Abstract
Light provides a major source of information from the environment during plant growth and development. Light perception is mediated through the action of several photoreceptors, including the phytochromes. Recent results demonstrate that light responses involve the regulation of several thousand genes. Some of the key events controlling this gene expression are the translocation of the phytochrome photoreceptors into the nucleus followed by their binding to transcription factors. Coupled with these events, the degradation of positively acting intermediates appears to be an important process whereby photomorphogenesis is repressed in darkness. This review summarizes our current knowledge of these processes.
Introduction
Plants utilize light as a source of energy and as a source of information about their environment. Dark-grown (etiolated) seedlings display an apical hook, closed and unexpanded cotyledons and elongated hypocotyls. This developmental programme (known as skotomorphogenesis) is necessary for newly germinating seedlings to grow through soil or fallen leaves to reach the light. Upon light exposure, seedlings undergo de-etiolation: cotyledons open, expand and begin to photosynthesize, hypocotyl elongation is inhibited and cell differentiation is initiated in vegetative meristems. These events are known as photomorphogenesis and result largely from light-mediated alterations in gene expression (Ma et al., 2001; Tepperman et al., 2001; Schroeder et al., 2002).
The phytochromes
In plants, light-dependent responses are controlled by a series of photoreceptors that can be classified into three known groups—the phytochromes, cryptochromes and phototropins (Quail, 2002a).
Phytochromes are typically encoded by small multigene families, e.g. PHYA-PHYE in Arabidopsis (Møller et al., 2002; Nagy and Schäfer, 2002; Quail, 2002a,b). Each forms a homodimer of ∼240 kDa and light sensitivity is conferred by the presence of a tetrapyrrole chromophore covalently bound to the N-terminal half of each monomer (Montgomery and Lagarias, 2002). Dimerization domains are located within the C-terminal half of the proteins, as are other domains involved in the activation of signal transduction (Quail et al., 1995; Quail, 2002a). Each phytochrome can exist in two photointerconvertible conformations, denoted Pr (a red light-absorbing form) and Pfr (a far red light-absorbing form) (Figure 1A). Because sunlight is enriched in red light (compared with far red light), phytochrome is predominantly in the Pfr form in the light, and this can convert back to the Pr form during periods of darkness by a process known as dark reversion. Photoconversion back to Pr can also be mediated by pulses of far red light.
Figure 1.
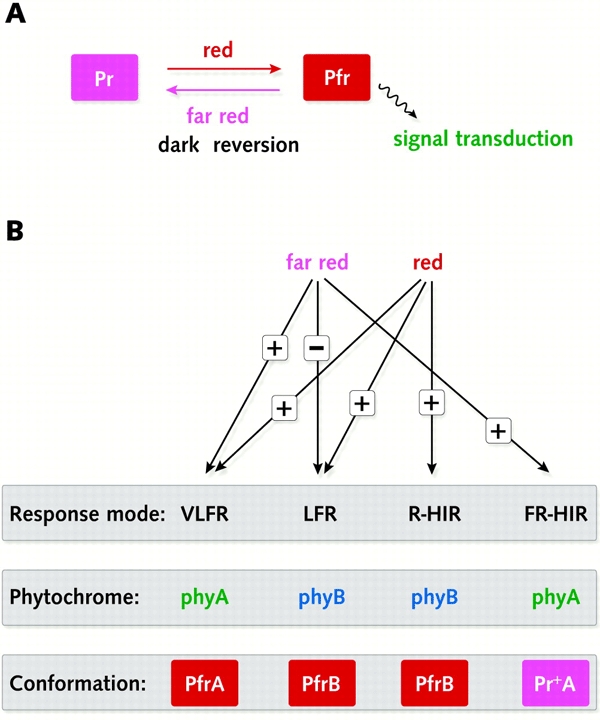
Phytochrome response modes. (A) The phytochrome photocycle. Pr and Pfr denote the red and far red light-absorbing conformations of phytochrome, respectively, which are reversible depending on light conditions. Pfr can also be converted to Pr in a light-independent process known as dark reversion (Nagy and Schäfer, 2002). (B) The different phytochrome response modes. The influence of red and far red light on each response mode is shown, together with the phytochrome principally involved in initiating the response.
In dark-grown seedlings, phyA is the most abundant phytochrome, although it is rapidly degraded upon exposure to light (Møller et al., 2002; Nagy and Schäfer, 2002). Expression of the PHYA gene is also repressed in the light. The other PHY genes are expressed at much lower levels and their expression is not strongly influenced by light. phyB is more abundant than phyC–E, but its mRNA levels are typically only ∼1% of those of phyA mRNA in the dark (Quail et al., 1995). In sunlight, however, phyB is the most abundant phytochrome due to phyA degradation. The physiological functions of phyA and phyB are the best characterized (see below), and only recently has some information been obtained about the roles of phyC–E (Møller et al., 2002).
Classically, phytochrome responses have been defined by their wavelength and fluence or fluence-rate (i.e. intensity) requirements into three groups—very low fluence responses (VLFR), low fluence responses (LFR) and high irradiance responses (HIR) (Figure 1B). HIRs are now further subdivided into red (R)- and far red (FR)-HIRs (Nagy and Schäfer, 2002). Studies with phytochrome-deficient mutants grown in defined VLFR, LFR and HIR light environments have revealed that specific phytochromes can be ascribed to individual responses (Figure 1B), although significant redundancy exists (Møller et al., 2002; Nagy and Schäfer, 2002). FR-HIR responses are, however, specifically mediated by phyA due to the unusual spectral properties of this phytochrome. It has been proposed that a novel form of PrA (denoted Pr+A), which has photocycled through Pfr, is responsible (Shinomura et al., 2000).
Phytochrome localization
In the dark, de novo synthesized phytochrome is in the Pr form and is localized within the cytoplasm. Upon conversion to Pfr each of the Arabidopsis phytochromes have been found to translocate to the nucleus and to form discrete speckles (Kircher et al., 2002; Nagy and Schäfer, 2002). The physiological significance of these observations can be inferred, at least for phyA and phyB, from the correlations that have been found between nuclear translocation and response, e.g. phyB localizes to the nucleus in R-HIR conditions whereas phyA nuclear localization is most effective in FR-HIR conditions.
Phytochrome localization to the nucleus is a highly significant finding given that many phytochrome responses are dependent upon changes in gene expression. However, it should be noted that phytochrome translocation is rather slow, except for phyA, and that the majority of the intracellular Pfr pool is not translocated to the nucleus (Nagy and Schäfer, 2002). These and other observations (see below) suggest that phytochromes may activate signalling pathways in both the cytoplasm and the nucleus.
Dissection of phytochrome signal transduction pathways
Photomorphogenic mutants. Many putative light signal transduction intermediates have been identified from mutant screens aimed principally at isolating mutants insensitive to light or mutants displaying constitutive photomorphogenesis in darkness (Møller et al., 2002). The most severe white light-insensitive mutants include photoreceptor mutants, as well as one mutated in a gene encoding a bZIP transcription factor known as HY5 (Oyama et al., 1997). The severity of this mutant demonstrates that HY5 plays a key role in the control of photomorphogenesis (see below). While hy5 mutants are affected in both phyA and phyB signalling, other mutants are affected in one or the other, the majority being specific for phyA (Møller et al., 2002; Nagy and Schäfer, 2002; Quail, 2002a).
Most phyA signalling mutants are defective in genes encoding nuclear-localized proteins, e.g. FHY1, FHY3, SPA1, FAR1, LAF1 and EID1 (see Møller et al., 2002; Nagy and Schäfer, 2002; Quail, 2002a,b; Wang and Deng, 2002). However, some have mutations in genes encoding the cytoplasmic proteins PAT1, FIN219 and SUB1. Genetic analyses suggest that, apart from SPA1, EID1 and SUB1, all of these proteins act as positive elements in the pathway, i.e. the mutants display reduced sensitivity to far red light (Nagy and Schäfer, 2002). Interestingly, SPA1 and EID1 play distinct roles in phyA signalling, SPA1 being primarily involved in VLFR and EIDI in FR-HIR signalling (Zhou et al., 2002).
Constitutively photomorphogenic mutants are defective in genes encoding the COP/DET/FUS family of proteins. Eleven loci have been assigned to this group (det1, cop1, cop8, cop9, cop10, cop11, cop16, fus5, fus8, fus11 and fus12) (Hardtke and Deng, 2000). The phenotypes of these mutants indicate that these genes encode negative regulators of light signalling, all of which are localized predominantly within the nucleus (see below).
Phytochrome-interacting proteins. Attempts to identify phytochrome-interacting partners have been most successful using the yeast two-hybrid approach. Due to the difficulties of generating a chromophore-reconstituted phytochrome in yeast cells, most yeast two-hybrid experiments have used the C-terminal region. This is far from ideal because the C-termini of phyA and phyB have been shown to be functionally interchangeable (Quail et al., 1995; Quail, 2002a,b), and because the absence of the N-terminal chromophore-binding domain presumably creates a rather artificial bait. Nevertheless, a number of proteins have been identified (Table 1). In some cases, these proteins interact with different domains within the phytochrome, but whether they can interact simultaneously has not yet been addressed. As with the genetic approaches, these two-hybrid screens have identified mostly nuclear-localized proteins (see below).
Table 1.
Phytochrome-interacting proteins
| Interacting partner | Method | Phy A or B? | Pfr or Pr? | Cellular localization | Proposed role in phy signalling | Reference |
|---|---|---|---|---|---|---|
| ARR4 | Y2H Pull-down Co-IP | B | Both | N, C | Positive regulator of B | Sweere et al. (2001) |
| Aux/IAA | Pull-down | A | Both | N | Cross-talk with auxin | Colón-Carmona et al. (2000) |
| Cry1 | Y2H Pull-down | A | ? | N | Photoreceptor co-action | Ahmad et al. (1998) |
| Cry2 | Co-IP FRET | B | ? | N | Photoreceptor co-action | Màs et al. (2000) |
| ELF3 | Y2H Pull-down | B | Both | N | Positive regulator of B in circadian clock | Liu et al. (2001) |
| NDPK2 | Y2H Pull-down | A | Pfr>Pr | C, N | Positive regulator of A and B | Choi et al. (1999) |
| PIF3 | Y2H Pull-down | B(A) | Pfr | N | Positive regulator of B (and A) | Ni et al. (1999) |
| PIF4 | Pull-down | B | Pfr | N | Negative regulator of B | Huq and Quail (2002) |
| PKS1 | Y2H Pull-down | A, B | Both | C | Negative regulator of B | Fankhauser et al. (1999) |
| ZTL/ADO1 | Y2H | B | ? | N, C | Positive regulator of B and cry1 in circadian clock | Jarillo et al. (2001) |
Abbreviations: Y2H, yeast two-hybrid; Co-IP, co-immunoprecipitation; FRET, fluorescence resonance energy transfer; N, nucleus; C, cytoplasm.
In all screens for phytochrome-interacting molecules it should be kept in mind that phytochromes (especially phyA) are very sticky and can interact non-specifically with a range of proteins and RNA in vitro (Quail, 1994). Therefore, the physiological relevance of an interaction should be confirmed both biochemically in planta and by genetic approaches. Although only a few interactions have so far been demonstrated in vivo (e.g. Màs et al., 2000; Sweere et al., 2001), circumstantial evidence does indicate that many of these proteins are indeed involved in phytochrome signalling, e.g. plants mutated in the relevant genes have light-insensitive or hypersensitive phenotypes in a particular light regime (e.g. Fankhauser et al., 1999).
Phytochrome signalling in the cytoplasm
The vast majority of phytochrome-interacting proteins are localized to the nucleus (Table 1). The only partner that is constitutively cytoplasmic is PKS1, a protein of unknown function (Fankhauser et al., 1999).
Interestingly, PKS1 can be phosphorylated by oat phyA. The C-terminal half of phytochrome, in fact, contains two regions with similarity to bacterial histidine kinases (Quail, 2002a,b). This may be significant because bacterial phytochromes function as true sensory histidine kinases, which relay a phosphogroup to an aspartate residue of a response regulator that is a transcriptional activator (Montgomery and Lagarias, 2002). However, the only plant phytochrome that has been demonstrated to have protein kinase activity to date is oat phyA. Furthermore, phyA autophosphorylation and the phosphorylation of PKS1 occur on Ser/Thr residues (Fankhauser et al., 1999; Møller et al., 2002) and mutant analysis has not yet elucidated the significance of these reactions (Krall and Reed, 2000). A loss-of-function pks1 mutant shows enhanced responsiveness to red light, implying that it may negatively regulate phyB signalling (Fankhauser et al., 1999). Given that phyB does not appear to have protein kinase activity, these results are difficult to reconcile. Nonetheless, it has been proposed that the function of PKS1 may be to negatively regulate phytochrome nuclear translocation, perhaps by anchoring it within the cytoplasm (Fankhauser et al., 1999).
Other evidence for cytoplasmically-localized phytochrome signalling events comes from biochemical and pharmacological studies that have implicated the involvement of G-proteins, cGMP, calcium and calmodulin in the control of phytochrome-dependent gene expression (e.g. Shacklock et al., 1992; Bowler et al., 1994). Reverse genetics approaches have subsequently provided further support for the involvement of G-proteins (Okamoto et al., 2001). Conversely, a role for calcium in light signalling has been reinforced by the identification of SUB1, a cytoplasmically-localized calcium-binding protein that appears to negatively regulate cryptochrome and phyA responses (Guo et al., 2001).
Phytochrome signalling in the nucleus
Phytochrome-interacting partners. Because most phytochrome-interacting proteins localize to the nucleus (Table 1), the majority of recent work on phytochrome signalling has focused on events within this compartment. In particular, phytochrome-interacting factor 3 (PIF3) has been intensively studied (Quail, 2002a,b). This protein is a basic helix–loop–helix (bHLH) transcription factor that has been shown to bind G-boxes (Martinez-Garcia et al., 2000), functionally important cis-elements within the promoters of some light-regulated genes (Quail, 2000). This implies that the signalling pathway between nuclear-localized phytochrome and transcriptional regulation is very short. Furthermore, the phytochrome/PIF3 interaction is red/far red light-reversible, and only occurs when phytochrome is in the Pfr form (Ni et al., 1999). Modulation of PIF3 activity in Arabidopsis suggests that it is preferentially (although not only) a positive regulator of phyB signalling (Quail, 2002a,b). However, the weak phenotypes observed in these plants indicate that PIF3 is responsible for mediating only a subset of phytochrome responses.
PIF3 can bind to the G-boxes within the promoters of the LHY and CCA1 genes (Martinez-Garcia et al., 2000), which encode Myb transcription factors that are thought to play key roles in the control of circadian rhythms. This suggests that PIF3 may provide a link between phyB and the circadian clock, rather than being central to the control of acute light responses.
HFR1 is another bHLH protein initially identified in a screen for mutants insensitive to far red light (Fairchild et al., 2000). Although HFR1 does not bind directly to phytochrome, it can dimerize with PIF3 in yeast (Fairchild et al., 2000). Furthermore, the observation that HFR1 mRNA is 30-fold more abundant in plants exposed to far red light than those in red light may explain the specificity of this factor for phyA signalling. The fact that plants contain several bHLH proteins could provide PIF3 with a range of interacting proteins to fine-tune phytochrome responses (Quail, 2002b). This hypothesis has been supported by the identification of another bHLH protein, PIF4, which can also bind directly to phytochrome (Huq and Quail, 2002).
The extreme N-terminal fragment of phyB has been shown to interact with a nuclear protein with similarity to response regulators (denoted ARR4) (Sweere et al., 2001). ARR4 also binds to phyB in vivo and stabilizes it in the physiologically active Pfr form, thereby increasing its responsiveness. This has also been confirmed by in vivo spectroscopic data, which revealed a decrease in the rate of rapid dark reversion of phyB to the Pr form (Nagy and Schäfer, 2002). ARR4-overexpressing lines indeed show enhanced light responsiveness, and an ARR4 knockout line showed reduced responsiveness to red light (M. Lexa and K. Harter, unpublished data). However, in spite of its similarities to bacterial response regulators, there is no evidence that ARR4 is phosphorylated by phytochrome, although a point mutation that prevents its phosphorylation (D95N) has a dominant-negative phenotype in transgenic seedlings (V. Rodado, K. Harter and E. Schäfer, unpublished data).
Finally, several lines of evidence indicate that phyA and phyB can interact with the cryptochromes cry1 and cry2 (Ahmad et al., 1998; Màs et al., 2000). Like the phytochromes, both cry1 and cry2 appear to be localized to the nucleus in the light (Christie and Briggs, 2001), and cry2 is phosphorylated in blue light (Shalitin et al., 2002). This physical interaction under conditions in which both photoreceptors are capable of signalling is important considering the physiological evidence of cooperation between phytochrome and cryptochromes in some light conditions (Nagy and Schäfer, 2002).
Downstream events. Genetic approaches have implicated a range of nuclear-localized proteins downstream of phytochrome and its physically-interacting partners that are involved in phytochrome signalling. Some of these are now quite well characterized, most notably the COP9 signalosome, COP1, and HY5.
The group of COP/FUS proteins that are now known to comprise the COP9 signalosome (CSN) were initially identified in mutant screens for constitutive photomorphogenic phenotypes (see above) (Hardtke and Deng, 2000). In fact, in 8 of the 11 cop/det/fus mutants, the COP9 signalosome is absent. Sequence analysis of the core components and associated proteins suggests an evolutionary relationship with the lid subcomplex of the 19S regulatory particle of the 26S proteasome, which degrades polyubiquitinated proteins (Hardtke and Deng, 2000). This finding suggests that the COP9 signalosome could be involved in the degradation of a select set of substrate proteins by functioning as an alternative lid subcomplex of the 19S regulatory particle (Hardtke and Deng, 2000). Such substrates could include both positive regulators of photomorphogenesis in the dark and negative regulators in the light, although there is currently no evidence for the latter. Rather, the pleiotropic phenotypes of mutants lacking the COP9 signalosome suggest a multifaceted role in controlling plant development, which is also supported by recent findings linking it to auxin responses and pathogen defence (Schwechheimer et al., 2001; Azevedo et al., 2002; Hellmann and Estelle, 2002).
The COP1 and COP10 proteins are not intrinsically associated with the COP9 signalosome but also appear to play a role in regulating protein degradation (Hardtke and Deng, 2000; Hellmann and Estelle, 2002; Suzuki et al., 2002). COP10 resembles a ubiquitin-conjugating E2 enzyme (Suzuki et al., 2002), whereas COP1 has been proposed to be an E3 ubiquitin ligase containing several recognizable domains, such as a RING-finger zinc-binding domain, a coiled-coil domain and a WD-40 repeat motif (Hardtke and Deng, 2000). The total amount of cellular COP1 protein is not affected by light, although in darkness the protein is predominantly localized in the nucleus, whereas light causes its slow redistribution into large cytoplasmic aggregates (Hardtke and Deng, 2000).
Further studies have demonstrated that the COP1 protein interacts directly with the HY5 protein (Ang et al., 1998). HY5 encodes a constitutively nuclear bZIP transcription factor that is thought to positively regulate photomorphogenesis by binding to G-boxes within the promoters of light-inducible genes (Hardtke and Deng, 2000). Removal of the COP1-interacting domain of HY5 results in exaggerated photoresponsiveness when this mutant form is overexpressed in transgenic plants. This suggests that COP1 could negatively regulate photomorphogenesis by mediating the polyubiquitination of HY5, thus marking it for subsequent degradation by the proteasome (Osterlund et al., 2000). The homology of the COP9 signalosome and the proteasome subcomplex makes it an obvious candidate for mediating this degradation (Hellmann and Estelle, 2002), a hypothesis that is supported by the fact that null mutations within COP9 signalosome components prevent HY5 degradation (Hardtke and Deng, 2000). COP10 may also be involved in HY5 degradation (Suzuki et al., 2002).
COP1 can also interact with a number of other proteins (Hardtke and Deng, 2000) including putative transcription factors (e.g. Holm et al., 2002), and is thought to regulate their activity in a similar way to HY5. Furthermore, COP1 has recently been found to bind both cry1 and cry2 (Wang et al., 2001; Yang et al., 2001), which may be instrumental in the release of HY5 from COP1 and the subsequent activation of light-inducible genes.
Further evidence for the importance of protein degradation in phyA signalling has been provided by the eid1 mutant, which displays an extremely enhanced sensitivity to FR-HIR (Dieterle et al., 2001). EID1 encodes a nuclear localized F-box protein, which probably acts by targeting activated components of the phyA signalling pathway to the ubiquitin-dependent degradation pathway (Dieterle et al., 2001; Hellmann and Estelle, 2002). Whether COP1, COP10, or the COP9 signalosome can directly or indirectly interact with EID1 has not yet been established. Although it has been known for several decades that activated phyA is degraded in a ubiquitin-dependent process, it is curious that there is as yet no evidence that any of the proteasome-related components described above are involved.
A further level of regulation may be mediated by the DET1 protein. Like cop/fus mutants, plants defective in DET1 display constitutive de-etiolation in darkness, implying that DET1 also plays a key role in the repression of light-inducible genes (Quail, 2002a). DET1 does not appear to participate in the regulation of proteolysis but rather binds to nucleosome core particles via an interaction with the N-terminal tail of histone H2B (Benvenuto et al., 2002). Furthermore, DET1 is also part of a complex that contains UV–DDB1, which in animal cells is part of histone acetyltransferase complexes (Schroeder et al., 2002). The significance of these findings is reinforced by the observation that DET1 binds preferentially to non-acetylated H2B tails (Benvenuto et al., 2002), suggesting that it binds to the nucleosomes of light-inducible genes and is subsequently displaced by a light-dependent acetylation of the histone tails, thus permitting gene expression. The phenotypes of the cop/det/fus mutants suggest that chromatin remodelling is equally important in controlling light-dependent gene expression as protein degradation, although the links between the two processes have yet to be identified.
Concluding remarks
From the data available, a model for phytochrome signalling has been presented in Figure 2. Key aspects of the model are: (i) phytochrome can initiate signal transduction from both the nucleus and the cytoplasm; (ii) activated nuclear-localized phytochromes can interact with bHLH transcription family members and thereby rapidly activate transcription; (iii) response regulators may represent an ancestrally-inherited mechanism whereby the Pfr form is stabilized in the nucleus; (iv) HY5 is a key transcription factor that activates light-responsive genes; (v) proteasome-dependent degradation of positively acting regulatory factors represses light-responsive gene expression in the dark; (vi) phytochromes and cryptochromes directly interact to regulate co-action of these two classes of photoreceptors; (vii) chromatin remodelling via DET1/DDB1 provides epigenetic control of light-responsive gene expression; and (viii) light-regulated trancriptional control converges most frequently on G-boxes.
Figure 2.
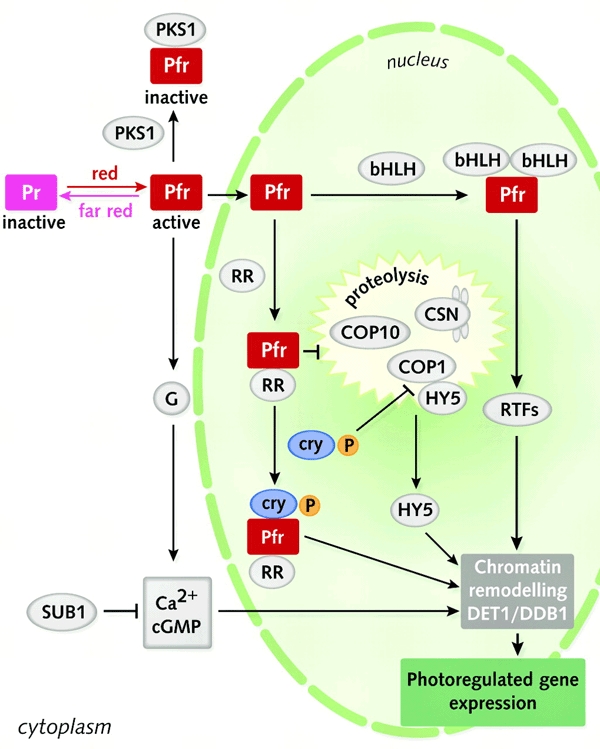
A model of phytochrome signal transduction. Activated phytochrome (Pfr) is proposed to regulate transcription through several parallel pathways. A rapid response involves Pfr translocation to the nucleus, where it binds transcription factors of the bHLH family (in particular PIF3). Key regulatory transcription factors (RTFs) that are responsible for inducing a range of light-regulated genes are subsequently activated. In a second nuclear-localized pathway, phytochromes are proposed to bind response regulators (RR), which stabilize them in the activated form and can induce light-regulated gene expression by inhibiting COP1-, COP10- and CSN-dependent proteolysis of the HY5 transcription factor and by binding to activated cryptochromes (cry). In all cases, regulation of the genes responsible for photomorphogenesis is predicted to require chromatin remodelling mediated by the DET1/DDB1 nucleosome-binding complex. In the cytoplasm, phytochrome may activate gene expression through G-proteins (G), calcium and cGMP-dependent pathways, which are regulated by SUB1. In addition, phytochromes may be sequestered away from the signalling-competent pool by PKS1. Elements involved in signalling from specific photoreceptors or controlling specific responses have not been included.
These different regulatory mechanisms probably act in parallel pathways that provide ample opportunities for cross-talk between them and with other inputs, e.g. with cytokinin signalling via response regulators (Hwang and Sheen, 2001) and with auxin signalling via ubiquitin-dependent reactions (Schwechheimer et al., 2001; Hellmann and Estelle, 2002). However, the subcellular localization and timing in each pathway is likely to vary. For example, genomic expression profiles have shown that a large percentage of genes induced early following light stimulation encode transcription factors (Tepperman et al., 2001; Quail, 2002a,b). These results suggest that phytochromes initially activate a set of key transcription factors that in turn induce downstream target genes.
In conclusion, it is possible that the key events regulating phytochrome-mediated photoperception and signal transduction have now been elucidated. However, the complexity of these responses indicates that an enormous amount of work is still required to understand how they function together and how specific subsets of responses are controlled. This will necessitate not only whole genome expression profiling in different mutants, but will also require information on how cellular responses are integrated into the whole plant context over time. Because phytochrome signalling has been so well studied, it is likely to provide breakthrough technologies for addressing these issues and will continue to attract the interest of plant scientists for the foreseeable future.


Acknowledgments
We apologize that many of the original research articles could not be included owing to space restrictions. Research on phytochrome signal transduction in our laboratories is currently supported by the European Commission (QLK5-CT-2000-00357) (C.B. and E.S.), the Italian Ministry of Agriculture (MiPAF) (C.B.), and the Italian National Research Council Target and Strategic Projects in Biotechnology (C.B.).
References
- Ahmad M., Jarillo J.A., Smirnova O. and Cashmore A.R. (1998) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A. Mol. Cell, 1, 939–948. [DOI] [PubMed] [Google Scholar]
- Ang L.-H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A. and Deng X.-W. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell, 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Sadanandom A., Kitagawa K., Freialdenhoven A., Shirasu K. and Schulze-Lefert P. (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science, 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Benvenuto G., Formiggini F., Laflamme P., Malakhov M. and Bowler C. (2002) The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol., 12, 1529–1534. [DOI] [PubMed] [Google Scholar]
- Bowler C., Neuhaus G., Yamagata H. and Chua N.-H. (1994) Cyclic GMP and calcium mediate phytochrome phototransduction. Cell, 77, 73–81. [DOI] [PubMed] [Google Scholar]
- Choi G., Yi H., Lee J., Kwon Y.-K., Soh M.S., Shin B., Luka Z., Hahn T.-R. and Song P.s. (1999) Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature, 401, 610–613. [DOI] [PubMed] [Google Scholar]
- Christie J.M. and Briggs W.R. (2001) Blue light sensing in higher plants. J. Biol. Chem., 276, 11457–11460. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., Chen D.L., Yeh K.-C. and Abel S. (2000) Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol., 124, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M., Zhou Y.-C., Schäfer E. and Kretsch T. (2001) EID1, an F-box protein involved in phytochrome Aspecific light signaling. Genes Dev., 15, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A. and Quail P.H. (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev., 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Yeh K.C., Lagarias J.C., Zhang H., Elich T.D. and Chory J. (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science, 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Guo H., Mockler T., Duong H. and Lin C. (2001) SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science, 291, 487–490. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S. and Deng X.-W. (2000) The cell biology of the COP/DET/FUS proteins. Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol., 124, 1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H. and Estelle M. (2002) Plant development: regulation by protein degradation. Science, 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J. and Deng X.-W. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev., 16, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E. and Quail P.H. (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J., 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I. and Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature, 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Jarillo J.A., Capel J., Tang R.-H., Yang H.-Q., Alonso J.M., Ecker J.R. and Cashmore A.R. (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature, 410, 487–490. [DOI] [PubMed] [Google Scholar]
- Kircher S. et al. (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell, 14, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall L. and Reed J.W. (2000) The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl Acad. Sci. USA, 97, 8169–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.L., Covington M.F., Fankhauser C., Chory J. and Wagner D.R. (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell, 13, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.G., Li J.M., Qu L.J., Hager J., Chen Z., Zhao H.Y. and Deng X.-W. (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell, 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia J.F., Huq E. and Quail P.H. (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science, 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Màs P., Devlin P.F., Panda S. and Kay S.A. (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature, 408, 207–211. [DOI] [PubMed] [Google Scholar]
- Møller S.G., Ingles P.J. and Whitelam G.C. (2002) The cell biology of phytochrome signalling. New Phytol., 154, 553–590. [DOI] [PubMed] [Google Scholar]
- Montgomery B.L. and Lagarias J.C. (2002) Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci., 7, 357–366. [DOI] [PubMed] [Google Scholar]
- Nagy F. and Schäfer E. (2002) Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol., 53, 329–355. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M. and Quail P.H. (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature, 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Matsui M. and Deng X.-W. (2001) Overexpression of the heterotrimeric G-protein asubunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell, 13, 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N. and Deng X.-W. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature, 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y. and Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev., 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (1994) Phytochrome genes and their expression. In Kendrick, R.E. and Kronenberg, G.H.M. (eds), Photomorphogenesis in Plants. Kluwer Academic, Dordrecht, pp. 71–104. [Google Scholar]
- Quail P.H. (2000) Phytochrome-interacting factors. Semin. Cell Dev. Biol., 11, 457–466. [DOI] [PubMed] [Google Scholar]
- Quail P.H. (2002a) Photosensory perception and signalling in plant cells: new paradigms? Curr. Opin. Cell Biol., 14, 180–188. [DOI] [PubMed] [Google Scholar]
- Quail P.H. (2002b) Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol., 3, 85–93. [DOI] [PubMed] [Google Scholar]
- Quail P.H., Boylan M.T., Parks B.M., Short T.W., Xu Y. and Wagner D. (1995) Phytochromes: photosensory perception and signal transduction. Science, 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Schroeder D.F., Gahrtz M., Maxwell B.B., Cook R.K., Kan J.M., Alonso J.M., Ecker J.R. and Chory J. (2002) De-etiolated1 (DET1) and damaged DNA binding protein1 (DDB1) interact to regulate Arabidopsis photomorphogenesis. Curr. Biol., 12, 1462–1472. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino G., Callis J., Crosby W.L., Lyapina S., Deshaies R.J., Gray W.M., Estelle M. and Deng X.-W. (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science, 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Shacklock P.S., Read N.D. and Trewavas A.J. (1992) Cytosolic free calcium mediates red light-induced photomorphogenesis. Nature, 358, 753–755. [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C. and Lin C. (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature, 417, 763–767. [DOI] [PubMed] [Google Scholar]
- Shinomura T., Uchida K. and Furuya M. (2000) Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol., 122, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G., Yanagawa Y., Kwok S.F., Matsui M. and Deng X.-W. (2002) Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev., 16, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere U., Eichenberg K., Lohrmann J., Mira-Rodado V., Bäurle I., Kudla J., Nagy F., Schäfer E. and Harter K. (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science, 294, 1108–1111. [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Zhu T., Chang H.s., Wang X. and Quail P.H. (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl Acad. Sci. USA, 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. and Deng X.-W. (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J., 21, 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y. and Deng X.-W. (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in mediation of photomorphogenic development. Science, 294, 154–158. [DOI] [PubMed] [Google Scholar]
- Yang H.-Q., Tang R.-H. and Cashmore A.R. (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell, 13, 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.-C., Dieterle M., Büche C. and Kretsch T. (2002) The negatively acting factors EID1 and SPA1 have distinct functions in phytochrome Aspecific light signaling. Plant Physiol., 128, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]


