Abstract
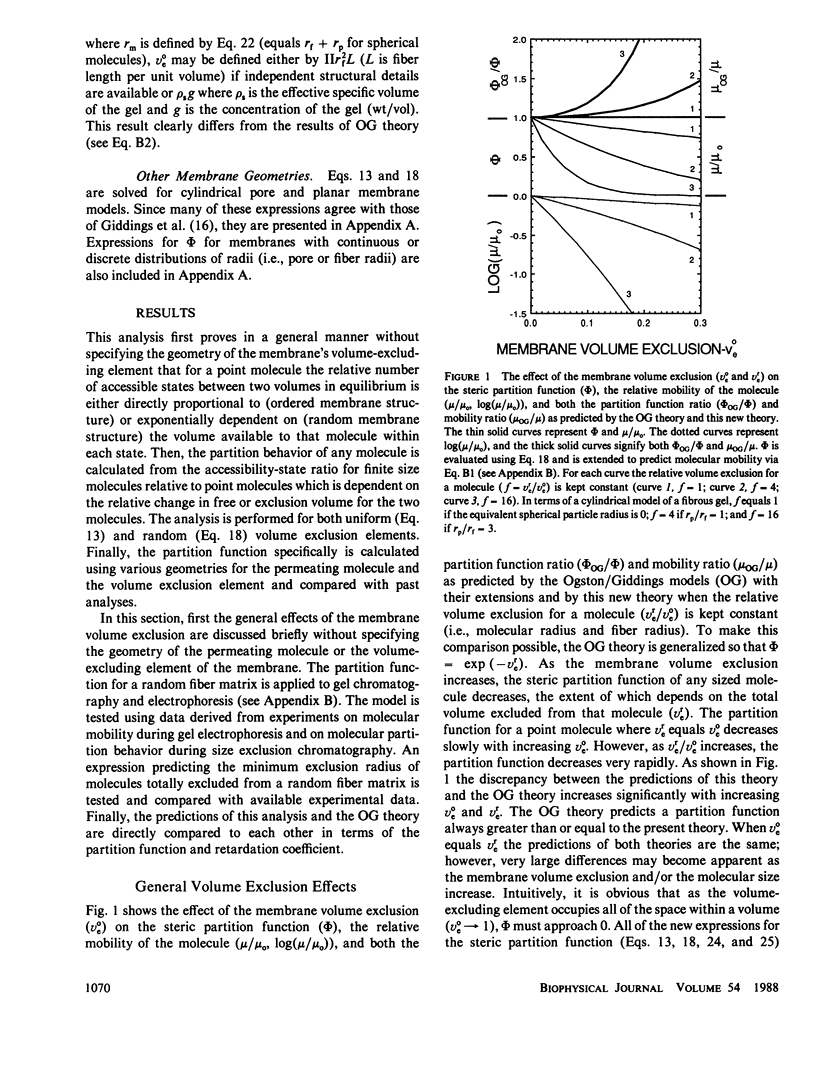
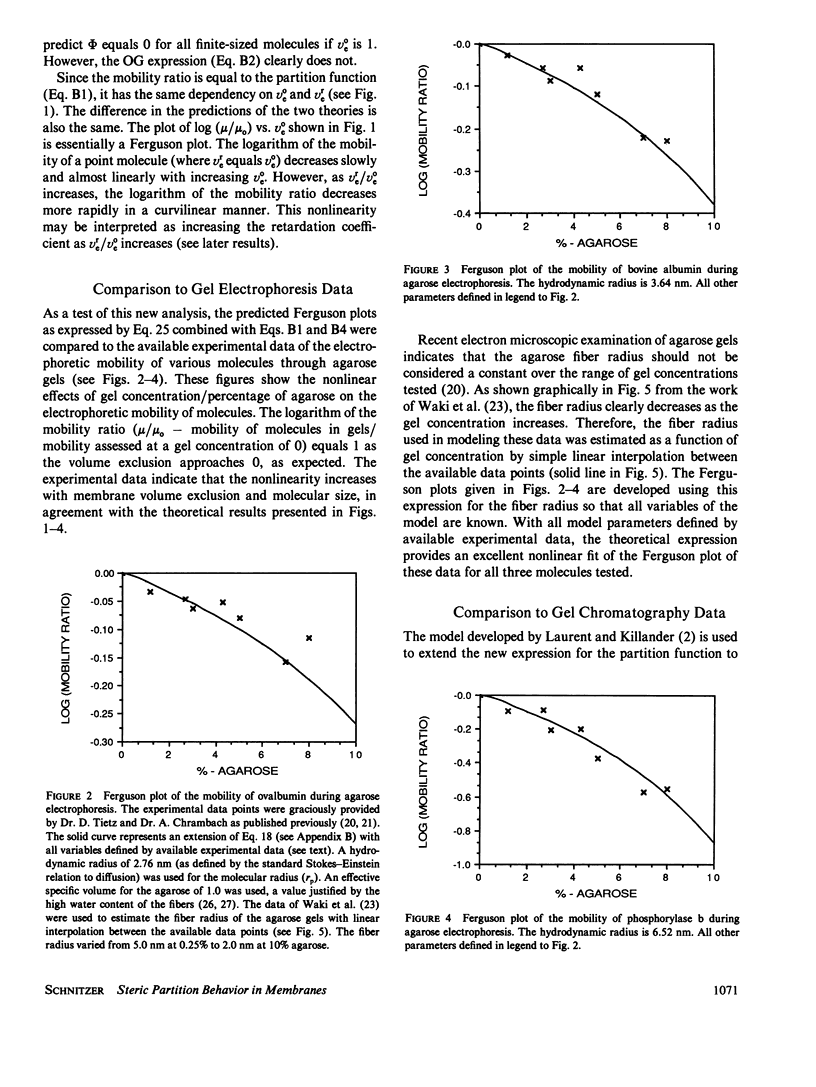
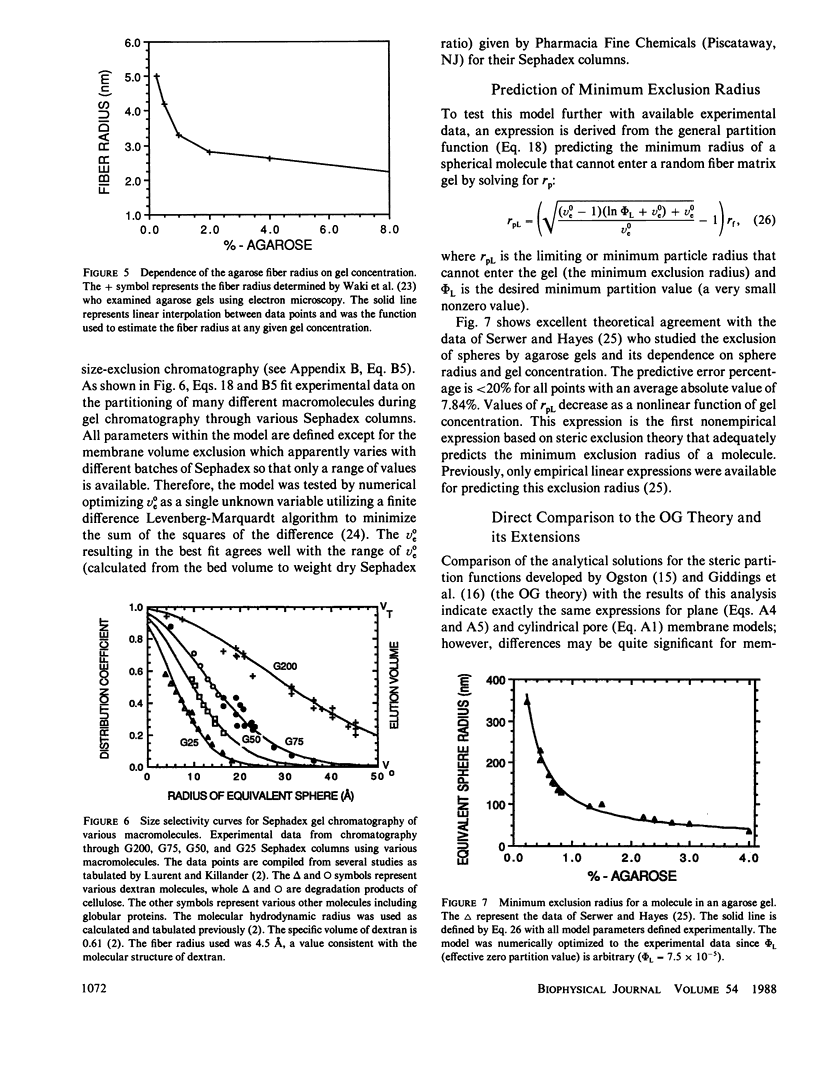
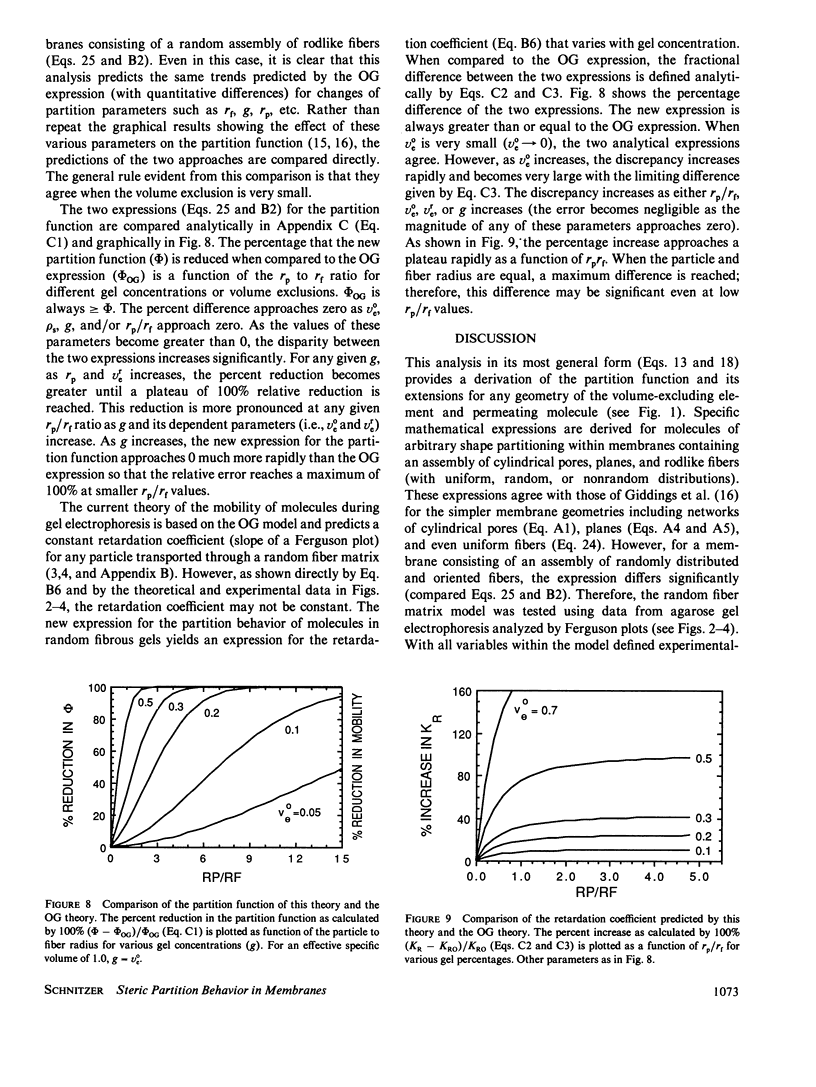
The principles of statistical physics are used to formulate general expressions for the steric partition behavior of molecules in both random and ordered membrane structures that may be applied to any shape of the solute and/or the volume-excluding element of the membrane. These expressions fully define partitioning in terms of the volume excluded to point molecules and to finite-sized molecules. The mean effective exclusion volume for a molecule is calculated as a function of a global interaction energy, which varies with position, conformation, and orientation of the molecule. It allows consideration of electrostatic and other nonsteric factors. To test the model, specific partition functions are derived for several simple geometries describing the membrane and solute. Frequently, the derived expressions agree with past analyses; however, a new expression describing partitioning within an random network of fibers is derived. It agrees with past results only in the limit of low exclusion volumes. With greater volume exclusions, past results greatly overestimate the partition function. It is applied to gel electrophoresis and chromatography and survives testing with available experimental data. Unlike past analyses, it predicts nonlinear Ferguson plots for agarose gel electrophoresis. In addition, an analytical expression predicting the minimum radius of a sphere excluded from a random fiber matrix is derived, tested, and found to agree with experimental data.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Curry F. E., Michel C. C. A fiber matrix model of capillary permeability. Microvasc Res. 1980 Jul;20(1):96–99. doi: 10.1016/0026-2862(80)90024-2. [DOI] [PubMed] [Google Scholar]
- Laurent T. C. Determination of the structure of agarose gels by gel chromatography. Biochim Biophys Acta. 1967 Mar 22;136(2):199–205. doi: 10.1016/0304-4165(67)90064-5. [DOI] [PubMed] [Google Scholar]
- Laurent T. C. The interaction between polysaccharides and other macromolecules. 9. The exclusion of molecules from hyaluronic acid gels and solutions. Biochem J. 1964 Oct;93(1):106–112. doi: 10.1042/bj0930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro A. M., Babich M. A., Taylor W. D., Keith A. D. Diffusion of a small molecule in the cytoplasm of mammalian cells. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3414–3418. doi: 10.1073/pnas.81.11.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner-White E. J., Poet R. R. Computer graphics of large macromolecules. Biochem Soc Trans. 1985 Aug;13(4):793–795. doi: 10.1042/bst0130793. [DOI] [PubMed] [Google Scholar]
- Polefka T. G., Garrick R. A., Redwood W. R., Swislocki N. I., Chinard F. P. Solute-excluded volumes near the Novikoff cell surface. Am J Physiol. 1984 Nov;247(5 Pt 1):C350–C356. doi: 10.1152/ajpcell.1984.247.5.C350. [DOI] [PubMed] [Google Scholar]
- Rodbard D., Chrambach A. Estimation of molecular radius, free mobility, and valence using polyacylamide gel electrophoresis. Anal Biochem. 1971 Mar;40(1):95–134. doi: 10.1016/0003-2697(71)90086-8. [DOI] [PubMed] [Google Scholar]
- Rodbard D., Chrambach A. Unified theory for gel electrophoresis and gel filtration. Proc Natl Acad Sci U S A. 1970 Apr;65(4):970–977. doi: 10.1073/pnas.65.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer J. E. Glycocalyx electrostatic potential profile analysis: ion, pH, steric, and charge effects. Yale J Biol Med. 1988 Sep-Oct;61(5):427–446. [PMC free article] [PubMed] [Google Scholar]
- Serwer P., Hayes S. J. Exclusion of spheres by agarose gels during agarose gel electrophoresis: dependence on the sphere's radius and the gel's concentration. Anal Biochem. 1986 Oct;158(1):72–78. doi: 10.1016/0003-2697(86)90591-9. [DOI] [PubMed] [Google Scholar]
- Swanson J. A., McNeil P. L. Nuclear reassembly excludes large macromolecules. Science. 1987 Oct 23;238(4826):548–550. doi: 10.1126/science.2443981. [DOI] [PubMed] [Google Scholar]
- TISELIUS A., PORATH J., ALBERTSSON P. A. Separation and fractionation of macromolecules and particles. Science. 1963 Jul 5;141(3575):13–20. doi: 10.1126/science.141.3575.13. [DOI] [PubMed] [Google Scholar]
- Tietz D., Chrambach A. Computer simulation of the variable agarose fiber dimensions on the basis of mobility data derived from gel electrophoresis and using the Ogston theory. Anal Biochem. 1987 Mar;161(2):395–411. doi: 10.1016/0003-2697(87)90468-4. [DOI] [PubMed] [Google Scholar]
- Waki S., Harvey J. D., Bellamy A. R. Study of agarose gels by electron microscopy of freeze-fractured surfaces. Biopolymers. 1982 Sep;21(9):1909–1926. doi: 10.1002/bip.360210917. [DOI] [PubMed] [Google Scholar]


