Abstract
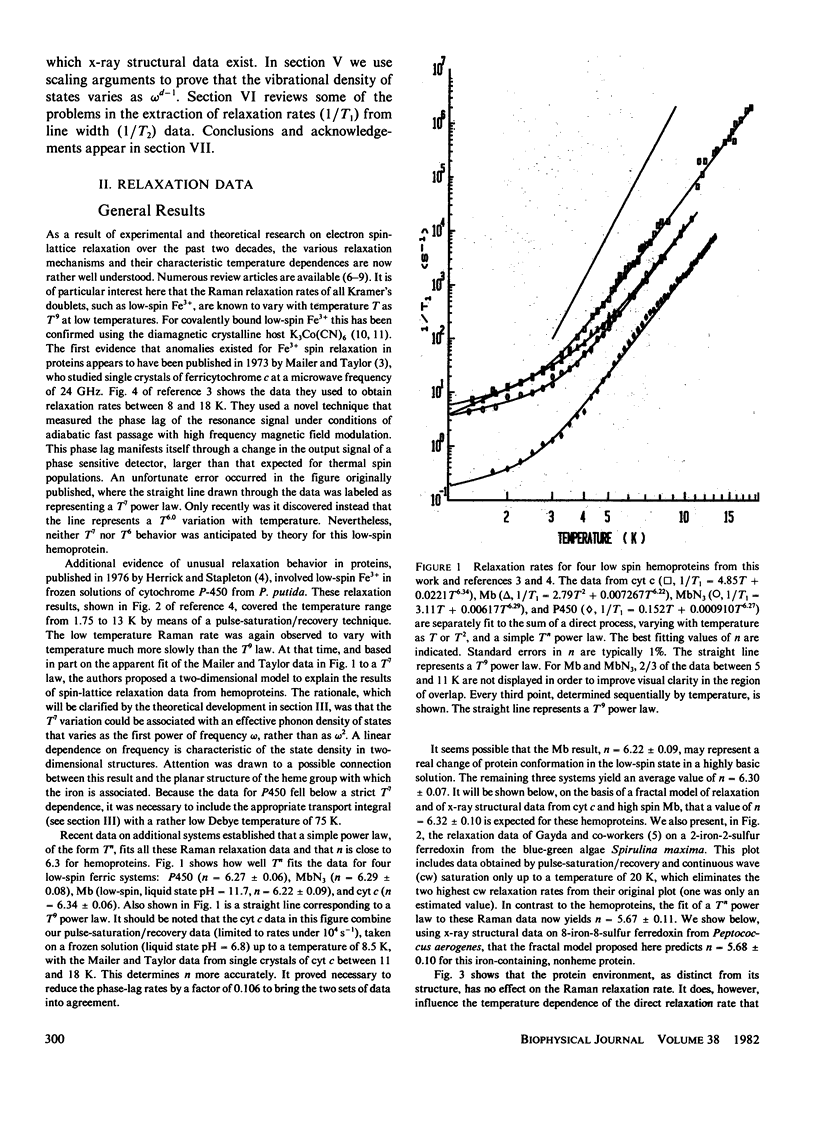
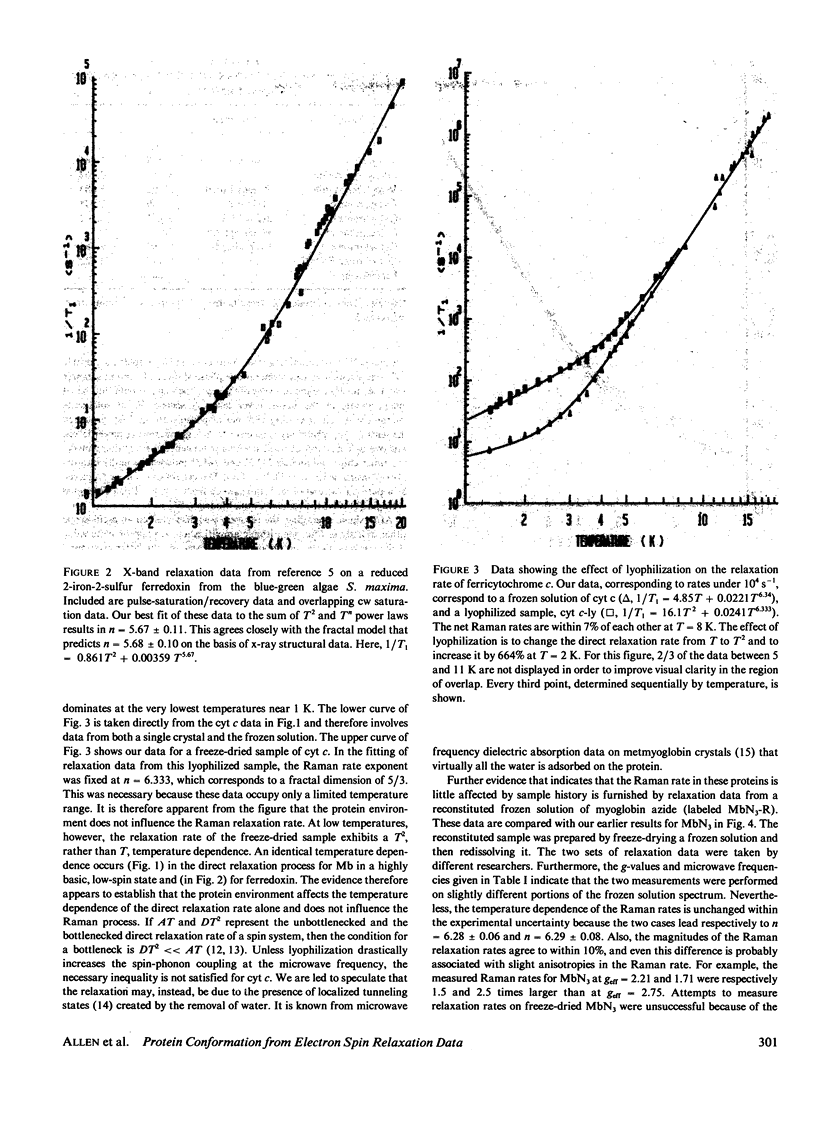
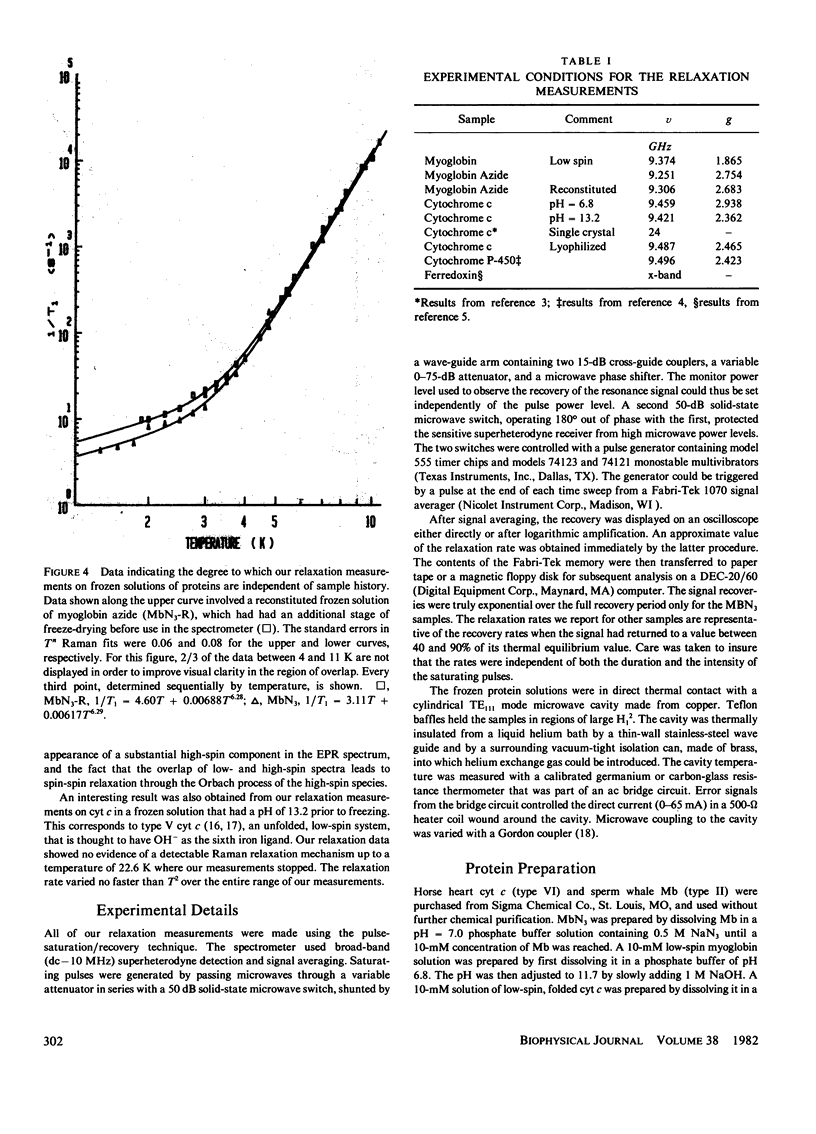
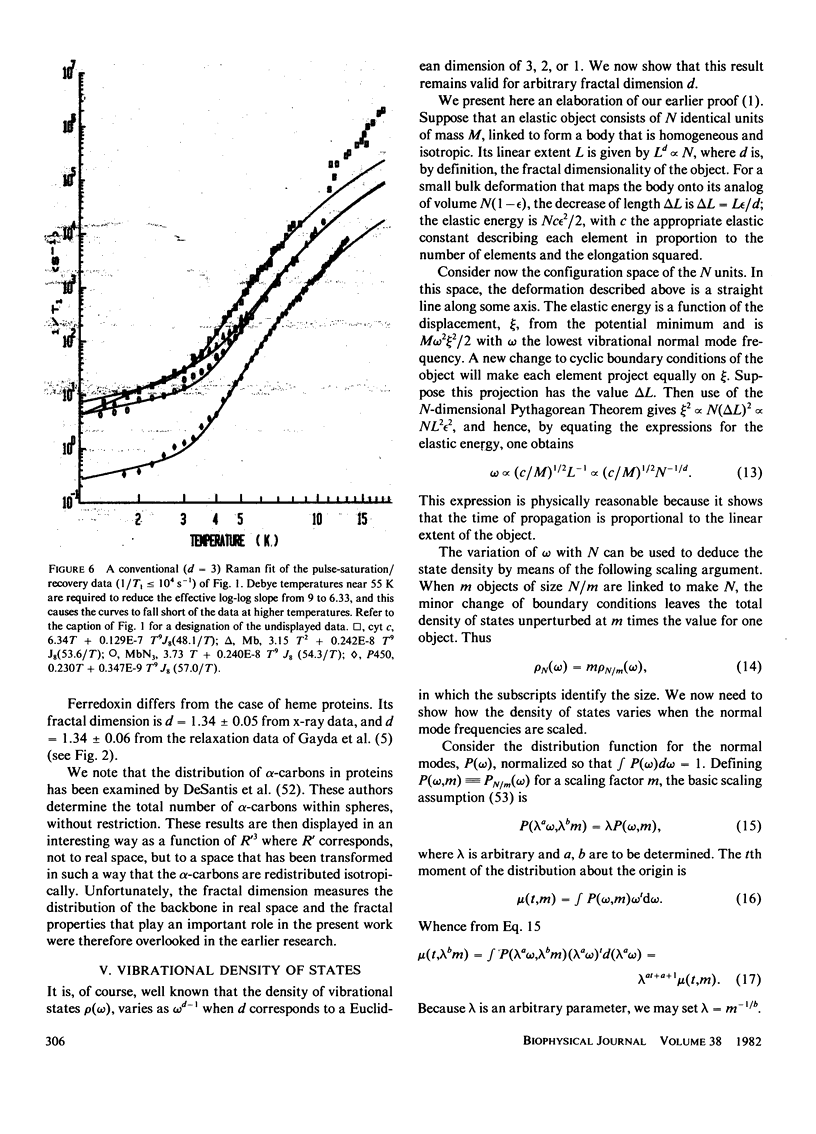
Electron spin relaxation data from five ferric proteins are analyzed in terms of the fractal model of protein structures. Details of this model are presented. The results lead to a characterization of protein structures by a single parameter, the fractal dimension, d. This structural parameter is shown to determine the temperature dependence of the Raman electron spin relaxation rate, which varies as T3 + 2d. Computations of d are made using x-ray data for 17 proteins. The results range from d = 1.76 for lysozyme to d = 1.34 for ferredoxin. These values are compared with values of d obtained from the present electron spin relaxation data on five ferric proteins. Typical results are d = 1.34 +/- 0.06 from relaxation data and 1.34 +/- 0.05 from x-ray data for ferredoxin; d = 1.67 +/- 0.03 from relaxation data and 1.66 +/- 0.05 from x-ray data for ferricytochrome c. The data thus support the theoretical model. Applications of this spin resonance technique to the study of changes in protein conformation are discussed.
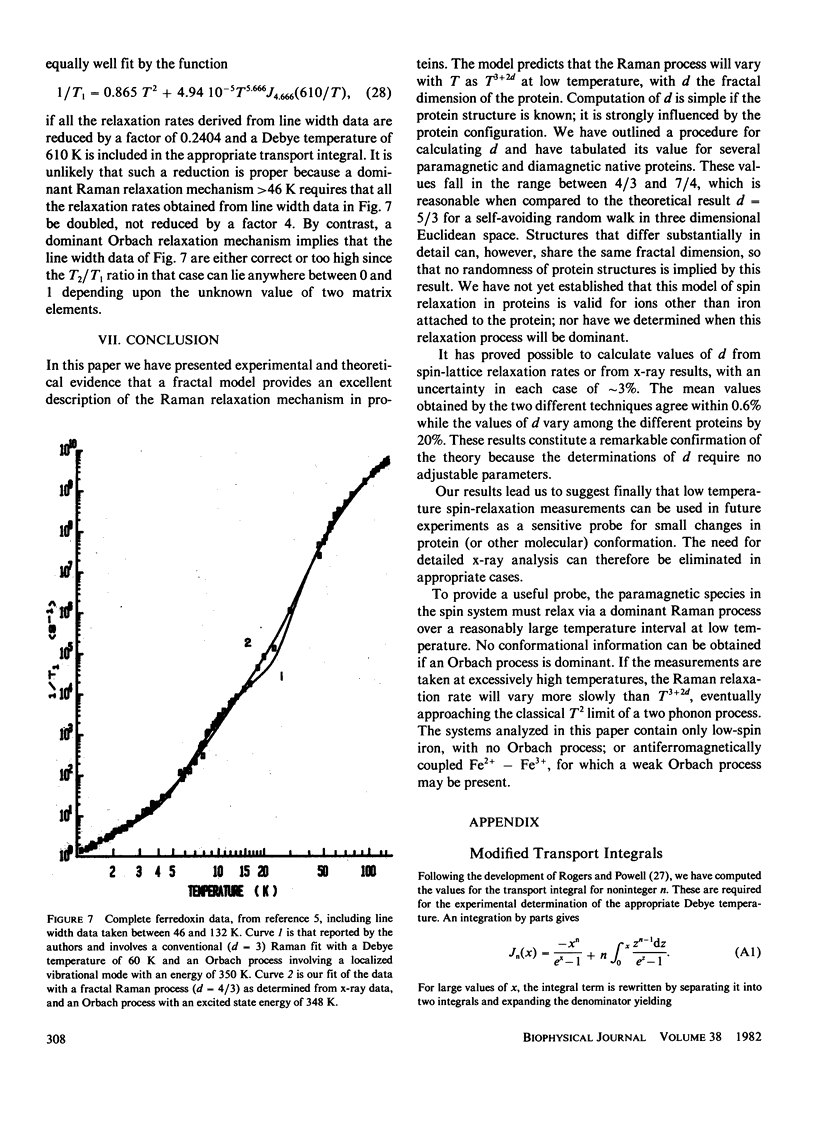
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T., Siefker L. C., Jensen L. H. Structure of Peptococcus aerogenes ferredoxin. Refinement at 2 A resolution. J Biol Chem. 1976 Jun 25;251(12):3801–3806. doi: 10.2210/pdb1fdx/pdb. [DOI] [PubMed] [Google Scholar]
- Almassy R. J., Dickerson R. E. Pseudomonas cytochrome c551 at 2.0 A resolution: enlargement of the cytochrome c family. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2674–2678. doi: 10.1073/pnas.75.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972 Jul 21;68(2):187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr, Kraut J., Freer S. T., Nguyen-Huu-Xuong, Alden R. A., Bartsch R. G. Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein. J Biol Chem. 1974 Jul 10;249(13):4212–4225. [PubMed] [Google Scholar]
- Diamond R. Real-space refinement of the structure of hen egg-white lysozyme. J Mol Biol. 1974 Jan 25;82(3):371–391. doi: 10.1016/0022-2836(74)90598-1. [DOI] [PubMed] [Google Scholar]
- Drenth J., Jansonius J. N., Koekoek R., Swen H. M., Wolthers B. G. Structure of papain. Nature. 1968 Jun 8;218(5145):929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- Fehlhammer H., Bode W. The refined crystal structure of bovine beta-trypsin at 1.8 A resolution. I. Crystallization, data collection and application of patterson search technique. J Mol Biol. 1975 Nov 15;98(4):683–692. doi: 10.1016/s0022-2836(75)80004-0. [DOI] [PubMed] [Google Scholar]
- Gayda J. P., Bertrand P., Deville A., More C., Roger G., Gibson J. F., Cammack R. Temperature dependence of the electronic spin-lattice relaxation time in a 2-iron-2-sulfur protein. Biochim Biophys Acta. 1979 Nov 23;581(1):15–26. doi: 10.1016/0005-2795(79)90216-2. [DOI] [PubMed] [Google Scholar]
- Gurd F. R., Falk K. E., Malmström B. G., Vänngård T. A magnetic resonance study of sperm whale ferrimyoglobin and its complex with 1 cupric ion. J Biol Chem. 1967 Dec 25;242(24):5724–5730. [PubMed] [Google Scholar]
- Ladner R. C., Heidner E. J., Perutz M. F. The structure of horse methaemoglobin at 2-0 A resolution. J Mol Biol. 1977 Aug 15;114(3):385–414. doi: 10.1016/0022-2836(77)90256-x. [DOI] [PubMed] [Google Scholar]
- Lambeth D. O., Campbell K. L., Zand R., Palmer G. The appearance of transient species of cytochrome c upon rapid oxidation or reduction at alkaline pH. J Biol Chem. 1973 Dec 10;248(23):8130–8136. [PubMed] [Google Scholar]
- Mailer C., Taylor C. P. Rapid adiabatic passage EPR of ferricytochrome c: signal enhancement and determination of the spin-lattice relaxation time. Biochim Biophys Acta. 1973 Oct 18;322(2):195–203. doi: 10.1016/0005-2795(73)90293-6. [DOI] [PubMed] [Google Scholar]
- Mathews F. S., Argos P., Levine M. The structure of cytochrome b 5 at 2.0 Angstrom resolution. Cold Spring Harb Symp Quant Biol. 1972;36:387–395. doi: 10.1101/sqb.1972.036.01.050. [DOI] [PubMed] [Google Scholar]
- Morton R. A., Bohan T. L. The electron paramagnetic resonance spectrum at 77 degrees K of lyophilized and frozen solutions of horse heart ferricytochrome c. Can J Biochem. 1971 Mar;49(3):328–331. doi: 10.1139/o71-048. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Lipscomb W. N. Carboxypeptidase A: a protein and an enzyme. Adv Protein Chem. 1971;25:1–78. doi: 10.1016/s0065-3233(08)60278-8. [DOI] [PubMed] [Google Scholar]
- Salemme F. R., Freer S. T., Xuong N. H., Alden R. A., Kraut J. The structure of oxidized cytochrome c 2 of Rhodospirillum rubrum. J Biol Chem. 1973 Jun 10;248(11):3910–3921. doi: 10.2210/pdb1c2c/pdb. [DOI] [PubMed] [Google Scholar]
- Segal D. M., Cohen C. H., Davies D. R., Powers J. C., Wilcox P. E. The stereochemistry of substrate binding to chymotrypsin A . Cold Spring Harb Symp Quant Biol. 1972;36:85–90. doi: 10.1101/sqb.1972.036.01.014. [DOI] [PubMed] [Google Scholar]
- Swanson R., Trus B. L., Mandel N., Mandel G., Kallai O. B., Dickerson R. E. Tuna cytochrome c at 2.0 A resolution. I. Ferricytochrome structure analysis. J Biol Chem. 1977 Jan 25;252(2):759–775. [PubMed] [Google Scholar]
- Timkovich R., Dickerson R. E. The structure of Paracoccus denitrificans cytochrome c550. J Biol Chem. 1976 Jul 10;251(13):4033–4046. doi: 10.2210/pdb155c/pdb. [DOI] [PubMed] [Google Scholar]
- Yonetani T., Schleyer H. Electromagnetic properties of hemoproteins. II. The effect of physical states on electron paramagnetic resonance parameters of hemoproteins. J Biol Chem. 1967 Sep 10;242(17):3926–3933. [PubMed] [Google Scholar]


