Abstract
We describe the use of modified fluorescent-labeled oligonucleotide probes in the sequence-specific detection of messenger RNAs in live human cells. To make this detection possible, we developed a previously undescribed probe design that combines earlier quenched autoligation chemistry with a previously undescribed fluorescence resonance energy transfer (FRET) strategy to lower background signals. The probe pairs consisted of a nucleophilic 3′-phosphorothioate probe carrying a Cy5 FRET acceptor, and an electrophilic probe containing the combination of a 5′ end electrophile/quencher and a fluorescein FRET donor. Probes were introduced to HL-60 cells by use of the streptolysin O pore-forming peptide. Signals from three different messenger RNAs, as well as 28S ribosomal RNA, could be detected and quantitated by flow cytometry. Probes targeted to ribosomal sequences and β-actin mRNA also could be detected over background by confocal fluorescence microscopy. Varying the target site and probe backbone chemistry were found to have large effects on signal. The data suggest that quenched autoligating probes may be of general utility as biological tools in following localization, transcription, and processing of eukaryotic cellular messages and may have applications in diagnostic or prognostic analysis of disease-related RNAs in human tissues.
Keywords: fluorescence, imaging, oligonucleotide
The detection and imaging of biomolecules in native human cells is a rapidly growing field of research with important implications in biology and medicine. The varied molecular targets under development include proteins (1, 2), nucleic acids (3–5), and small molecular species (6–8). In addition to work in native cells, there has been rapid development of genetic tools for modifying cells for specific tracking of biomolecules; a prominent example is the engineering of fluorescent protein conjugates with native proteins (9). However, it is often desirable to detect species in genetically unmodified cells. For example, detection of molecular components in clinical samples from human patients would be most conveniently done directly, without prior genetic manipulation of the cells.
Our specific goal is the detection of RNA sequences in intact cells at single-nucleotide resolution by fluorescence. If it can be developed successfully, important uses for mRNA detection include imaging of localization, quantification of specific transcripts, and identification of disease-related RNAs for clinical application. Messenger RNAs are expected to be considerably more difficult to detect than ribosomal RNAs because they exist in smaller quantities in the cell (10). Moreover, because their sequences vary widely and their structures are generally unknown, it is difficult to find an optimal target site (11–13). In addition, there is the issue of breaching the barrier of the cell membrane in live human cells (14, 15). Finally, live cells also contain numerous enzymatic activities that degrade oligonucleotide-based probes.
No method has yet been successful enough to gain wide application for imaging mRNA in living cells. However, a small number of molecular approaches are under development (3, 4, 16–26). Prominent among these approaches are quenched probe strategies, including molecular beacon (MB) probes (19, 22–24) and quenched autoligating (QUAL) probes (25–28). Quenched probe strategies offer the advantage of decreased background signal in the absence of the RNA target (29). Recent reports have shown evidence for detection by fluorescence microscopy of messenger RNAs by the MB class of probes in intact human cells (3, 16–19, 22–24). However, MB probes have shown strong interference by nonspecific signals, which may be due in part to protein binding of the probes (21, 30). Other classes of quenched nucleic acid probes are also under development but have not yet been applied in cells (31, 32). Finally, a number of approaches for imaging mRNAs in genetically engineered cells have been reported recently (33, 34); although such a strategy can be a useful tool for biological study, it is not applicable for imaging native RNAs in unmodified cellular specimens.
Another aim of the present work is the automated detection of RNAs by fluorescence. Flow cytometry (FC) would seem to be an ideal platform for such automation. Although FC methodologies are broadly useful in analysis of cellular components, there exist few prior reports of the use of FC in intact human cells for the detection of RNAs (35), and fluorescent probes have not been reported to detect native mRNAs by FC. Early studies with quenched probes have obtained signals by microscopy (23).
Our molecular strategy relies on the principle of the self-directed reaction of two oligonucleotide probes bound at adjacent positions on a target nucleic acid (36–39). The reaction of a nucleophilic phosphorothioate group on one probe with a 5′-electrophilic quenching dabsyl group on an adjacent probe causes displacement of the quencher, causing an appended fluorophore to emit signal (25). Because of the short length of these QUAL probes before autoligation, their binding to mismatched targets is weak, slowing ligation and yielding high selectivity for single-nucleotide differences (25). By this approach, high-copy-number 16S rRNA sequences in living bacterial cells were imaged with single-nucleotide discrimination (26). However, background signals were significant, and no prior studies exist on the application of QUAL probes in detection of less-abundant messenger RNAs. Moreover, no studies have been carried out with these probes in either fixed or intact human cells.
Here we report on a previously undescribed molecular strategy to lower background level dramatically in QUAL probes and the successful application of the probes in intact human cells. The approach takes advantage of a nontypical FRET pair system combined with quenching and signal amplification with a recently reported universal linker (28) (Fig. 1). By using this previously undescribed QUAL-FRET probe design, we show that messenger RNAs, as well as 28S ribosomal RNA, in human HL-60 cells could be detected by FC and imaged by confocal microscopy.
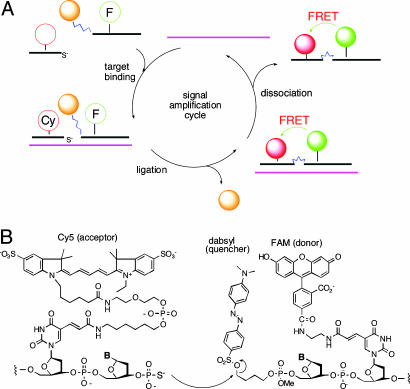
Fig. 1.
Structures and molecular mechanism of QFRET probe autoligations. (A) Illustration of the binding, ligation, and dissociation cycle, which can generate multiple signals per target. (B) Molecular structure of labeled nucleophile and electrophile probes, which carry a quenched FRET donor (FAM) and a FRET acceptor (Cy5).
Results
Design of QUAL-FRET Probes. The quenched autoligation-FRET (QFRET) approach (Fig. 1 A) was developed to address a number of problems of RNA imaging in cells. First, there are problems of low signal resulting from low abundance and/or short lifetime of mRNAs as compared with ribosomal RNAs that had been detected previously in bacteria (26). This low signal was addressed by adopting a recently reported universal linker to carry the dabsyl group, which can lead to as much as two orders of magnitude of signal amplification (28). Second, there is the problem of background signal, which in the case of previous QUAL probes can arise from hydrolysis of the dabsyl group, from off-template ligation, and from incomplete quenching by dabsyl. QFRET probes were designed to ameliorate most of the background signal. QFRET probe pairs (Fig. 1B) consist of two linear DNA probes, where the nucleophilic phosphorothioate probe has a Cy5 FRET acceptor, and the electrophilic dabsyl linker probe has a fluorescein (FAM) FRET donor. Upon binding at adjacent sites, the nucleophilic phosphorothioate group attacks the electrophilic dabsyl linker, forming the ligated product and releasing the dabsyl group, and causing the FAM donor to become unquenched. As a result, a FRET signal appears from Cy5. This previously undescribed FRET-based approach is designed to eliminate unwanted signal from FAM when it simply hydrolyzes without binding target and to minimize signal from incomplete quenching of FAM by the dabsyl group.
The atypical FAM-Cy5 FRET pair (40) was chosen to further suppress background signal from spectral overlap. The two labels are well separated in their absorption wavelengths (FAM and Cy5 have maxima at 490 and 649 nm, respectively). This strategy is expected to greatly lower background fluorescence levels as compared with more typical FRET pairs.
However, little information about the relationship between the distance and FRET efficiency of the FAM–Cy5 pair has been reported. To investigate this relationship, we tested a series of oligonucleotide sequences containing both dyes, where the distance between FAM and Cy5 gradually increases (n = 0, 3, 7, 11 nt apart). These strands were analyzed in duplex form, in the presence of a complementary DNA oligonucleotide (see Figs. 6–8, which are published as supporting information on the PNAS web site). When excited at 490 nm, all sequences showed an emission peak at 649 nm, interpreted as a FRET signal. The highest FRET efficiencies were observed within a 3- to 7-nt distance (Fig. 7B). We used this separation distance range for our subsequent QFRET probe designs.
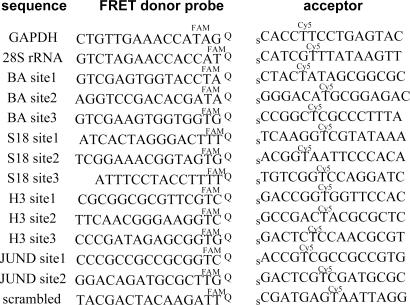
The current probe design started with natural DNA backbones, which might be expected to undergo significant intracellular cleavage (41). Such degradation would not be expected to lead to false-positive (background) signals in unreacted probes or ligated products because degradation would not lead to a FRET signal. However, degraded probes before ligation would have lower ability to generate correct positive signal, and ligated products also could lose their positive signal. To inhibit possible cleavage, we prepared probes with 2′-O-methyl nucleosides (41) and probes with a natural DNA backbone for a comparative evaluation. Sequences of the initial probes are shown in Fig. 2.
Fig. 2.
QFRET probe sequences for detection and imaging of cellular RNAs. In GAPDH and 28S rRNA cases, QFRET probes were tested both with natural DNA or 2′-OMe RNA backbone. In all other case, 2′-OMe RNA backbones were used. Q, dabsyl universal linker; s, 3′-phosphorothioate group.
Preparation of probes was carried out as described in Materials and Methods. The electrophilic dabsyl-linker probes were readily assembled by using phosphoramidite reagents and required no steps after synthesis (28). The Cy5-labeled phosphorothioate probes required one off-synthesizer step (carried out while the probe was still on solid support) for conjugation with the Cy5 dye but required no steps after the oligonucleotide was removed from the column.
QUAL-FRET Ligation in Solution. Before testing the probes in human cells, we evaluated their performance in solution, using synthetic DNA targets. The QFRET probe pairs consisted of a 9-mer dabsyl-FAM-electrophile probe and a 12-mer Cy5-phosphorothioate probe. For comparison, we tested simple FRET autoligation (IFRET) probes lacking a quencher (39), where a 5′-terminal iodide (instead of dabsyl) is used to activate ligation. Ligations were carried out in buffer containing 10 mM MgCl2 (pH 7.0) at 25°C. Extents of reaction were compared at equimolar target DNA and probe concentrations (500 nM each), and the reaction progress with QFRET as well as IFRET probes was followed in solution by changes in fluorescence spectra (Fig. 6B).
When no nucleic acid target was present, QFRET probes showed much lower background fluorescence levels at 667 nm (the Cy5 maximum) than did the nonquenched IFRET probes. In the latter case, the large unquenched FAM emission yielded a significant signal at 667 nm despite the large spectral separation of FAM and Cy5. Immediately after the addition of target DNA, both probes increased FRET signal (see Fig. 6), apparently due to the close approximation of the FAM and Cy5 labels with adjacent binding of the probes on the target. Interestingly, the QFRET probes afforded a higher FRET signal (by a factor of 1.5) despite the quenching of the FAM donor. Thus, the data show that a quencher plays an important role in increasing the signal-to-background ratio by suppressing background, and thus its loss in the specific probe ligation plays a significant role in enhancing the desired signal.
Next, we followed a time course for ligation over short times to evaluate relative initial rates (Fig. 8). Probes and target were held at equimolar concentrations (500 nM each), and the reaction progress was monitored in solution by the increase in fluorescence signal at 667 nm (with excitation at 490 nm). After an initial rapid increase, the FRET signals gradually increased further as the ligation reaction proceeded. On the initial increase, 2′-OMe probes gave a 2.5-fold stronger signal than natural DNA probes did. However, both types of probes reached almost the same intensity after 140 min. In both cases the signal/background ratio was 800-fold greater than the case with no target. Thus, the data indicated that the QFRET probes offer much greater sensitivity to low target concentrations (by lowering background signal) than previous non-FRET designs and that at 6 h, similar results are seen in solution whether the probe backbone consisted of DNA or 2′-O-methyl-RNA.
Detection of RNAs in HL-60 Cells by FC. Although the method had not been previously reported to be used with quenched probes in living human cells, FC offers the possibility of rapid analysis of a large number of cells and quantitative evaluation of an average over all RNA signals. Thus, we evaluated the feasibility of FC analysis of RNAs by QFRET in HL-60 cells.
The initial targets we chose were GAPDH mRNA and 28S rRNA, both of which are abundant in the cell (42). We prepared 15-mer nucleophile and electrophile probes, and made both the all-DNA and 2′-O-methyl-RNA variants (Fig. 2). To introduce the probes to cells, cells were permeabilized with streptolysin O (SLO) (43) in the presence of the probes. After 30-min incubation, cells were resealed with cell culture media containing CaCl2 and incubated for 1 h at 37°C. As negative control experiments, we tested the crossed QFRET pair of 28S rRNA electrophile probe with GAPDH acceptor probe, which should not ligate because the probes would be localized on different RNAs. For each experiment 50,000 cells were analyzed.
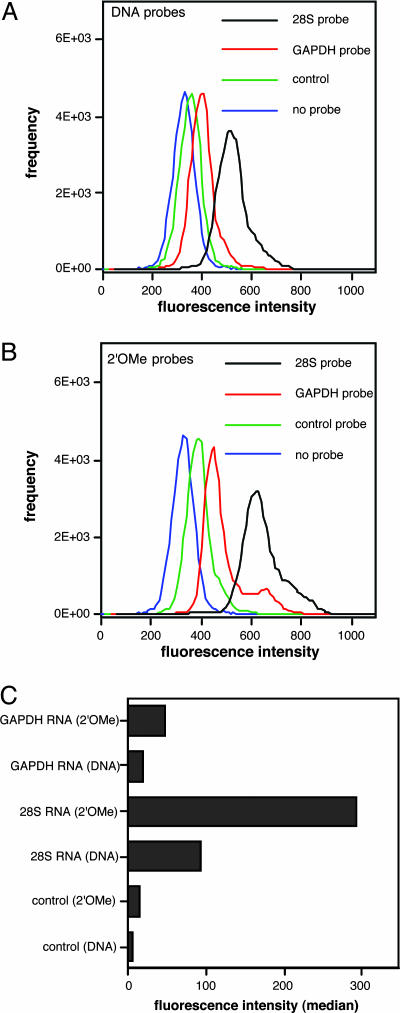
The resulting data are summarized in Fig. 3. The histogram data indicate cell populations with varied FRET signal intensity (emission range 660∼670 nm), which corresponds to 28S rRNA, GADPH mRNA, control probe, and no probe. The median numbers of FRET intensity after subtraction of median background autofluorescence (no probe) was plotted in Fig. 3C. Both GAPDH and 28S rRNA probes offered significantly higher intensity than did the controls. This result clearly showed that cellar endogenous GAPDH mRNA can be detected by QFRET probes. The 28S rRNA signals were 8.6-fold (2′-OMe probe) and 6.8 fold (natural probe) higher than those of GAPDH where background (control) was subtracted from each signal. This quantitative result is consistent with the notion that 28S rRNA is likely to be the most abundant RNA in the cell (21). Compared with natural DNA probes, the 2′-OMe probes gave generally stronger signals, by a factor of 2.6–3.2 (Fig. 3C). Finally, to test reproducibility, we performed the 28S rRNA probing in triplicate; data showed a standard deviation of ±9% in signal by FC (data not shown). Thus, we confirmed that QFRET probes could detect mRNA or rRNA signals by FC analysis and that 2′-OMe probes afforded a measurable advantage over DNA probes in this application, presumably from their greater resistance to degradation.
Fig. 3.
Detection of GAPDH mRNA and 28S rRNA in living HL-60 cells by FC. QFRET probe sequences are shown. The control probe pair consisted of donor probe for 28S rRNA and acceptor probe for GAPDH. HL-60 cells permeabilized by SLO were incubated with QFRET probes (200 nM) in PBS buffer (pH 7.0) for 1.5 h. The resulting cell suspension (n = 50,000) was directly analyzed by FC as described in Materials and Methods.(A and B) Histograms showing cell-count frequency vs. FRET intensity for each probe sequence. (C) Medians of FRET intensity calculated from the histograms. The data were corrected by cell autofluorescence background values (no probes).
Target Site Accessibility in Four Different mRNAs. As is observed with other in vivo genetic targeting strategies such as antisense and siRNA, target site accessibility within a given mRNA is expected to vary significantly as secondary structure varies (13). To evaluate the extent to which signal varies from site to site for QFRET probes, we used FC to quantitatively evaluate signals at multiple sites. Moreover, signal is also expected to vary with copy number of the RNA being targeted. We prepared probes for four different mRNAs and for up to three different sites on each mRNA (Fig. 2). The mRNA targets were β-actin mRNA (BA), ribosomal protein S18 mRNA (S18), histone 3 mRNA (H3), and JUN-D mRNA (JUND). Estimated copy numbers for these RNAs are 7,000, 11,000, 9,000, and 5,000, respectively (44). All probes were prepared with 2′-OMe backbone. We prepared scrambled-sequence probes as controls, which were used to define background signal levels. For initial target site choices, the secondary structures of these mRNAs were calculated from the primary sequences by the mfold program (45) (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). External loop sites were chosen as targets.
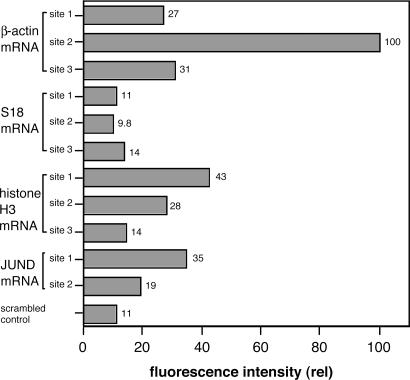
The resulting FC data, monitoring Cy5 signal with excitation of FAM, are summarized in Fig. 4. The results show that for three of four mRNA targets (BA, H3, JUND), significant signals were observed that surpassed background levels, whereas for S18, signals intensity fell within the range of background. The highest signals were observed for BA, where site2 probes gave a signal 9-fold higher than background. The second-highest signals, on average, were observed for H3. The overall order of signal intensity was BA > H3 > JUND > S18.
Fig. 4.
Dependence of QFRET signals on varied RNAs and varied target sites in intact HL-60 cells. Probe sequences are shown in Fig. 2. All probes had 2′-OMe RNA backbone. Cells were treated with QFRET probes (200 nM) and analyzed by FC under the same conditions as in Fig. 3. Relative FRET intensities were calculated from medians from 50,000 cells. The median with scrambled probes was used as a reference standard.
Examination of the signals within a given RNA showed that intensities varied significantly depending on target site. For example, in the BA case, signal increased in the order: site2 > site3 > site1, with a 3.7-fold advantage for the site2 probes over the site3 case. Similar variation was seen for the H3 and JUND cases (Fig. 4).
Previous reports of in situ hybridization with bacterial RNAs have shown that in folded ribosomal RNA targets, probe accessibilities can be enhanced by addition of unlabeled “helper” oligonucleotides that bind near the probe binding sites (26). Thus, we tested the ability of helper DNAs to enhance signal in the BA target. However, signal did not increase as compared with the case lacking helper DNA (see Fig. 9, which is published as supporting information on the PNAS web site). Together, the results showed that signal intensity for QFRET probes depends on which mRNA target is chosen (presumably in part because of differences in copy number) and that signal also varies significantly with target site (likely because accessibility varies with folded structure).
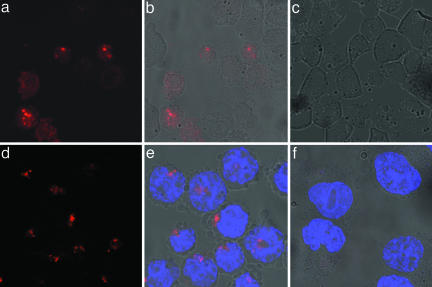
Imaging 28S rRNA and BA mRNA by Confocal Fluorescence Microscopy. Fluorescence microscopy is the most widely used method for imaging of biomolecules in cellular studies. Thus, we tested whether QFRET probes could be visualized in a sequence-dependent manner by this method. First, imaging 28S rRNA was carried out by using 2′-OMe QFRET probes as for the previous experiments (see Fig. 2 for sequences). After 3-h incubation, the cell suspension was directly spotted on glass slides without any washing step and was imaged by confocal microscope. Specific FRET signals were observed by excitation at 488 nm and collecting 650-nm Cy5 emission using a long-pass filter. Fig. 5 shows images of cells incubated with the 28S rRNA probe (a and b) or control probe (c). A bright signal was observed only with the 28S rRNA probe, with little or none from the control. The majority of bright spots were located in the nucleus, which is in fair agreement with previous results by FISH in fixed specimens (46).
Fig. 5.
Imaging of 28S rRNA and BA in HL-60 cells by laser confocal microscopy. (a–c) 28S RNA probes and control. (d–f) BA site2 probes and control. a and d show Cy5 signals only. b and e show overlay with bright field image. c and f show overlay of signal from control (scrambled) probe with bright field image. FRET signals from Cy5 are shown in red; Hoechst 33342 stain (blue) was used as a reference showing localization of nuclear DNA in the BA probes case.
Next, we tested the BA site2 mRNA probe and a scrambled control (Fig. 5 d–f). Again, Cy5 signals were observed from BA site2 probes (d and e), whereas the control probe gave little or no observable signal (f). For the active probes, the most intense signals tended to lie outside the nucleus in this case.
Discussion
The present data establish that QFRET probes can be used to detect and image specific RNAs in human cells. We have established FC as a useful approach for measuring signals from these probes. This method offers the advantages of rapid analysis of large numbers of cells and accurate evaluation of quantitative signals. The analysis of large cell numbers is useful because imaging of small numbers of cells can be confused by the fact that uptake of probes can be quite variable from cell to cell. Moreover, FC is commonly used in initial diagnosis of diseases such as leukemia (47); the present results suggest the possibility that future QFRET probes might be developed to genetically type cells before clinical treatment.
Our results show that the use of the FAM–Cy5 pair in a quenched FRET format offers considerably lower background signals in RNA detection than previous IFRET or QUAL approaches (26, 28, 38). Because modern laser excitation and sensitive cameras can measure very small quantities of signal, the main limiting factor in fluorescence in living cell applications appears to be the level of background signal relative to positive signal. In the case of QFRET probes, background signals can arise from off-target ligations and from indirect excitation of the acceptor dye (Cy5) by the laser source at 488 nm. Although the former depends on concentrations of the two probes and might in the future be modulated by dosing concentration or delivery technique, the latter is strongly suppressed by the large wave-length separation between the FAM donor and Cy5 acceptor. Moreover, our data show that the quenching by dabsyl also enhances signal over background substantially. Thus, for the current application, QFRET probes are more sensitive than prior nonquenched autoligating probes (39) and are likely to have advantages over nonligating FRET probe pairs (35) as well. However, we were not able to detect S18 mRNA with these probes, so more work will be needed to increase signal-to-noise ratios further for detection of moderate-to-low copy number RNAs.
A few recent reports have described the use of beacon-type or linear FRET probes for microscopic imaging of mRNAs in eukaryotic cells (19–24). A comparison of the present results with those prior ones offers some useful insights. The earliest studies attempting to image cellular RNAs used single MBs (19, 22, 23), linear FRET probes (20), or linear 2′-O-methyl probes (21) microinjected into cells. The chief difficulty in those early studies was a large background signal that obscured results and led to conclusions characterized as “uncertain” (18, 19, 23). An important source of background signal in the quenched-probe cases was binding of probes by cellular proteins, which can lead to false signal quite readily (21, 30), because many proteins with affinity for nucleic acid are present in the cellular medium.
An important recent development that has helped to overcome this issue of background signal has been the use of dual beacon probes that use FRET signals (23, 24). Although beacon probes may still bind cellular proteins and individually yield signals, it is much less likely that they will bind near one another and yield a FRET signal. Like that dual beacon strategy, the current QFRET probes also make use of quenching and FRET in combination. However, because ligation is required to generate the main signal, nonspecific signal is highly unlikely to occur even if both half-probes were to bind to a given protein, because very close approach of the reactive ends is required for nucleophilic attack. A second difference with the present probes is the choice of a nontypical FRET pair (FAM–Cy5) that has large optical separation. When typical FRET pairs such as FAM–TMR are used, a false signal from overlapping excitation of the acceptor, and from non-FRET excitation of the acceptor by donor emission, occurs. This false signal has led to the need for digital subtraction methodologies for recent dual-beacon studies (23), whereas our approach was carried out without the need for such postimaging manipulation. Because we were able to acquire signal directly, the QFRET probes could be applied both to microscopy and to FC; such a digital correction may not be as readily applied to this latter analytical method. Beacon probes have not yet been reported to be detected by FC with human cells.
Although multiple methods exist for delivery of oligonucleotides into cells, we adopted the SLO peptide as the delivery mode (43). Nearly all previous studies of quenched probes in cells relied on microinjection for delivery of probes (19, 22, 23), and one recent study with MB probes established the use of the SLO peptide for delivery (24). One of primary goals is identification of disease-related RNAs in clinical samples, where microinjection would not be practical. The present results show successful RNA detection using SLO. It remains to be seen whether other methods of probe delivery would also be successful.
Materials and Methods
Synthesis of Probes. Oligonucleotides were synthesized with β-cyanoethyl phosphoroamidite chemistry and purified by reverse-phase HPLC. The dabsyl linker phosphoroamidite was prepared as described in ref. 28, and probes containing it were deprotected with K2CO3 in MeOH under ultra mild conditions (Glen Research, Sterling, VA). For synthesis of nucleophilic probes carrying a Cy5-label and a 3′-phosphorothioate group, an asymmetric branching amidite (Biosearch) was used. The oligonucleotide sequence then was added following standard procedures. The Cy5 dye then was introduced by removing the column from the synthesizer, deprotecting the levulinyl group (48), placing the column back on the synthesizer, and coupling Cy5 Linker Phosphoroamidite (Biosearch). Deprotection was done with K2CO3 in methanol. Details are given in Supporting Materials and Methods.
Target Sites. Target sites were chosen as described in the text, using secondary structures calculated by mfold (45) for mRNAs and the known secondary structure of 28S rRNA (49). Calculated mRNA structures and target sites are given in Figs. 10–14, which are published as supporting information on the PNAS web site.
Autoligation Reactions in Solution. Ligations were performed in Na·Pipes buffer (70 mM, pH 7.0) containing 10 mM MgCl2 and 50 μM DTT with target nucleic acid (500 nM), dabsyl FAM-labeled probe (500 nM), and Cy5-labeled phosphorothioate probe (500 nM) at 25°C for 6 h. Reactions were observed by fluorescence spectrometry (excitation at 490 nm). Details are given in Supporting Materials and Methods.
Cell Culture. HL-60 cells were grown in DMEM containing 10% FBS. See Supporting Materials and Methods for details.
Cell Permeabilization and Probe Delivery. SLO was purchased from Sigma-Aldrich and activated as described by Faria et al. (43). To introduce probes into cells, HL-60 cells were washed with Mg2+/Ca2+-free PBS twice and then incubated with 300 μl of Mg2+/Ca2+-free PBS buffer solution containing dabsyl-FAM-labeled probe (200 nM), Cy5-labeled-phosphorothioate probe (200 nM), calf thymus DNA (1 μg/ml), and Hoechst 33342 stain (0.2 μg/ml) (Molecular Probes). SLO then was added (30 units/ml final concentration). After 30 min, cells were resealed by the addition of 1 ml of DMEM containing CaCl2 (0.2 g/liter) and incubated for 1 h at 37°C. Details are given in Supporting Materials and Methods.
FC Analysis. The live cell suspension was directly analyzed without any washing step, using a FACScan instrument (Becton Dickinson). FRET signals were observed under excitation by argon laser at 488 nm and emission at 660∼675 nm. Forward angle light scatter (FSC), side angle light scatter (SSC), and fluorescence (FRET) data were recorded, and for each measurement, 50,000 events were stored. Data were analyzed with the flowjo program (Version 4.6.2; Tree Star, Ashland, OR). FRET intensity was determined as the median of FRET value of single cells lying in a gate defined in a FSC vs. SSC dot plot, in which almost 95% of cells were gated. FRET signals were corrected by subtraction of background fluorescence of negative control (no probe).
Confocal Microscope Imaging. Cell suspensions were concentrated to 20-μl volumes. After reaction with QUAL-FRET probes for 3 h, live cells were directly spotted onto a glass slide without any washing step and covered with a glass cover slide. Fluorescence images were obtained through a Zeiss LSM 510 confocal laser scanning microscope equipped with Plan-Apo 63× objective lens. FRET Cy5 imaging was done by excitation with an argon laser (488 nm); emission was measured with a 650 LP filter; pinhole, 886 μm. The resulting raw data were analyzed by LSM image browser (Zeiss). Details are given in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM068122 and the Army Research Office. H.A. is the recipient of postdoctoral fellowships from the Japan Society for the Promotion of Science and the National Institutes of Health/Stanford Quantitative Chemical Biology program.
Author contributions: E.T.K. designed research; H.A. performed research; H.A. analyzed data; and E.T.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: BA, β-actin mRNA; FC, flow cytometry; H3, histone 3 mRNA; JUND, JUN-D mRNA; MB, molecular beacon; QFRET, quenched autoligation-FRET; QUAL, quenched autoligating; SLO, streptolysin O; S18, ribosomal protein S18 mRNA.
References
- 1.Patel, N. H. (1994) in Drosophila melanogaster: Practical Uses in Cell Biology, Methods in Cell Biology, eds. Goldstein, L. S. B. & Fyrberg, E. (Academic, San Diego), Vol. 44, pp. 445-487. [Google Scholar]
- 2.Hayhurst, A. & Georgiou, G. (2001) Curr. Opin. Chem. Biol. 5, 683-689. [DOI] [PubMed] [Google Scholar]
- 3.Tyagi, S., Bratu, D. P. & Kramer, F. R. (1998) Nat. Biotechnol. 16, 49-53. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi, S., Marras, S. A. & Kramer, F. R. (2000) Nat. Biotechnol. 18, 1191-1196. [DOI] [PubMed] [Google Scholar]
- 5.Levsky, J. M. & Singer, R. H. (2003) J. Cell Sci. 116, 2833-2838. [DOI] [PubMed] [Google Scholar]
- 6.Adams, S. R., Harootunian, A. T., Buechler, Y. J., Taylor, S. S. & Tsien, R. Y. (1991) Nature 349, 694-697. [DOI] [PubMed] [Google Scholar]
- 7.Kojima, H., Nakatsubo, N., Kikuchi, K., Kawahara, S., Kirino, Y., Nagoshi, H., Hirata, Y. & Nagano, T. (1998) Anal. Chem. 70, 2446-2453. [DOI] [PubMed] [Google Scholar]
- 8.Cook, B. N. & Bertozzi, C. R. (2002) Bioorg. Med. Chem. 10, 829-840. [DOI] [PubMed] [Google Scholar]
- 9.Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3, 906-918. [DOI] [PubMed] [Google Scholar]
- 10.Lewin, B. (1997) Gene (Oxford Univ. Press, New York), p. 162.
- 11.Giles, R. V. & Tidd, D. M. (1992) Nucleic Acids Res. 20, 763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu, G. C., Cao, Q. N., Zhou, F. & Israel, Y. (1998) J. Biol. Chem. 273, 25125-25131. [DOI] [PubMed] [Google Scholar]
- 13.Sohail, M. & Southern, E. M. (2000) Adv. Drug Delivery Rev. 44, 23-34. [DOI] [PubMed] [Google Scholar]
- 14.Bijsterbosch, M. K., Manoharan, M., Rump, E. T., DeVrueh, R. L. A., vanVeghel, R., Tivel, K. L., Biessen, E. A. L., Bennett, C. F., Cook, P. D. & vanBerkel, T. J. C. (1997) Nucleic Acids Res. 25, 3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojanasakul, Y. Y. (1996) Adv. Drug Delivery Rev. 18, 115-131. [Google Scholar]
- 16.Tyagi, S. & Kramer, F. R. (1996) Nat. Biotechnol. 14, 303-308. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi, S. & Alsmadi, O. (2004) Biophys. J. 87, 4153-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo, T. (1998) Biochim. Biophys. Acta 1379, 178-184. [DOI] [PubMed] [Google Scholar]
- 19.Sokol, D. L., Zhang, X., Lu, P. & Gewirtz, A. M. (1998) Proc. Natl. Acad. Sci. USA 95, 11538-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuji, A., Koshimoto, H., Sato, Y., Hirano, M., Sei-Iida, Y., Kondo, S. & Ishibashi, K. (2000) Biophys. J. 78, 3260-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molenaar, C., Marras, S. A., Slats, J. C. M., Truffert, J. C., Lemaitre, M., Raap, A. K., Dirks, R. W. & Tanke, H. J. (2001) Nucleic Acids Res. 29, e89/1-e89/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlette, J. & Tan, W. (2001) Anal. Chem. 73, 5544-5550. [DOI] [PubMed] [Google Scholar]
- 23.Bratu, D. P., Cha, B.-J., Mhlanga, M. M., Kramer, F. R. & Tyagi, S. (2003) Proc. Natl. Acad. Sci. USA 100, 13308-13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santangelo, P. J., Nix, B., Tsourkas, A. & Bao, G. (2004) Nucleic Acids Res. 32, e57/1-e57/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sando, S. & Kool, E. T. (2002) J. Am. Chem. Soc. 124, 2096-2097. [DOI] [PubMed] [Google Scholar]
- 26.Sando, S., Abe, H. & Kool, E. T. (2004) J. Am. Chem. Soc. 126, 1081-1087. [DOI] [PubMed] [Google Scholar]
- 27.Sando, S. & Kool, E. T. (2002) J. Am. Chem. Soc. 124, 9686-9687. [DOI] [PubMed] [Google Scholar]
- 28.Abe, H. & Kool, E. T. (2004) J. Am. Chem. Soc. 126, 13980-13986. [DOI] [PubMed] [Google Scholar]
- 29.Silverman, A. P. & Kool, E. T. (2005) Trends Biotechnol. 23, 225-230. [DOI] [PubMed] [Google Scholar]
- 30.Fang, X. H., Li, J. J. & Tan, W. H. (2000) Anal. Chem. 72, 3280-3285. [DOI] [PubMed] [Google Scholar]
- 31.Sando, S., Sasaki, T., Kanatani, K. & Aoyama, Y. (2003) J. Am. Chem. Soc. 125, 15720-15721. [DOI] [PubMed] [Google Scholar]
- 32.Cai, J., Li, X., Yue, X. & Taylor, J. S. (2004) J. Am. Chem. Soc. 126, 16324-16325. [DOI] [PubMed] [Google Scholar]
- 33.Hasegawa, S., Jackson, W. C., Tsien, R. Y. & Rao, J. (2003) Proc. Natl. Acad. Sci. USA 100, 14892-14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isaacs, F. J., Dwyer, D. J., Ding, C. M., Pervouchine, D. D., Cantor, C. R. & Collins, J. J. (2004) Nat. Biotechnol. 22, 841-847. [DOI] [PubMed] [Google Scholar]
- 35.Ishibashi, K. & Tsuji, A. (2003) Anal. Chem. 75, 2715-2723. [DOI] [PubMed] [Google Scholar]
- 36.Xu, Y. Z. & Kool, E. T. (1997) Tetrahedron Lett. 38, 5595-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, Y. & Kool, E. T. (1998) Nucleic Acids Res. 26, 3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Y. & Kool, E. T. (1999) Nucleic Acids Res. 27, 875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, Y., Karalkar, N. B. & Kool, E. T. (2001) Nat. Biotechnol. 19, 148-152. [DOI] [PubMed] [Google Scholar]
- 40.Takatsu, K., Yokomaku, T., Kurata, S. & Kanagawa, T. (2004) Nucleic Acids Res. 32, e60/1-e60/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue, H., Hayase, Y., Imura, A., Iwai, S., Miura, K. & Ohtsuka, E. (1987) Nucleic Acids Res. 15, 6131-6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatia, P., Taylor, W. R., Greenberg, A. H. & Wright, J. A. (1994) Anal. Biochem. 216, 223-226. [DOI] [PubMed] [Google Scholar]
- 43.Faria, M., Spiller, D. G., Dubertret, C., Nelson, J. S., White, M. R. H., Scherman, D., Helene, C. & Giovannangeli, C. (2001) Nat. Biotechnol. 19, 40-44. [DOI] [PubMed] [Google Scholar]
- 44.Eisenberg, E. & Levanon, E. Y. (2003) Trends Genet. 19, 362-365. [DOI] [PubMed] [Google Scholar]
- 45.Zuker, M. (2003) Nucleic Acids Res. 31, 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paillasson, S., van de Corput, M., Dirks, R. W., Tanke, H. J., Robert-Nicoud, M. & Ronot, X. (1997) Exp. Cell Res. 231, 226-233. [DOI] [PubMed] [Google Scholar]
- 47.Jennings, C. D. & Foon, K. A. (1997) Blood 90, 2863-2892. [PubMed] [Google Scholar]
- 48.Lyttle, M. H., Walton, T. A., Dick, D. J., Carter, T. G., Beckman, J. H. & Cook, R. M. (2002) Bioconjugate Chem. 13, 1146-1154. [DOI] [PubMed] [Google Scholar]
- 49.Gorski, J. L., Gonzalez, I. L. & Schmickel, R. D. (1987) J. Mol. Evol. 24, 236-251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.