Abstract
Germ cell tumors (GCTs) of the testis are the predominant cancer among young men. We analyzed gene expression profiles of 50 GCTs of various subtypes, and we compared them with 443 other common malignant tumors of epithelial, mesenchymal, and lymphoid origins. Significant differences in gene expression were found among major histological subtypes of GCTs, and between them and other malignancies. We identified 511 genes, belonging to several critical functional groups such as cell cycle progression, cell proliferation, and apoptosis, to be significantly differentially expressed in GCTs compared with other tumor types. Sixty-five genes were sufficient for the construction of a GCT class predictor of high predictive accuracy (100% training set, 96% test set), which might be useful in the diagnosis of tumors of unknown primary origin. Previously described diagnostic and prognostic markers were found to be expressed by the appropriate GCT subtype (AFP, POU5F1, POV1, CCND2, and KIT). Several additional differentially expressed genes were identified in teratomas (EGR1 and MMP7), yolk sac tumors (PTPN13 and FN1), and seminomas (NR6A1, DPPA4, and IRX1). Dynamic computation of interaction networks and mapping to existing pathways knowledge databases revealed a potential role of EGR1 in p21-induced cell cycle arrest and intrinsic chemotherapy resistance of mature teratomas.
Keywords: testicular cancer, unknown primary tumors, DNA microarrays, molecular interaction networks
Human germ cell tumors (GCTs) are a diverse group of neoplasms that most commonly arise in the gonads, particularly in the testis. They account for up to 60% of all malignancies diagnosed in men between 20 and 40 years of age. Their incidence (6-11 per 100,000) has increased among Caucasians in recent decades, with an annual increase of 3-6% (1).
The histopathological classification of GCTs has been controversial because of the different concepts of histogenesis of these neoplasms, as well as the pluripotent nature of transformed primordial germ cells. GCTs can mimic normal patterns of embryonic segregation and differentiation, giving rise to structures resembling embryonic (endoderm, mesoderm, ectoderm) and extra-embryonic (yolk sac, trophoblast) derivatives. On the basis of the presence of those elements, they are further divided into pure GCTs (seminoma, embryonal carcinoma, teratoma, choriocarcinoma, and yolk sac tumor) or mixed GCTs, if more than one element is present.
Compared with most cancers of adults, GCTs are highly sensitive to chemotherapy (2). Even with metastases, 80% of GCT patients can be cured by cisplatin-based combination chemotherapy, followed by secondary resection of residual tumor lesions, which can contain necrotic cells, viable malignant cells, or mature teratoma. In contrast to the other histological subtypes, mature teratomas show a less aggressive clinical behavior, but are unresponsive to chemotherapy. The biological bases for the chemosensitivity of GCTs and the clinical behavior of mature teratomas are unclear (3).
In this study, we have performed a comparative analysis of genome-wide gene expression profiles of GCTs. In addition to the analysis of different histological subtypes of GCTs, we sought to identify molecular profiles that distinguished these tumors from other human malignancies, and to further our knowledge about genes involved in GCT development and progression, their interaction partners, and their regulatory modules.
Methods
Tumor Specimens. A total of 23 specimens (21 testicular and two mediastinal GCTs) were collected under Institutional Review Board-approved guidelines from patients diagnosed with GCTs who underwent surgery at Indiana University Cancer Center between 1998 and 2000. Three of these specimens were from primary tumors (one testicular, two mediastinal) and 20 were from residual tumor resections of metastases after chemotherapy for testicular GCTs. Following their excision, samples were immediately frozen in liquid nitrogen and stored at -80°C until use. These included 13 teratomas, 2 seminomas, 2 yolk sac tumors, 3 teratomas with transformation into sarcoma, and 3 mixed tumors (teratoma with the elements of yolk sac tumor). Data from an additional 27 GCTs from the public Stanford Microarray Database (SMD) (4), comprising 23 seminomas, 2 yolk sac tumors, and 2 embryonal cell carcinomas, were reanalyzed together with the 23 new specimens, for a total of 50 GCTs. Finally, data from 443 other common malignant tumors in the SMD (71 prostate, 103 gastric, 17 pancreatic, 38 renal, 82 liver, 41 soft tissue, 67 lung, and 24 lymphoma), were used for a comparative analysis of GCT vs. other malignancies (Table 2, which is published as supporting information on the PNAS web site).
Microarray Preparation. Total RNA was isolated by using TRIzol (Invitrogen) followed by mRNA purification using an Oligotex Midi mRNA Isolation kit (Qiagen). Cy5-labeled cDNA, synthesized by using tumor sample mRNA as a template, and Cy3-labeled common reference cDNA, synthesized by using a pool of mRNAs derived from a panel of 11 cell lines (5), were hybridized to cDNA microarrays printed at the Stanford Functional Genomics Facility, consisting of 42,174 elements representing 25,672 unique UniGene cluster IDs. Microarrays were scanned with GenePix 4000 (Axon Instruments), and raw data were archived in the SMD.
Data Filtering and Transformation. Nonflagged elements with signal-to-noise ratio of at least 1.2 in both channels were included in subsequent analysis. The same criteria were used for the retrieval of the raw data from the already published data sets available in SMD. Data normalization was performed essentially as described in ref. 6, with some modifications. Global intensity normalized data for each array were renormalized by fitting a LOWESS curve to the data points corresponding to the housekeeping gene set consisting of 269 clones (Table 3, which is published as supporting information on the PNAS web site). These clones were selected from the single 115-array set representing 35 normal human tissues (7). Local intensity-based normalization was applied first to each of these arrays. After dividing the average fluorescence range into 30 segments, a maximum of 10 housekeeping genes with the smallest variance were chosen from each segment by computing the mean and its 95% confidence interval, and selecting only those genes for which the confidence interval encompasses zero. The normalized data matrix was further mean centered by columns and rows, and clones with at least 80% good data across all of the samples were selected for further analysis. This selection resulted in 11,921 clones, used in subsequent comparisons.
Unsupervised and Supervised Analysis. Unsupervised hierarchical clustering was performed in cluster and visualized in treeview (8). Multidimensional scaling was done in matlab (The Math-Works, Natick, MA). Two statistical software packages, Significance Analysis of Microarrays (SAM) (9), and Prediction Analysis of Microarrays (PAM) (10), were used for the supervised data analysis.
Interaction Networks and Functional Analysis. These data were generated through the use of Ingenuity Pathways Analysis, a web-delivered application that enables discovery, visualization, and exploration of biological interaction networks. Detailed information about this analysis software can be found at www.ingenuity.com. Gene ontology analysis was also executed by using Ingenuity Pathways Analysis tools, and significance for the enrichment of the genes with particular biological function was determined by righttailed Fisher's exact test with α = 0.05 and the whole database as a reference set. Functional information and virtual tissue Northern blot data, representing the mRNA expression of the gene through relative frequencies of ESTs from cDNA libraries from various tissues, were also obtained from SOURCE (11).
Real-Time RT-PCR. Expression of MMP7, EGR1, AFP, FN1, POU5F1, IRX1, and NR6A1 were confirmed in 12 specimens for which sufficient amount and quality of RNA was available, by using the Applied Biosystems Prism 7900HT Sequence Detection System and SYBR Green PCR Master Mix according to manufacturer's instructions. For primer sequences and reaction conditions see Fig. 4, which is published as supporting information on the PNAS web site.
Results and Discussion
GCT Class Predictors. DNA microarrays are widely used to study gene expression signatures of human neoplasms. However, such studies are typically organ-specific and do not address variations between different types of tumors. Here we present an integrated gene expression analysis of 50 GCTs and 443 other malignant tumors, corresponding to eight additional common tumor types. Several recent large-scale microarray meta-analyses have demonstrated the robustness of critical gene expression patterns (12, 13). The fact that all analyzed arrays in this study were produced by the Stanford Functional Genomics Facility, and hybridized according to the same experimental protocol using essentially the same common reference RNA, further supports the feasibility of our approach.
Using two-class, unpaired SAM, we have identified 683 clones as being differentially expressed in GCTs as compared with the eight other tumor types (304 overexpressed and 379 underexpressed), when a 2-fold change cutoff and minimal false discovery rate (median number of falsely discovered genes <1) are selected (the complete list is available as Table 4, which is published as supporting information on the PNAS web site). To determine the biological relevance of the identified genes, and to provide a framework for the prioritization of numerous molecular alterations evident in GCT transcriptome, we have systematically explored the functions of these 683 clones, mapped to 511 unique UniGene Cluster IDs, by using the gene ontology analysis feature of the Ingenuity Pathway Analysis tools. The most dominant cellular functions in the analyzed gene list are cell cycle progression (42 genes, P = 0.008), cell proliferation (66 genes, P = 0.019), and cell death (73 genes, P = 0.008).
Among cell cycle regulating genes, CCND2 was notably overexpressed in GCTs compared with other cancers. Aberrant expression of cyclin D2 is an early event in human germ cell tumorigenesis, and possibly one of the critical alterations during its progression (14). CCND2 resides in the short arm of chromosome 12, gain of which is a consistent feature of testicular GCTs, usually in the form of isochromosome 12p. Another overexpressed gene in the 12p region was LDHB, the most important lactate dehydrogenase isoenzyme in patients with testicular GCTs (15). In addition to CCND2 and LDHB, our GCT gene expression signature includes 17 other overexpressed genes mapped to 12p (Table 1), some of which have been previously implicated in alterations of 12p (16). We performed detailed exploratory analysis of the data available in the Ingenuity Pathways Knowledge Base and SOURCE database for the differentially expressed 12p genes. This analysis has revealed several strong candidates potentially involved in the regulation of cell cycle in GCTs, including NOL1, DDX11, and FOXM1, a Forkhead box transcription factor that is required for normal S-M coupling, and whose overexpression accelerates progression through G2/M (17). In addition, C1QDC1, CMAS, AEBP2, PHC1, DDX47, and TEAD4 are associated with various developmental processes and may be important determinants of specific histological features of these tumors. All but one of these 20 clones are also shown to be overexpressed in either embryonic stem cells or gonads on the basis of virtual tissue Northern blot data. These findings support the notion of regional clustering of genes with similar functions in the human genome, and they suggest that numerical and structural alterations of chromosomes contribute to tumor behavior not just by affecting the expression of a few key oncogenes and tumor suppressor genes, but also by altering the expression levels of multiple co-localized genes, some of which may act in a synergistic fashion. Similar effects are widespread in the genomes of other tumor types and can be exemplified by coamplification and coexpression of multiple genes involved in the ERBB2 amplicon in breast cancers (18).
Table 1. Chromosomal location, virtual tissue Northern blot normalized expression levels, and rank of chromosome 12p genes overexpressed in GCTs compared with other tumors.
| Name | Location | Virtual tissue | Expression, % | Rank |
|---|---|---|---|---|
| C1ODC1 | 12p11 | Embryonic stem cells | 9.45 | 2 |
| DDX11 | 12p11 | Embryonic stem cells | 33.90 | 1 |
| OVOS2 | 12p11 | Testis | 20.03 | 3 |
| CMAS | 12p12 | Pluripotent cell line | 10.93 | 3 |
| LDHB | 12p12 | Embryonic stem cells | 8.93 | 1 |
| AEBP2 | 12p12 | Embryonic stem cells | 11.09 | 1 |
| DERA | 12p12 | Embryonic stem cells | 6.50 | 3 |
| CCND2 | 12p13 | Embryonic stem cells | 13.67 | 2 |
| PHC1 | 12p13 | Embryonic stem cells | 55.87 | 1 |
| NOL1 | 12p13 | Embryonic stem cells | 10.53 | 1 |
| FOXM1 | 12p13 | Embryonic stem cells | 8.05 | 2 |
| M6PR | 12p13 | Small intestine | 7.37 | 1 |
| FLJ22662 | 12p13 | Embryonic stem cells | 14.21 | 1 |
| DDX47 | 12p13 | Embryonic stem cells | 11.50 | 1 |
| TEAD4 | 12p13 | Pluripotent cell line | 15.80 | 1 |
| SLC2A3 | 12p13 | Embryonic stem cells | 32.58 | 1 |
| SLC2A14 | 12p13 | Testis | 40.19 | 1 |
| CLEC2 | 12p13 | Testis | 20.43 | 2 |
| KIAA1238 | 12p13 | Embryonic stem cells | 15.86 | 1 |
Gene expression changes of multiple other genes (MYCN, KIT, MYBL2, AFP) among the GCTs are consistent with previously published reports (19) and support the biological relevance of the gene expression signature that we have ascertained. Newly identified associations involve genes regulating various cellular functions, of which global transcription factors and DNA-modifying enzymes (MCM5, MCM6, RECQL4, DNMT3A, RIF1) deserve particular attention.
We further explored the differential gene expression of GCTs relative to a group of eight other tumor types, by using the nearest shrunken centroid classifier implemented in the PAM package (10). This analysis was motivated by the clinical importance of GCT in the differential diagnosis of unknown primary cancers (UPC). UPCs are biopsy-proved malignancies for which the anatomic origin remains unidentified after history, physical examination, and standard laboratory studies. The reported incidence of UPC ranges from 2% to 5% of all patients who are diagnosed with cancer (20). Males with the extragonadal germ cell syndrome represent a highly treatable subset of UPC, and development of molecular markers for the identification of that subset is an important ongoing effort.
In our GCT vs. other malignancies classification, all of the samples were first randomly split into training and validation sets, comprising 247 and 246 samples. During training, the optimal number of clones was selected on the basis of prediction accuracy estimated by 10-fold cross-validation, and the resulting predictor was further tested during class prediction in the validation set. The optimal GCT class predictive accuracy (100% on the training set and 96% on the validation set) was obtained with an 84-clone predictor, which also enabled clear separation of GCTs by multidimensional scaling, as visualized in Fig. 1. The final predictor consisted of 10 12p genes, as well as 55 other genes with known function that we previously detected by SAM to be differentially expressed in GCTs. The relatively high number of required clones can be explained by diverse features of the GCT class. If only seminomas are included in the training set, and misclassification is estimated by cross-validation, the optimal predictor consists of only seven clones. The single most important gene in that predictor is NR6A1, an orphan member of the nuclear receptor superfamily, initially described as the murine germ cell nuclear factor, GCNF, essential for normal embryonic and germ cell development in mice (21, 22). Another component is DPPA4, identified as an OCT-4-related gene necessary for the establishment of developmental pluripotency in mouse embryos (23). Although limited data exist about the functional properties of these genes, their germ cell restricted expression pattern, as well as particularly high scores in the identified predictors, warrant the performance of additional validation studies of these potentially useful diagnostic markers.
Fig. 1.
GCT class predictor. Supervised GCT class prediction analysis using nearest shrunken centroids (PAM) identified a subset of 84 clones that distinguish GCTs from all other analyzed tumors. (a) The centroid values in GCTs (class 2) and other tumors (class 1) are plotted. (b) Cross-validated error curves from the classifier (Upper) and the confusion matrix obtained after prediction in the test set (Lower) are displayed. (c) Multidimensional scaling was performed on the complete sample set by using the clones from the PAM classifier.
Gene Expression Signatures of Histological Subtypes of GCT. Although germ cell tumors represent a unique pathological and clinical entity with a common cell of origin, they vary in their phenotypic characteristics and are further subdivided into several subtypes based on the line of differentiation of the transformed primordial germ cell. Gene expression profiling approaches have already been applied to the study of underlying transcriptomic alterations in GCTs. However, these studies were mainly performed on a limited number of samples, with only one GCT subtype dominating the whole data set (24, 25). In contrast, our analysis was performed on a dataset with a balanced number of seminomas and nonseminomas, which enables more accurate determination of differentially expressed genes across these two groups.
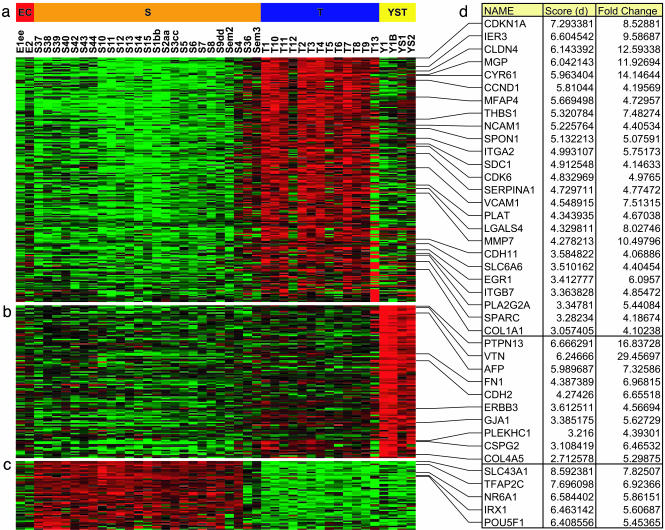
Pearson correlation-based hierarchical clustering of 5,808 clones with 80% good data and at least 4-fold change in at least one tumor, compared with the mean expression across all of the GCTs, clearly separates four histological subtypes, reflecting at the transcriptomic level their significant phenotypic differences (Fig. 5, which is published as supporting information on the PNAS web site). Further selection of the most important changes in teratomas, yolk sac tumors, and seminomas was performed by the use of two-class SAM, with less than one falsely discovered gene as a significance cutoff. Observed changes were further prioritized on the basis of fold-change values, and clones with a minimal 4-fold change were examined in more detail (Fig. 2). Detailed supervised analysis of embryonal carcinomas was not performed, because only two samples were available. We have also performed quantitative RT-PCR validation of the microarray data on a subset of available tumors, and we confirmed the expression levels for the six genes detected to be differentially up-regulated in teratomas (MMP7, EGR1), yolk sac tumors (AFP, FN1), and seminomas (POU5F1, IRX1, NR6A1) (Fig. 4). The high correlation of microarray and quantitative RT-PCR data further supports the significance of the observed changes in expression.
Fig. 2.
Differentially expressed genes in GCT histological subtypes. (a-c) Clones with statistically significant 4-fold overexpression in teratomas (a), yolk sac tumors (b), and seminomas (c) were identified by using SAM. Expression profiles are displayed as a pseudocolor map, where shades of green and red represent under- and overexpression, respectively, relative to the mean expression of each clone in all of the GCTs. Clones are ordered on the basis of their statistical significance, and samples, on the basis of the histological subtype. (d) Names, d scores, and fold change values of selected genes.
Teratomas. Two hundred and eight clones were highly overexpressed in teratomas in comparison to all other nonmixed GCTs (Table 5, which is published as supporting information on the PNAS web site). The list is dominated by genes encoding integral membrane proteins (CDH11, NCAM1, VCAM1, CLDN4, ITGA2, ITGB7, SDC1, SLC6A6), extracellular matrix components or associated proteins (CYR61, COL1A1, COL3A1, COL6A3, THBS1, LGALS4, MFAP4, MGP, SPARC, SPON1), as well as tissue remodeling enzymes and regulators (MMP7, SERPINA1, SERPINA3, PLA2G2A, PLAT), reflecting the complex tissue structure of these tumors and their differentiation along all three lines of differentiation. To explore further how the overexpressed genes in the teratoma signature are related, they were placed in the context of present knowledge about their molecular interactions by using Ingenuity Pathways Analysis tools. The 208 clones were first mapped to 110 nonredundant elements in the Ingenuity knowledge base, and networks of interacting genes and their products were dynamically computed on the basis of individually modeled known relationships. The score was then determined for each network based on the P value, which indicates the likelihood of the input genes in the network being found together because of random chance. Scores of 2 or higher are considered significant with 99% confidence. The top-scoring network (Fig. 3) consists of 35 interacting genes involved in the regulation of cell death. A particularly important node in that network is CDKN1A (p21, Waf1/Cip1), the top-scoring overexpressed gene in the initial list of 208 clones, linked with its known interaction partners CDK6 and CCND1. It has been hypothesized that the activation of p21 and an ability to undergo p21-induced cell cycle arrest may, together with up-regulation of various factors involved in drug export (P-glycoprotein, MRP2, BCRP) and transformation (GST), explain the intrinsic chemotherapy resistance of mature teratomas (3). Their intact G1/S checkpoint control might allow DNA damage repair to occur, instead of activation of the apoptotic cascade that usually occurs in other GCT subtypes upon drug exposure. p21 is induced by both p53-dependent and -independent mechanisms (26). Because no correlation was found between the number of p53-positive cells and the apoptotic index in a small series of mature teratomas (3), it is possible that other, p53-independent, factors play a critical role in the regulation of p21 in these tumors. The teratoma-associated network is linked with EGR1, early growth response 1, a C2H2-type zinc-finger protein that functions as an important transcriptional regulator involved in various cellular functions. EGR1 has not been previously implicated as a molecular determinant of teratoma chemoresistance. However, it has been shown that EGR1 expression prevents apoptosis and promotes cell survival after DNA damage caused by ionizing radiation in several different cell types (27). This effect seems to be independent of p53 status, and in the doubly expressing human fibrosarcoma cell line exposed to UV radiation, the survival effect of EGR1 was dominant over the apoptotic effect of p53 (28).
Fig. 3.
Functional network analysis of the teratoma-overexpressed genes. The top-scoring network consisted of 35 nodes, representing genes, with their shape indicating the functional class of the gene product, and multiple edges indicating the biological relationships between the nodes (see the key on the right). Nodes are color-coded in red according to the d score value of the corresponding clones as determined by SAM. Gene ontology analysis was performed by using a right-tailed Fisher exact test, and statistically significant high-level functions are reported in the table. Edges between CDKN1A- and CDKN1A-interacting genes are colored in blue.
Yolk Sac Tumors. When the same thresholds as above were used, 109 clones were identified to be overexpressed in this histological subtype of GCTs (Table 6, which is published as supporting information on the PNAS web site). Our statistical approach correctly identified AFP as one of the most highly overexpressed genes in that group of tumors. This gene encodes α-fetoprotein, a glycoprotein expressed mostly by yolk sac tumor elements of GCTs, and routinely used as a diagnostic marker for these cancers. The top-scoring gene in the yolk sac signature is PTPN13, also known as Fas-associated phosphatase-1, FAP-1, which was found to interact with Fas receptor and inhibit Fas-mediated programmed cell death (29). Although all GCTs are positive for FAP-1, teratomas and spermatocytic seminomas are the only ones also positive for Fas (30). Particularly high expression of FAP-1 in yolk sac tumors may have other effects, because this protein was also shown to have a key role in the apoptotic process in human breast cancer cells independent of Fas (31), and to interact with GTPase-activating proteins (32), and thus may function as a regulator of the Rho signaling pathway, involved in signal transduction processes leading to cytoskeletal-dependent responses, including cell migration, invasion, and metastasis. Several other genes directly involved in cell adhesion processes are highly expressed in yolk sac tumors (FN1, VTN, CSPG2, COL4A5, PLEKHC1, CDH2, GJA1). Interestingly, these tumors also show overexpression of ERBB3. Members of the ErbB receptor tyrosine kinases, especially ERBB2, ERBB3, and their ligand neuregulin-β, control the growth and survival of primordial germ cells in genital ridges of developing mouse, and their specific time-dependent expression pattern is necessary for normal development of gonads (33).
Seminomas. Fifty-one clones were highly overexpressed in seminomas (Table 7, which is published as supporting information on the PNAS web site). The highest significance was observed for SLC43A1, also known as prostate cancer overexpressed gene 1, POV1. Very limited data exist about the function of this gene. It was originally described in clinically aggressive prostate cancers (34) and was recently implicated as an early-onset gene in the development of seminomas (35). Other interesting genes with high expression in seminomas include the transcription factors POU5F1, TFAP2C, NR6A1, and IRX1. POU5F1 (OCT3/4) was previously reported as a master regulator of pluripotency, with increased expression controlling embryonic differentiation into primitive endoderm and mesoderm. In contrast, repression of POU5F1 induces loss of pluripotency and dedifferentiation to trophectoderm. This gene has also been suggested as a marker of pluripotent human germ cell tumors, expressed in seminomas and embryonal carcinomas, but not in the various types of differentiated nonseminomas (36). A similar pattern of expression in GCTs has been described for another gene in our seminoma signature, TFAP2C (37), transcription factor AP-2-γ, a member of the AP-2 transcription factor family, which plays important roles in the development and differentiation of the neural tube, neural crest derivatives, skin, heart, and urogenital tissues (38). Two genes have not been previously implicated in the development and progression of GCTs, but they have critical roles in various developmental processes. These include NR6A1, the single most important gene in the seven-clone seminoma predictor, as well as IRX1, one of the Iro homeobox transcription factors required at early stages of development to define large territories and to further subdivide neural tube and heart (39). Their altered pattern of expression could be a characteristic of seminomas and could be involved in the determination of their pluripotent nature.
Conclusions
GCTs have many unique properties in comparison with other solid tumors, and limited data exist about molecular mechanisms underlying such unique biology. In this study, we have analyzed transcriptomic changes in a set of 50 GCTs. Key alterations previously described in the literature as GCT-specific, including routinely used diagnostic and prognostic markers, were confirmed as highly expressed by the appropriate GCT subtypes in our study. In addition, we have developed a gene expression-based GCT class predictor of high predictive accuracy and identified a set of genes that could potentially be used in the diagnosis of tumors of unknown primary origin. We have also detected a set of genes highly overexpressed in teratomas, yolk sac tumors, and seminomas, and have further explored their most important functional interactions and expression modules. These findings may be useful in studying the biology of GCTs, and in developing novel therapeutic targets or diagnostic markers for these diseases.
Supplementary Material
Acknowledgments
We thank Brian Francisco and Christianna Stuber for their excellent technical assistance and the Stanford Functional Genomics Facility for its support. This research was supported by National Institutes of Health Grants R33 CA 89830 (to B.I.S.), R01 CA 102283 (to R.A.H.), and P01 CA 74295 (to L.H.E.) from the U.S. Public Health Service.
Author contributions: D.J., R.A.H., L.H.E., and B.I.S. designed research; D.J., S.S., R.Y., Y.W., and G.E.D. performed research; R.A.H. contributed new reagents/analytic tools; D.J., S.S., R.Y., Y.W., R.T., L.H.E., and B.I.S. analyzed data; and D.J. and B.I.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: GCT, germ cell tumor; PAM, Prediction Analysis of Microarrays; SAM, Significance Analysis of Microarrays; SMD, Stanford Microarray Database.
References
- 1.Oosterhuis, J. W. & Looijenga, L. H. (2005) Nat. Rev. Cancer 5, 210-222. [DOI] [PubMed] [Google Scholar]
- 2.Einhorn, L. H. (2002) Proc. Natl. Acad. Sci. USA 99, 4592-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer, F., Stoop, H., Scheffer, G. L., Scheper, R., Oosterhuis, J. W., Looijenga, L. H. & Bokemeyer, C. (2003) Clin. Cancer Res. 9, 767-773. [PubMed] [Google Scholar]
- 4.Gollub, J., Ball, C. A., Binkley, G., Demeter, J., Finkelstein, D. B., Hebert, J. M., Hernandez-Boussard, T., Jin, H., Kaloper, M., Matese, J. C., et al. (2003) Nucleic Acids Res. 31, 94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perou, C. M., Jeffrey, S. S., van de Rijn, M., Rees, C. A., Eisen, M. B., Ross, D. T., Pergamenschikov, A., Williams, C. F., Zhu, S. X., Lee, J. C., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 9212-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson, D. L., Buckley, M. J., Helliwell, C. A. & Wilson, I. W. (2003) Bioinformatics 19, 1325-1332. [DOI] [PubMed] [Google Scholar]
- 7.Shyamsundar, R., Kim, Y. H., Higgins, J. P., Montgomery, K., Jorden, M., Sethuraman, A., van de Rijn, M., Botstein, D., Brown, P. O. & Pollack, J. R. (2005) Genome Biol. 6, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibshirani, R., Hastie, T., Narasimhan, B. & Chu, G. (2002) Proc. Natl. Acad. Sci. USA 99, 6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diehn, M., Sherlock, G., Binkley, G., Jin, H., Matese, J. C., Hernandez-Boussard, T., Rees, C. A., Cherry, J. M., Botstein, D., Brown, P. O. & Alizadeh, A. A. (2003) Nucleic Acids Res. 31, 219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., Barrette, T., Pandey, A. & Chinnaiyan, A. M. (2004) Proc. Natl. Acad. Sci. USA 101, 9309-9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal, E., Friedman, N., Koller, D. & Regev, A. (2004) Nat. Genet. 36, 1090-1098. [DOI] [PubMed] [Google Scholar]
- 14.Houldsworth, J., Reuter, V., Bosl, G. J. & Chaganti, R. S. (1997) Cell Growth Differ. 8, 293-299. [PubMed] [Google Scholar]
- 15.von Eyben, F. E., Liu, F. J., Amato, R. J. & Fritsche, H. A. (2000) Acta Oncol. 39, 509-517. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez, S., Jafer, O., Goker, H., Summersgill, B. M., Zafarana, G., Gillis, A. J., van Gurp, R. J., Oosterhuis, J. W., Lu, Y. J., Huddart, R., et al. (2003) Oncogene 22, 1880-1891. [DOI] [PubMed] [Google Scholar]
- 17.Leung, T. W., Lin, S. S., Tsang, A. C., Tong, C. S., Ching, J. C., Leung, W. Y., Gimlich, R., Wong, G. G. & Yao, K. M. (2001) FEBS Lett. 507, 59-66. [DOI] [PubMed] [Google Scholar]
- 18.Kauraniemi, P., Barlund, M., Monni, O. & Kallioniemi, A. (2001) Cancer Res. 61, 8235-8240. [PubMed] [Google Scholar]
- 19.Skotheim, R. I. & Lothe, R. A. (2003) APMIS 111, 136-151. [DOI] [PubMed] [Google Scholar]
- 20.Glover, K. Y., Varadhachary, G. R., Lenzi, R., Raber, M. N. & Abbruzzese, J. L. (2004) in Clinical Oncology, ed. Abeloff, M. D. (Elsevier Churchill Livingstone, Philadelphia), pp. 2627-2643.
- 21.Chen, F., Cooney, A. J., Wang, Y., Law, S. W. & O'Malley, B. W. (1994) Mol. Endocrinol. 8, 1434-1444. [DOI] [PubMed] [Google Scholar]
- 22.Chung, A. C., Katz, D., Pereira, F. A., Jackson, K. J., DeMayo, F. J., Cooney, A. J. & O'Malley, B. W. (2001) Mol. Cell. Biol. 21, 663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bortvin, A., Eggan, K., Skaletsky, H., Akutsu, H., Berry, D. L., Yanagimachi, R., Page, D. C. & Jaenisch, R. (2003) Development (Cambridge, U.K.) 130, 1673-1680. [DOI] [PubMed] [Google Scholar]
- 24.Sugimura, J., Foster, R. S., Cummings, O. W., Kort, E. J., Takahashi, M., Lavery, T. T., Furge, K. A., Einhorn, L. H. & Teh, B. T. (2004) Clin. Cancer Res. 10, 2368-2378. [DOI] [PubMed] [Google Scholar]
- 25.Sperger, J. M., Chen, X., Draper, J. S., Antosiewicz, J. E., Chon, C. H., Jones, S. B., Brooks, J. D., Andrews, P. W., Brown, P. O. & Thomson, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 13350-13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gartel, A. L. & Tyner, A. L. (2002) Mol. Cancer Ther. 1, 639-649. [PubMed] [Google Scholar]
- 27.Hallahan, D. E., Dunphy, E., Virudachalam, S., Sukhatme, V. P., Kufe, D. W. & Weichselbaum, R. R. (1995) J. Biol. Chem. 270, 30303-30309. [DOI] [PubMed] [Google Scholar]
- 28.de Belle, I., Huang, R. P., Fan, Y., Liu, C., Mercola, D. & Adamson, E. D. (1999) Oncogene 18, 3633-3642. [DOI] [PubMed] [Google Scholar]
- 29.Sato, T., Irie, S., Kitada, S. & Reed, J. C. (1995) Science 268, 411-415. [DOI] [PubMed] [Google Scholar]
- 30.Kersemaekers, A. M., van Weeren, P. C., Oosterhuis, J. W. & Looijenga, L. H. (2002) J. Pathol. 196, 423-429. [DOI] [PubMed] [Google Scholar]
- 31.Bompard, G., Puech, C., Prebois, C., Vignon, F. & Freiss, G. (2002) J. Biol. Chem. 277, 47861-47869. [DOI] [PubMed] [Google Scholar]
- 32.Saras, J., Franzen, P., Aspenstrom, P., Hellman, U., Gonez, L. J. & Heldin, C. H. (1997) J. Biol. Chem. 272, 24333-24338. [DOI] [PubMed] [Google Scholar]
- 33.Toyoda-Ohno, H., Obinata, M. & Matsui, Y. (1999) Dev. Biol. 215, 399-406. [DOI] [PubMed] [Google Scholar]
- 34.Chuaqui, R. F., Englert, C. R., Strup, S. E., Vocke, C. D., Zhuang, Z., Duray, P. H., Bostwick, D. G., Linehan, W. M., Liotta, L. A. & Emmert-Buck, M. R. (1997) Urology 50, 302-307. [DOI] [PubMed] [Google Scholar]
- 35.Skotheim, R. I., Monni, O., Mousses, S., Fossa, S. D., Kallioniemi, O. P., Lothe, R. A. & Kallioniemi, A. (2002) Cancer Res. 62, 2359-2364. [PubMed] [Google Scholar]
- 36.Looijenga, L. H., Stoop, H., de Leeuw, H. P., de Gouveia Brazao, C. A., Gillis, A. J., van Roozendaal, K. E., van Zoelen, E. J., Weber, R. F., Wolffenbuttel, K. P., van Dekken, H., et al. (2003) Cancer Res. 63, 2244-2250. [PubMed] [Google Scholar]
- 37.Hoei-Hansen, C. E., Nielsen, J. E., Almstrup, K., Sonne, S. B., Graem, N., Skakkebaek, N. E., Leffers, H. & Meyts, E. R. (2004) Clin. Cancer Res. 10, 8521-8530. [DOI] [PubMed] [Google Scholar]
- 38.Hilger-Eversheim, K., Moser, M., Schorle, H. & Buettner, R. (2000) Gene 260, 1-12. [DOI] [PubMed] [Google Scholar]
- 39.Cavodeassi, F., Modolell, J. & Gomez-Skarmeta, J. L. (2001) Development (Cambridge, U.K.) 128, 2847-2855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.