Abstract
The retinoblastoma tumor-suppressor protein (Rb) plays a critical role in controlling cellular proliferation and apoptosis by regulating E2F transcription factors. Rb is a key target of oncoproteins expressed by DNA tumor viruses, but RNA viruses are not known to regulate Rb function. Here, we show that Rb abundance is negatively regulated in cells containing replicating genomic RNA from hepatitis C virus, a human virus strongly associated with hepatocellular carcinoma. The viral RNA-dependent RNA polymerase NS5B forms a complex with Rb, targeting it for degradation and resulting in reduction of Rb abundance, activation of E2F-responsive promoters, and cell proliferation. NS5B contains a conserved Leu-x-Cys/Asn-x-Asp motif that is homologous to Rb-binding domains in the oncoproteins of DNA viruses. This domain overlaps the polymerase active site, and mutations within it abrogate Rb binding and reverse the effects of NS5B on E2F promoter activation and cell proliferation. These findings suggest a unique link between an oncogenic RNA virus implicated in the development of liver cancer and a critically important tumor-suppressor protein.
Keywords: cell cycle regulation, chronic hepatitis, E2F transcription factor, hepatocellular carcinoma, viral oncogenesis
Hepatitis C virus (HCV) infection is a leading cause of morbidity and mortality in many human populations (1). Persons with persistent HCV infection are at increased risk of developing cirrhosis and hepatocellular carcinoma (2). The strong association between hepatocellular carcinoma and HCV infection is particularly notable in that HCV is a positive-strand RNA virus, classified within the genus Hepacivirus of the family Flaviviridae. Its 9.6-kb genome replicates exclusively within the cytoplasm (3) and encodes a single, large polyprotein that is processed by cellular and viral proteases into only 10 individual structural and nonstructural viral proteins. Although inflammation associated with chronic hepatitis C is likely to contribute to the development of hepatocellular carcinoma, there is evidence that one or more of the proteins expressed by the virus contribute directly to carcinogenesis. Transgenic mice expressing a high abundance of the core protein develop hepatocellular carcinoma and steatosis (4). Liver cancer also developed in transgenic mice expressing a much lower abundance of the entire viral polyprotein but not in a companion transgenic lineage expressing a higher abundance of the structural proteins (core, E1, E2, and p7) only (5). Such data suggest a direct role for both structural and nonstructural proteins of HCV in oncogenesis.
Mechanisms that regulate cell-cycle progression are frequently disrupted in hepatocellular carcinomas associated with HCV infection. These regulatory mechanisms include the retinoblastoma protein (Rb) (6), which plays a major role in controlling the G1 to S-phase transition through a repressive effect on E2F transcription factors (7). Rb functions as a tumor suppressor, and the gene which encodes it (RB) is frequently mutated in various types of tumors, including retinoblastomas, small-cell lung carcinomas, and osteosarcomas (8). Rb is targeted by oncoproteins expressed by several DNA tumor viruses, including adenovirus E1A (9) and human papilloma-virus E7 (10). These oncoproteins interact directly with Rb through a conserved LxCxE motif and effectively abrogate its function (11).
Here, we demonstrate a direct link between HCV RNA replication and cellular expression of Rb. We show that the expression of Rb is down-regulated posttranslationally in cells containing replicating HCV RNA and that the viral NS5B polymerase is responsible for this regulation. Our data reveal a mechanism by which HCV, an oncogenic RNA virus, may promote hepatocellular carcinogenesis.
Materials and Methods
Cell Lines and Plasmids. The NNeo/C-5B and NNeo/3-5B replicon cells and their isoclonal IFN-cured counterparts have been described in refs. 12 and 13. See also Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Luciferase Reporter Promoter Assays. To determine the impact of NS5B expression on the activity of E2F-responsive promoters, we transfected cells with luciferase reporter plasmids in which luciferase was under the transcriptional control of the p107 or MAD2 promoters. Details are available in Supporting Materials and Methods.
Immunoblot and Coimmunoprecipitation Assays. The interaction of NS5B with cellular Rb was studied by using a combination of immunoblot and coimmunoprecipitation assays, as described in Supporting Materials and Methods.
In Vitro Rb-Binding Assay. Extracts of Huh7 cells expressing Flag-tagged NS5B proteins were mixed with anti-Flag antibodies (M2, Sigma) and protein G Sepharose 4 Fast Flow (Amersham Pharmacia) and incubated for 2 h at 4°C. Immune complexes were precipitated and washed with buffer to prepare NS5B resins. Bacterially expressed GST or GST-Rb (301-928) (2.5 μg) was mixed with each NS5B resin and further incubated for 2 h at 4°C. The resins were washed again with buffer, and binding proteins were eluted and assayed by immunoblot.
Further details can be found in Supporting Materials and Methods.
Results
Rb Is Negatively Regulated in Cells Supporting HCV RNA Replication. Until very recently, no cultured cell type has been shown to be fully permissive for HCV replication. We thus asked whether Rb expression was altered in Huh7 cells supporting the autonomous replication of dicistronic HCV RNA replicons (Fig. 1A). We studied two independent, Huh7 hepatoma cell lines, NNeo/C-5B clones 2-3 and 3, both of which contain replicating genome-length RNA expressing all of the proteins of the HCV-N strain (12). We compared the abundance of Rb in these cell lines with that in their progeny, designated 2-3c and 3c, respectively, in which HCV RNA had been eliminated by prior treatment with IFN-α2b (13). The abundance of Rb in each of the HCV RNA-containing cell lines was reduced compared with that in the cognate-cured cell line (Fig. 1B). The absolute abundance of Rb differed in the 2-3c and 3c cell lines, reflecting their clonally distinct nature, but, in both cases, the related parental cells containing the replicating viral RNA had a lower Rb abundance. Rb is a member of a family of three closely related proteins that include p107 and p130, the expression of which is regulated independently of Rb (14). In contrast to Rb, the expression of p107 was not reduced by the presence of replicating viral RNA, whereas p130 abundance was reduced in only the clone 3 cells (Fig. 1B). Thus, HCV RNA replication appears to specifically affect Rb abundance.
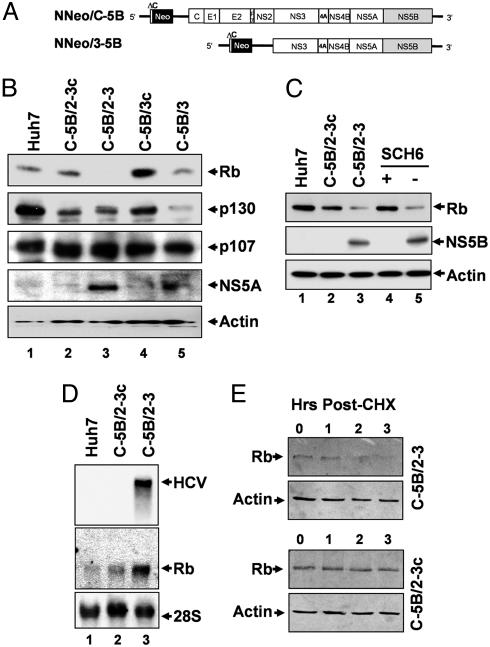
Fig. 1.
Rb is negatively regulated in HCV replicon cells. (A) Organization of the dicistronic genome-length NNeo/C-5B and subgenomic NNeo/3-5B HCV-N RNAs. (B) Immunoblot of retinoblastoma-family proteins (Rb, p130, and p107) in extracts of Huh7 cells, clone 2-3 and 3 replicon cells, and their cured 2-3c and 3c counterparts. (C) Immunoblots of Rb and NS5B in extracts of normal Huh7 cells, 2-3 and 2-3c cells, and 2-3 cells after 2-week treatment with SCH6 (+) or mock (-) treatment. (D) Northern blot of HCV RNA and Rb mRNA in Huh7 cells, 2-3 replicon cells, and cured 2-3c cells. (E) Immunoblot of Rb in extracts of cycloheximide (CHX)-treated 2-3 (Upper) and cured 2-3c cells (Lower). Duration of CHX treatment (Hrs) is indicated above the blots.
To further confirm this effect, we studied a third cell line containing a subgenomic HCV replicon, NNeo/3-5B(RG), which expresses only the NS3-NS5B region of the HCV polyprotein (Fig. 1 A) (12). Although its IFN-cured derivative, NNeo/3-5B(RG)c cells, had a relatively low abundance of Rb (see Fig. 5A, which is published as supporting information on the PNAS web site), Rb abundance was further reduced in the companion replicon cells. We also observed an increase in Rb abundance in 2-3 cells after treatment with SCH6, a peptidomimetic inhibitor of the NS3/4A protease that blocks replication of HCV RNA and leads to a reduction in viral-protein abundance (15) (Fig. 1C). Taken together, these data show that HCV RNA replication is associated with a reduction in Rb abundance in each of three clonally distinct hepatoma cell lines. Furthermore, the negative regulation of Rb in the subgenomic NNeo/3-5B(RG) cells suggests that this reduction in Rb abundance is related to expression of one or more of the nonstructural viral proteins.
We carried out a Northern blot analysis of Rb mRNA to determine whether the reduction in Rb abundance was transcriptionally regulated. A quantitative comparison with 28S RNA indicated that the Rb mRNA levels were not reduced, but rather were up-regulated in the HCV replicon cells (Fig. 1D), likely due to derepression of Rb mRNA expression, which is negatively regulated by its own gene product (16). We thus assessed the stability of the Rb protein in 2-3 cells and their cured 2-3c counterparts after treatment with cycloheximide, a protein-synthesis inhibitor. We found that the stability of Rb was decreased in 2-3 cells supporting HCV RNA replication (Figs. 1E and 5B). A pulse-chase labeling experiment generated similar results, suggesting a half-life for Rb of ≈2 h in the cured 2-3c cells, and only 1.2 h in the 2-3 cells containing the replicating viral RNA (Fig. 5C). Together, these data provide strong evidence that Rb is down-regulated posttranslationally by HCV RNA replication in Huh7 cells.
NS5B-Mediates the Regulation of Rb. To determine which HCV proteins are responsible for the negative regulation of Rb, we ectopically expressed viral proteins and determined Rb abundance in normal Huh7 cells. We limited these studies to the nonstructural proteins, because we found that the subgenomic replicon NNeo/3-5B(RG) negatively regulated Rb (Fig. 5A). NS5B-Flag expression markedly reduced the steady-state levels of endogenous Rb, whereas expression of NS3/4A-Flag, NS4B-Flag or NS5A-Flag did not (Fig. 2A). We next asked whether this reduction in Rb abundance reflects an interaction between Rb and NS5B. We found that NS5B, but not the viral core protein, coimmunoprecipitated with Rb from lysates of 2-3 cells (Fig. 2B). When we ectopically expressed NS5B by transfection of normal Huh7 cells with a vector encoding the viral polyprotein segment extending from NS3 to NS5B, we also found it to coimmunoprecipitate with endogenous Rb (Fig. 2C). Similarly, Rb coimmunoprecipitated with NS5B-Flag from lysates of transfected Huh7 cells (see Fig. 6, which is published as supporting information on the PNAS web site). These data suggest that NS5B forms a stable complex with Rb, whether expressed ectopically or from a replicating viral RNA.
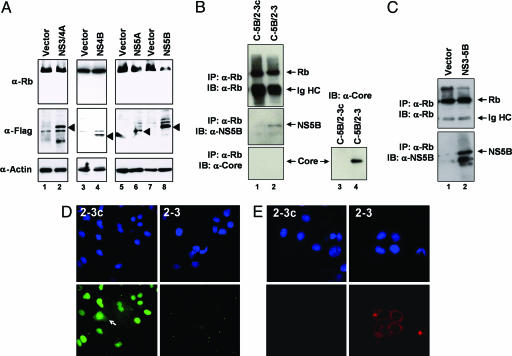
Fig. 2.
NS5B negatively regulates endogenous Rb abundance. (A) Immunoblots (IB) of Rb in extracts of normal Huh7 cells transfected with Flag-tagged HCV proteins: NS3/4A, NS4B, NS5A, NS5B, or empty expression vectors. Expression was monitored with Flag-specific antibody and Rb and actin abundances compared. (B) NS5B, but not the viral core protein, coimmunoprecipitates with endogenous Rb in lysates of C-5B/2-3 cells, which contain replicating viral RNA. Parallel immunoprecipitations were done with lysates of 2-3c cells, which do not contain replicating RNA or express NS5B. (Lower Right) A control immunoblot for core protein in comparable amounts of lysate from 2-3 and 2-3c cells. (C) Ectopically expressed NS5B coimmunoprecipitates with endogenous Rb. Normal Huh7 cells were transfected with either empty vector or an expression vector encoding the NS3-NS5B segment of the polyprotein under control of the CMV promoter. (D) Immunofluorescence detection of Rb in cured 2-3c (Left) and 2-3 replicon (Right) cells. (Top) DAPI stain (nuclei). (Bottom) Rb, visualized with a FITC-labeled antibody, was primarily localized to nuclei but present in the cytoplasm of a small proportion of 2-3c cells (arrow). Rb was below the threshold of detection in the 2-3 replicon cells. (E) Similar assays show that NS5B, visualized with a TRITC-labeled secondary antibody, is primarily localized to the perinuclear cytoplasm in 2-3 cells and not detectable in cured 2-3c cells.
NS5B is an RNA-dependent RNA polymerase and is essential for HCV RNA replication that occurs in the cytoplasm (17, 18). In contrast, Rb expression is predominantly nuclear (14). We confirmed by indirect immunofluorescence imaging that Rb had a primarily nuclear distribution in the cured 2-3c cells but was reduced to the point of nondetection in 2-3 replicon cells (Fig. 2D). Only in occasional cured cells was Rb also present within the cytoplasm (Fig. 2D, arrow). In contrast, NS5B localized primarily to the perinuclear cytoplasm in the replicon cells (Fig. 2E). Overall, these results suggest that NS5B interacts with Rb and likely targets it for degradation before its transport to the nucleus.
NS5B Active-Site Residues Are Involved in Binding and Regulating Rb Abundance. Most viral proteins that bind Rb share a common LxCxE motif (where x is variable) (14). We noted a conserved Leu-Val-(Cys/Asn)-Gly-Asp sequence in NS5B, (residues 314-318) with homology to the LxCxE motif (thus, an LxCxE homology motif or LH314-318) (Fig. 3A). Significantly, the sequence LxCxD can also function as an Rb-binding domain (19). The LH314-318 sequence is LVCGD in most strains of HCV, but LVNGD in HCV-N (from which the replicons we studied are derived) and a subset of other genotype 1b viruses. Interestingly, this otherwise-conserved sequence overlaps the GDD residues (residues 317-319) that are essential for the polymerase active site (20).
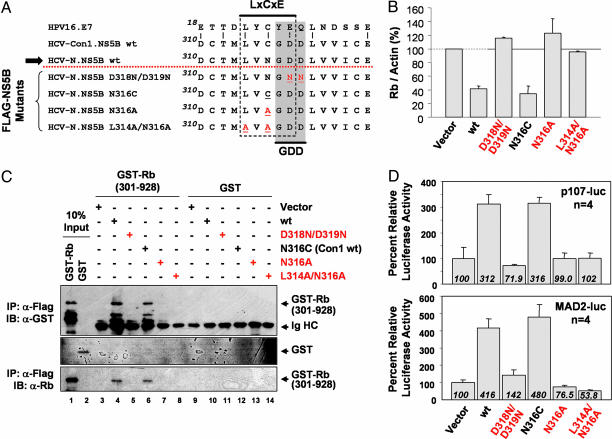
Fig. 3.
Mutations within the overlapping LH314-318 and GDD motifs of NS5B ablate regulation of Rb. (A) Alignment of the LxCxE domain of the HPV E7 protein and the LH314-318 motif in NS5B (residues 314-318) that overlaps the GDD sequence essential for polymerase activity. Shown above the dotted line are wt NS5B sequences of Con1 and HCV-N strains of HCV, and below the dotted line are the mutated HCV-N sequences constructed for this study. Mutations that disrupt the LxCxE motif are shown in red. The arrow indicates the sequence present in the replicons shown in Fig. 1A. (B) Quantitative analysis of Rb abundance normalized to actin levels in replicate immunoblots of extracts of cells expressing Flag-NS5B mutants. Data shown are means ± range (see also Fig. 8). (C) In vitro binding of Rb to wt and mutated NS5B. GST-Rb (301-928) that coimmunoprecipitated with Flag-tagged NS5B in Huh7 cell lysates was detected in immunoblots with an anti-GST (Top, B-14) or anti-Rb (Bottom, 4H1) antibody. Lanes 1 and 2 represent 10% of the GST-Rb (301-928) and GST input in the assays. Ig HC, Ig heavy chains. (D) Luciferase activity in normal Huh7 cells transfected with promoter reporter constructs in which luciferase expression is controlled by the human p107 (p107-luc, Upper) or MAD2 (MAD2-luc, Lower) promoters. Cells were cotransfected with wt NS5B, mutant NS5B, or empty expression vectors.
We conducted a mutational analysis to determine whether the LH314-318 motif contributes to the ability of NS5B to bind and negatively regulate Rb. We constructed a series of point mutations, designated D318N/D319N, N316C, N316A, and L314A/N316A, as depicted in Fig. 3A. The N316C substitution converts the LH314-318 motif of HCV-N to that of Con1 and most other strains of HCV, whereas the D318N/D319N, N316A, and L314A/N316A mutations disrupt the LH314-318 motif. We expressed these modified NS5B proteins in Huh7 cells, confirming equivalent expression levels of the wild-type (wt) and mutant proteins, and ascertained their ability to down-regulate Rb levels. Rb abundance was negatively regulated by both wt NS5B and N316C but little changed by L314A/N316A, D318N/D319N, or N316A (Fig. 3B; and see Fig. 7, which is published as supporting information on the PNAS web site). Thus, the LH314-318 motif appears to be essential for down-regulation of Rb in Huh7 cells.
We next compared the ability of wt and mutant NS5B proteins to bind Rb by using an in vitro binding assay. GST-Rb (301-928), a bacterially expressed fusion protein containing residues 301-928 of Rb (which includes the LxCxE-binding site) bound both wt and N316C variants but not the D318N/D319N, N316A, or L314A/N316A mutants with disrupted LH314-318 motifs (Fig. 3C). GST alone bound neither wt nor mutant NS5B. These results strongly suggest that the LH314-318 motif is important for Rb-binding activity, but they are surprising, however, because the crystal structure of the polymerase shows this site to be completely encircled and, thus, not accessible to Rb in the folded protein (20). If Rb were to interact with NS5B residues that contribute to the polymerase active site, we reasoned that Rb might negatively regulate its enzymatic activity. To test this hypothesis, we measured the polymerase activity of purified recombinant NS5B in the presence of Rb. We found that the addition of a ≈6-fold molar excess of a recombinant GST-Rb fusion protein resulted in a >50% inhibition of polymerase activity relative to that observed in the presence of a 26-fold molar excess of GST alone (see Fig. 8, which is published as supporting information on the PNAS web site). These results support an interaction between Rb and NS5B involving residues within the polymerase active site (see Discussion).
E2F-Responsive Promoters Are Activated by NS5B Expression. Because Rb is a corepressor of E2F transcription factors, we determined whether NS5B expression activates the expression of E2F-responsive genes. To make this determination, we used luciferase reporter constructs for the p107 and MAD2 promoters (p107-luc and MAD2-luc, respectively). p107, an Rb-family member, is expressed under the control of E2F (21), as is Mad2, a mitotic checkpoint protein (22). Ectopic expression of NS5B in Huh7 cells resulted in a 3- to 5-fold increase in the basal activity of both promoters (Fig. 3D). Control experiments using a p107 reporter mutated in each of two tandem E2F-binding sites (p107mt-luc) showed no NS5B-induced activation (data not shown), indicating that activation of the p107 promoter is E2F-specific. In separate experiments, when we cotransfected the wt p107 reporter plasmid with a vector expressing wt Rb, ectopic expression of NS5B enhanced promoter activity (Fig. 7B). However, NS5B did not activate the promoter when we coexpressed a mutant Rb (R661W) that does not bind E2F factors (23) (Fig. 7B). Together, these data suggest that NS5B reverses Rb-mediated repression of E2F-dependent transcription. Consistent with this notion, we found higher basal activity of the wt, but not the mutated p107 promoter, in 2-3 replicon cells than in cured 2-3c cells (Fig. 8C). Basal MAD2 promoter activity was also increased in 2-3 cells (data not shown).
To determine whether the regulation of E2F-responsive promoters by NS5B depends on the LH314-318 motif involved in Rb binding, we carried out similar promoter-activity assays using vectors expressing the NS5B mutants depicted in Fig. 3A. The N316C mutant (representing the LH314-318 sequence in Con1 and most other HCV strains) remained capable of activating E2F-responsive promoters but not other NS5B mutants in which the LH314-318 motif was disrupted (Fig. 3D). We conclude that NS5B reverses the Rb-mediated repression of E2F-dependent transcription in an LH314-318-motif-dependent fashion.
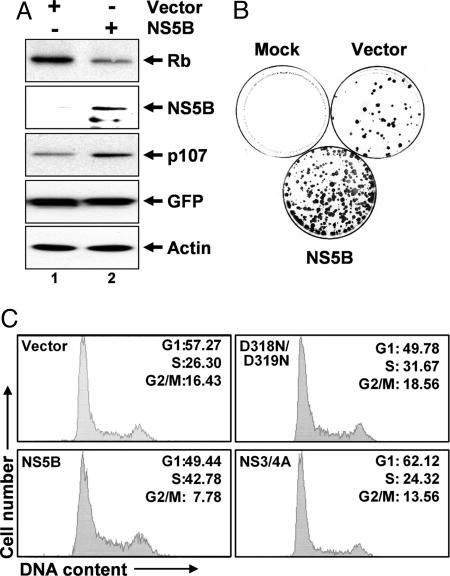
NS5B Regulates Cellular Proliferation and S-Phase Entry Through Its Interaction with Rb. Prior studies have failed to show any significant difference in the regulation of cell-cycle progression in the 2-3 replicon cells and their cured 2-3c counterparts (13). However, the Huh7 cells from which these cells were derived are known to contain a p53 mutation and express little p21 (24, 25). We, thus, assessed the influence of NS5B on cell-cycle progression in U2OS osteosarcoma cells that express wt p53 and Rb. First, we confirmed that ectopic expression of NS5B reduces the abundance of endogenous Rb in these cells (Fig. 4A). In contrast, p107 was up-regulated, suggesting activation of E2F-responsive genes. Colony-formation assays, in which we transfected U2OS cells with vectors expressing neomycin phosphotransferase (26), demonstrated a significantly greater number of cell colonies surviving G418 selection with simultaneous ectopic expression of NS5B (Fig. 4B; see also Fig. 9A, which is published as supporting information on the PNAS web site). This finding suggests that NS5B, by down-regulating Rb abundance and, thereby, activating E2F transcription factors, stimulates proliferation of U2OS cells and enhances the likelihood of transformation. Consistent with this interpretation, NS5B expression also resulted in a significant increase in the fraction of U2OS cells in S phase (Fig. 4C). This result was not observed with expression of the D318N/D319N mutant that does not bind Rb or with expression of the HCV NS3/4A protease. NS5B expression did not increase the S-phase fraction of Saos-2 osteosarcoma cells, which express a functionally inactive Rb (27) (Fig. 9B). Thus, NS5B appears to stimulate cell-cycle progression in an Rb-specific fashion, consistent with the activation of E2F transcription factors described above.
Fig. 4.
NS5B stimulates cell proliferation and S-phase entry. (A) Ectopic expression of NS5B down-regulates Rb in U2OS cells. Shown are immunoblots of Rb, p107, and NS5B in extracts of cells transfected with NS5B and empty expression vectors. GFP and actin were controls for transfection efficiency and loading, respectively. (B) Colony-formation assays of U2OS cells transfected with equal amounts of NS5B or empty expression vectors. Mock, mock transfected. (C) NS5B expression mediates an increase in the S-phase fraction of U2OS cells. Cells were transfected with vectors expressing wt NS5B, the D318N/D319N mutant that does not bind Rb, NS3/4A, or empty vector in addition to a GFP expression vector. GFP-positive cells were analyzed for cellular DNA content by flow cytometry. Percentages of cells in various phases of the cell cycle are indicated. Results are representative of two independent experiments.
Discussion
We have shown here that the abundance of the Rb tumor-suppressor protein is negatively regulated posttranslationally in Huh7 hepatoma cells supporting the replication of genome-length and subgenomic HCV RNAs derived from an infectious molecular clone of HCV. Elimination of the replicating RNA and the proteins it expresses by prior treatment with either IFN or a small-molecule NS3/4A protease inhibitor resulted in an increase in endogenous Rb abundance. We observed this HCV-specific down-regulation of Rb abundance in multiple replicon cell lines, indicating that it is not restricted to a particular clone of Huh7 cells. We have also documented this down-regulation in Huh7 cells containing a genome-length genotype 2a JFH-1 replicon (T.M. and S.M.L., unpublished data), indicating that Rb is negatively regulated by different strains of HCV that are widely divergent genetically. These findings differ significantly from those reported by Tsukiyama-Kohara et al. (28), who described an initially positive then negative effect on Rb after expression of HCV proteins in HepG2 cells. We found that the negative regulation of Rb abundance by replicating HCV RNA is due to the NS5B RNA-dependent RNA polymerase. Our data suggest that the polymerase forms a complex with Rb, thereby accelerating its degradation, activating E2F-responsive promoters, and stimulating cellular proliferation.
Several mutations involving residues within the GDD active-site motif of NS5B or the overlapping, immediately adjacent LH314-318 domain eliminated or severely reduced the ability of the polymerase to bind Rb and regulate its abundance (Fig. 3). This is a very surprising finding, because the residues of the LH314-318 motif are partially buried within the palm domain of the polymerase (20). Furthermore, crystallographic studies suggest that access to these residues is severely restricted by extensive interactions between the finger and thumb subdomains. Thus, if the LH314-318 motif of NS5B does, in fact, mediate a direct interaction with Rb in a manner analogous to the LxCxE pocket-binding domains of other Rb-binding proteins (14), as our data suggest, this interaction is likely to occur before the complete folding of NS5B or only after a very significant structural rearrangement of the polymerase. Further investigation will be required to distinguish between these possibilities. However, the fact that recombinant Rb specifically impairs the enzymatic activity of NS5B in a cell-free polymerase assay (Fig. 8) suggests that this interaction may occur even with a fully prefolded polymerase.
The down-regulation of Rb abundance by NS5B may favorably influence the cellular environment for HCV replication. We have shown that HCV RNA synthesis is stimulated during the S phase of the cell cycle (13). Moreover, compared with cap-dependent translation, the cap-independent translation of HCV RNA is enhanced in actively growing cells and reduced in resting cells (29). Thus, the stimulus to cell proliferation provided by down-regulation of Rb would be expected to enhance the replication of HCV, whereas increases in Rb expression would have the reverse effect. Although preliminary studies suggest that knock-down of Rb by RNA interference may enhance HCV RNA replication in Huh7 cells (T.M., K.L., and S.M.L., unpublished data), further studies are needed to determine whether this finding reflects the pleiotrophic effects of Rb on cellular proliferation or a reduction in Rb-mediated inhibition of the enzymatic function of NS5B.
Rb can be functionally inactivated in several ways (8). Our data are consistent with a mechanism in which NS5B negatively regulates Rb by targeting it for degradation, because we have shown that the stability of Rb is reduced in cells supporting HCV RNA replication. Supporting this hypothesis, we have observed increases in the abundance of Rb after treatment of replicon cells with proteasome inhibitors (T.M. and S.M.L., unpublished data). The human papilloma virus (HPV) E7 oncoprotein targets Rb through a similar mechanism, binding Rb through an LxCxE motif and resulting in destabilization of Rb in a 26S-proteasome-dependent manner (30). Like NS5B, E7 acts through degradation of Rb to promote cell proliferation that is favorable to viral replication. NS5B may also contribute to carcinogenesis through its ability to target Rb for degradation, again, in a fashion analogous to HPV E7. There is no potential for integration of HCV genes into host-cell chromosomes nor does there seem to be continued high-level expression of the Rb-binding protein leading directly to cellular transformation, as in HPV infection. However, the finding that NS5B negatively regulates steady-state levels of the Rb tumor suppressor suggests a unique molecular mechanism by which HCV infection might promote hepatocellular proliferation and subsequent development of liver cancer. These results also reveal how RNA viruses, in general, might mimic some features of well characterized DNA tumor viruses in causing cancer.
Supplementary Material
Acknowledgments
We thank Drs. C. Lesburg, B. Malcolm, and J. Jaeger for comments on the structure of NS5B and members of the Lemon laboratory for helpful discussions. This work was supported by National Institutes of Health Grants U19-AI40035 and R21-DA018054. K.L. is the John Mitchell Hemophilia of Georgia Liver Scholar of the American Liver Foundation.
Author contributions: T.M., Y.L., K.L., and S.M.L. designed research; T.M., M.N., and Y.L. performed research; M.N. and K.L. contributed new reagents/analytic tools; T.M., M.N., Y.L., K.L., and S.M.L. analyzed data; and T.M. and S.M.L. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HCV, hepatitis C virus; Rb, retinoblastoma tumor-suppressor protein; wt, wild type.
References
- 1.Alter, M. J., Mast, E. E., Moyer, L. A. & Margolis, H. S. (1998) Infect. Dis. Clin. North Am. 12, 13-26. [DOI] [PubMed] [Google Scholar]
- 2.Saito, I., Miyamura, T., Ohbayashi, A., Harada, H., Katayama, T., Kikuchi, S., Watanabe, Y., Koi, S., Onji, M., Ohta, Y., et al. (1991) Proc. Natl. Acad. Sci. USA 87, 6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gosert, R., Egger, D., Lohmann, V., Bartenschlager, R., Blum, H. E., Bienz, K. & Moradpour, D. (2003) J. Virol. 77, 5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moriya, K., Fujie, H., Shintani, Y., Yotsuyanagi, H., Tsutsumi, T., Ishibashi, K., Matsuura, Y., Kimura, S., Miyamura, T. & Koike, K. (1998) Nat. Med. 4, 1065-1067. [DOI] [PubMed] [Google Scholar]
- 5.Lerat, H., Honda, M., Beard, M. R., Loesch, K., Sun, J., Yang, Y., Okuda, M., Gosert, R., Xiao, S. Y., Weinman, S. A., et al. (2002) Gastroenterology 122, 352-365. [DOI] [PubMed] [Google Scholar]
- 6.Edamoto, Y., Hara, A., Biernat, W., Terracciano, L., Cathomas, G., Riehle, H. M., Matsuda, M., Fujii, H., Scoazec, J. Y. & Ohgaki, H. (2003) Int. J. Cancer 106, 334-341. [DOI] [PubMed] [Google Scholar]
- 7.Chellappan, S. P., Hiebert, S., Mudryj, M., Horowitz, J. M. & Nevins, J. R. (1991) Cell 65, 1053-1061. [DOI] [PubMed] [Google Scholar]
- 8.Chau, B. N. & Wang, J. Y. (2003) Nat. Rev. Cancer 3, 130-138. [DOI] [PubMed] [Google Scholar]
- 9.Whyte, P., Buchkovich, K. J., Horowitz, J. M., Friend, S. H., Raybuck, M., Weinberg, R. A. & Harlow, E. (1988) Nature 334, 124-129. [DOI] [PubMed] [Google Scholar]
- 10.Dyson, N., Howley, P. M., Munger, K. & Harlow, E. (1989) Science 243, 934-937. [DOI] [PubMed] [Google Scholar]
- 11.Lee, J. O., Russo, A. A. & Pavletich, N. P. (1998) Nature 391, 859-865. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda, M., Yi, M., Li, K. & Lemon, S. M. (2002) J. Virol. 76, 2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholle, F., Li, K., Bodola, F., Ikeda, M., Luxon, B. A. & Lemon, S. M. (2004) J. Virol. 78, 1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Classon, M. & Harlow, E. (2002) Nat. Rev. Cancer 2, 910-917. [DOI] [PubMed] [Google Scholar]
- 15.Foy, E., Li, K., Wang, C., Sumter, R., Ikeda, M., Lemon, S. M. & Gale, M., Jr. (2003) Science 300, 1145-1148. [DOI] [PubMed] [Google Scholar]
- 16.Shan, B., Chang, C. Y., Jones, D. & Lee, W. H. (1994) Mol. Cell. Biol. 14, 299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolykhalov, A. A., Mihalik, K., Feinstone, S. M. & Rice, C. M. (2000) J. Virol. 74, 2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivashkina, N., Wolk, B., Lohmann, V., Bartenschlager, R., Blum, H. E., Penin, F. & Moradpour, D. (2002) J. Virol. 76, 13088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalejta, R. F., Bechtel, J. T. & Shenk, T. (2003) Mol. Cell. Biol. 23, 1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesburg, C. A., Cable, M. B., Ferrari, E., Hong, Z., Mannarino, A. F. & Weber, P. C. (1999) Nat. Struct. Biol. 6, 937-943. [DOI] [PubMed] [Google Scholar]
- 21.Zhu, L., Zhu, L., Xie, E. & Chang, L. S. (1995) Mol. Cell. Biol. 15, 3552-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernando, E., Nahle, Z., Juan, G., Diaz-Rodriguez, E., Alaminos, M., Hemann, M., Michel, L., Mittal, V., Gerald, W., Benezra, R., et al. (2004) Nature 430, 797-802. [DOI] [PubMed] [Google Scholar]
- 23.Whitaker, L. L., Su, H., Baskaran, R., Knudsen, E. S. & Wang, J. Y. (1998) Mol. Cell. Biol. 18, 4032-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bressac, B., Galvin, K. M., Liang, T. J., Isselbacher, K. J., Wands, J. R. & Ozturk, M. (1990) Proc. Natl. Acad. Sci. USA 87, 1973-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koga, H., Sakisaka, S., Harada, M., Takagi, T., Hanada, S., Taniguchi, E., Kawaguchi, T., Sasatomi, K., Kimura, R., Hashimoto, O., et al. (2001) Hepatology 33, 1087-1097. [DOI] [PubMed] [Google Scholar]
- 26.Aprelikova, O. N., Fang, B. S., Meissner, E. G., Cotter, S., Campbell, M., Kuthiala, A., Bessho, M., Jensen, R. A. & Liu, E. T. (1999) Proc. Natl. Acad. Sci. USA 96, 11866-11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shew, J. Y., Lin, B. T., Chen, P. L., Tseng, B. Y., Yang-Feng, T. L. & Lee, W. H. (1990) Proc. Natl. Acad. Sci. USA 87, 6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukiyama-Kohara, K., Tone, S., Maruyama, I., Inoue, K., Katsume, A., Nuriya, H., Ohmori, H., Ohkawa, J., Taira, K., Hoshikawa, Y., et al. (2004) J. Biol. Chem. 279, 14531-14541. [DOI] [PubMed] [Google Scholar]
- 29.Honda, M., Kaneko, S., Matsushita, E., Kobayashi, K., Abell, G. A. & Lemon, S. M. (2000) Gastroenterology 118, 152-162. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez, S. L., Stremlau, M., He, X., Basile, J. R. & Munger, K. (2001) J. Virol. 75, 7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.