–0011.9. [DOI] [PMC free article] [PubMed]
- 36.Cotton W E. J Am Vet Med Assoc. 1919;55:504–528. [Google Scholar]
- 37.Elberg S S. A Guide to the Diagnosis, Treatment and Prevention of Human Brucellosis. Geneva: World Health Organization; 1981. [Google Scholar]
- 38.Paulsen I T, Nguyen L, Sliwinski M K, Rabus R, Saier M H., Jr J Mol Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 39.Wood D W, Setubal J C, Kaul R, Monks D E, Kitajima J P, Okura V K, Zhou Y, Chen L, Wood G E, Almeida N F, Jr, et al. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 40.Kingsley R A, Santos R L, Keestra A M, Adams L G, Baumler A J. Mol Microbiol. 2002;43:895–905. doi: 10.1046/j.1365-2958.2002.02805.x. [DOI] [PubMed] [Google Scholar]
- 41.Benz I, Schmidt M A. Infect Immun. 1989;57:1506–1511. doi: 10.1128/iai.57.5.1506-1511.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gay B, Mauss H, Sanchez-Teff S. Virchows Arch B Cell Pathol Incl Mol Pathol. 1986;52:169–176. doi: 10.1007/BF02889960. [DOI] [PubMed] [Google Scholar]
- 43.Carroll J A, Coleman S A, Smitherman L S, Minnick M F. Infect Immun. 2000;68:6750–6757. doi: 10.1128/iai.68.12.6750-6757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coleman S A, Minnick M F. Infect Immun. 2001;69:4373–4381. doi: 10.1128/IAI.69.7.4373-4381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell S J, Minnick M F. Infect Immun. 1995;63:1552–1562. doi: 10.1128/iai.63.4.1552-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Godfroid F, Taminiau B, Danese I, Denoel P, Tibor A, Weynants V, Cloeckaert A, Godfroid J, Letesson J J. Infect Immun. 1998;66:5485–5493. doi: 10.1128/iai.66.11.5485-5493.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foulongne V, Bourg G, Cazevieille C, Michaux-Charachon S, O'Callaghan D. Infect Immun. 2000;68:1297–1303. doi: 10.1128/iai.68.3.1297-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Koning-Ward T F, Ward A C, Hartland E L, Robins-Browne R M. Contrib Microbiol Immunol. 1995;13:262–263. [PubMed] [Google Scholar]
- 49.Clemens D L, Lee B Y, Horwitz M A. J Bacteriol. 1995;177:5644–5652. doi: 10.1128/jb.177.19.5644-5652.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jubier-Maurin V, Rodrigue A, Ouahrani-Bettache S, Layssac M, Mandrand-Berthelot M A, Kohler S, Liautard J P. J Bacteriol. 2001;183:426–434. doi: 10.1128/JB.183.2.426-434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.